Deep learning for rapid analysis of cell divisions in vivo during epithelial morphogenesis and repair
Figures
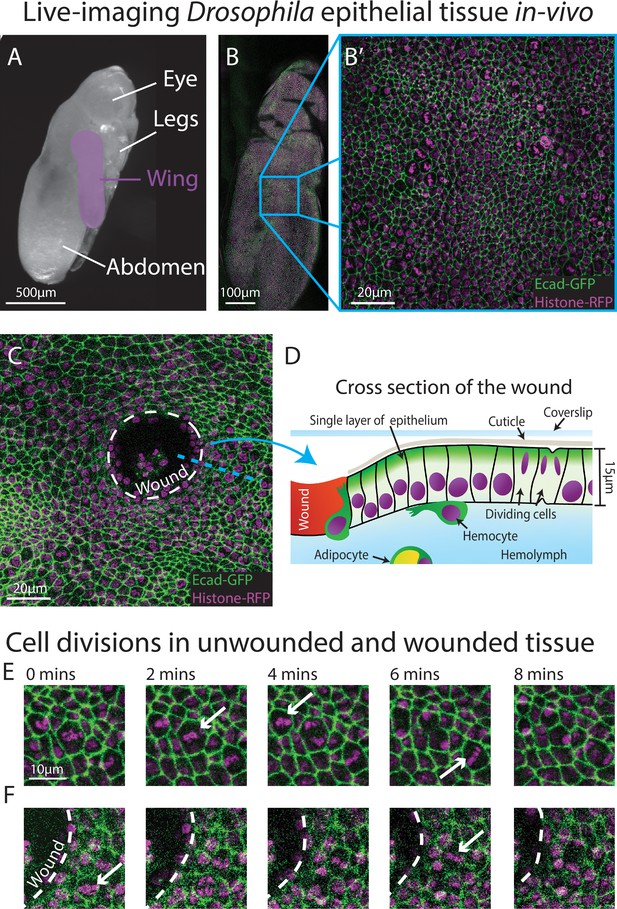
Live-imaging of Drosophila epithelial tissue dynamics in vivo.
(A) Translucent Drosophila pupa with the pupal wing highlighted in magenta. (B) The pupal wing (with magnified inset, B’, on the centre zone of the wing where we consistently image) with cell boundaries labelled using E-cadherin-GFP (green) and nuclei with Histone2Av-mRFP (magenta). (C) Magnified view of the pupal wing epithelium after wounding, with the white dashed line indicating the wound edge. (D) Schematic showing a cross-section through the upper layer of epithelium of the pupal wing, with haemolymph (insect blood containing haemocytes and adipocytes) beneath and rigid cuticle above (E) Multiple cell divisions (arrows) occur in the unwounded pupal wing epithelial tissue over the course of 8 min. (F) A cell division (arrow) occurs in a wounded epithelial tissue with the white dashed line indicating the wound edge.
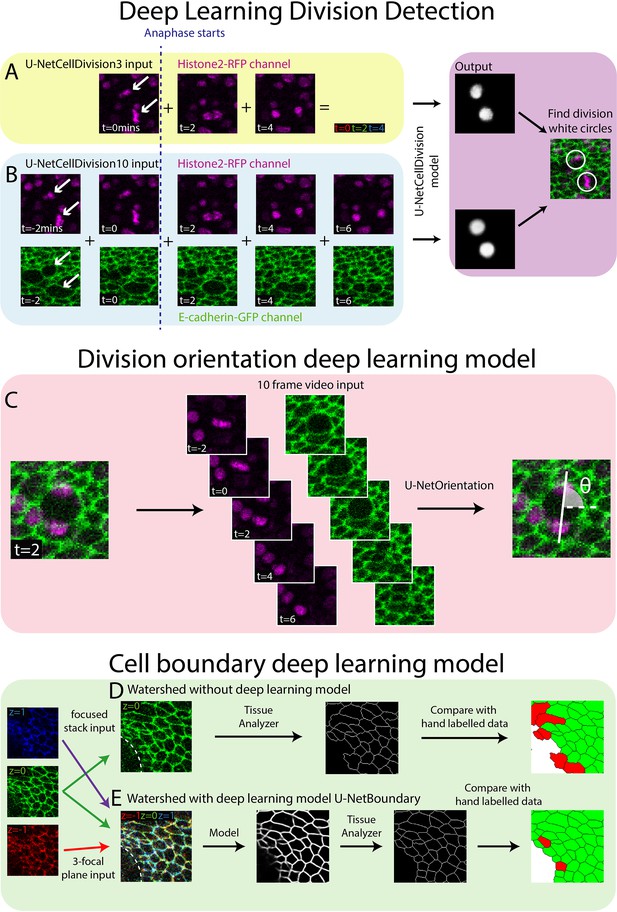
Deep learning detection of cell divisions, division orientation, and cell boundaries.
Four deep learning models were developed to analyse epithelial cell behaviours. (A) The first version of the division detection model receives three frames from the Histone2Av-mRFP channel, which can be combined into a single RGB image, as is standard for a U-Net model. (B) The second version of the model input has 10 frames, 5 each from the Histone2Av-mRFP and E-cadherin-GFP channels. The model produces a white circle (white spot) wherever it detects a division. (C) The cell division locations are then passed through the U-NetOrientation model to determine the division orientation. This model takes 10 frames of a division as the input. (D) Segmentation of the focussed cell boundaries without using a deep learning model. The focussed stack image is inputted to Tissue Analyzer for segmentation and the result is compared to a hand-labelled ground truth. Green cells are correctly segmented and red cells are incorrectly segmented. (E) The three-focal plane image is inputted into the U-NetBoundary model and then segmented using Tissue Analyzer; this result is then compared to the hand-labelled ground truth.
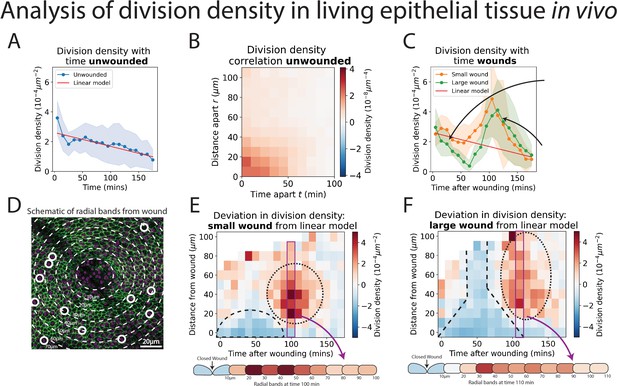
Analysis of cell division density in living epithelial tissue in vivo.
(A) The density of cell divisions in the unwounded tissue, with faded blue region showing the standard deviation. The red line is the line of ‘best fit’ of the unwounded data. (B) A heatmap of the division density correlation over distance and time in unwounded epithelial tissue. Red indicates positive, and blue negative correlation. (C) The density of cell divisions in the wounded tissue, with either small or large wounds, with faded regions showing associated standard deviation. The red line is the line of best fit of the unwounded data. The micrographs show representative divisions identified at two different timepoints post-wounding. (D) Diagram of the annular bands around a wound, each 10 m wide (white dashed line); white circles indicate cell divisions. (E, F) Heatmaps of the change in division density for small and large wounds compared with a best fit linear model of unwounded data. Red areas have more divisions, and blue less, than equivalent regions in the unwounded data. The dashed lines highlight areas in which cell divisions decrease and the dotted lines highlight areas in which divisions increase compared to unwounded data. Schematics below the heatmaps in E and F show the radial division densities 100 and 110 min after wounding, respectively (n = 14 unwounded, n = 8 small wounds, and n = 9 large wounds).
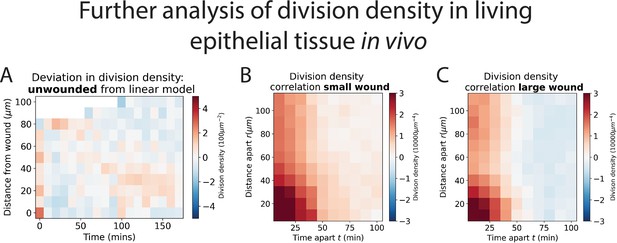
Further analysis of division density in living epithelial tissue.
(A) Heatmaps of the deviation in division density of unwounded tissue compared with a best fit linear model. The axes are time and distance from a virtual wound. Red areas have more division and blue less. (B, C) Heatmaps of the division density correlation for small and large wounds. Again, red areas have more divisions and blue less. Also see Figure 3.
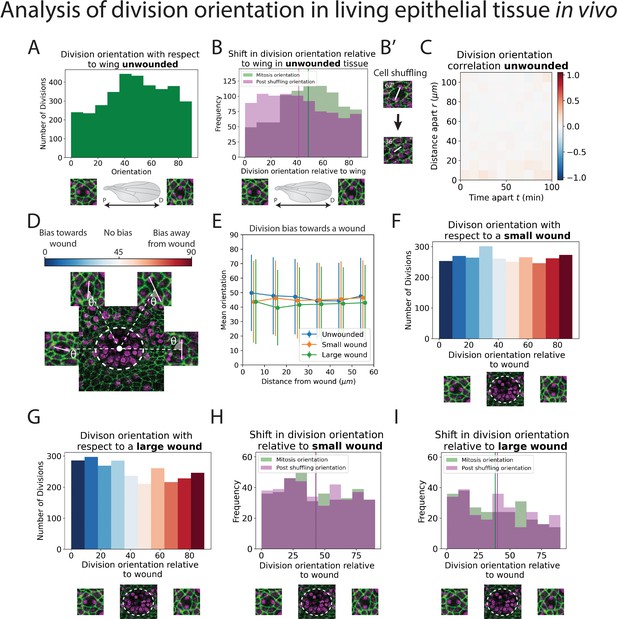
Analysis of division orientation in living epithelial tissue in vivo.
(A) Distribution of the division orientations with respect to the proximal–distal axis of the pupal wing in unwounded tissue. Cell division orientations of 0° and 90° are illustrated in the micrographs. (B) Distribution of the division orientations with respect to the wing in unwounded tissue (green) and the daughter cell orientations 20 min after dividing (magenta), with examples of the orientation of division before and after cell shuffling (B’). (C) Heatmap of the space–time correlation of division orientation. Red indicates positive correlation, blue negative, and white no correlation. (D) Diagram of cell division orientation with respect to a wound; lower values are dividing towards the wound and higher values away. (E) Mean division orientation towards the wound as a function of distance from wound for small and large wounds. For unwounded tissues an arbitrary point is chosen as a ‘virtual wound’. (F, G) Distribution of the division orientations with respect to small and large wounds. The spectrum of colours (same as in D) indicates the bias in orientation towards the wound. (H, I) Distribution of the division orientations with respect to the wound in small and large wounds (green), and the daughter cell orientation 20 min after dividing (magenta) (n = 14 unwounded, n = 8 small wounds, and n = 9 large wounds).
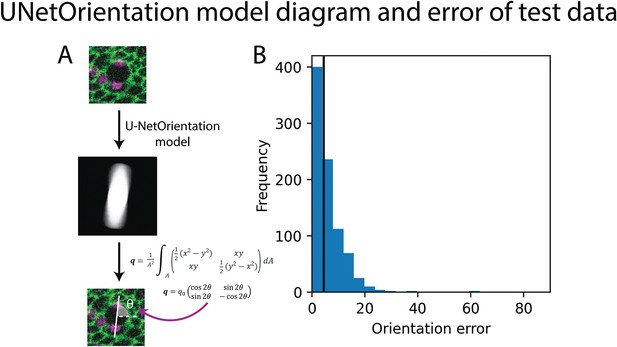
UNetOrientation model diagram and error of test data.
(A) Diagram of the output of U-NetOrientation. The oval is elongated in the same direction as the division, thus calculating its q-tensor tells us the orientation of the cell division. (B) The error of the U-NetOrientation model on the test dataset; black line shows median error. Also see Figure 4.
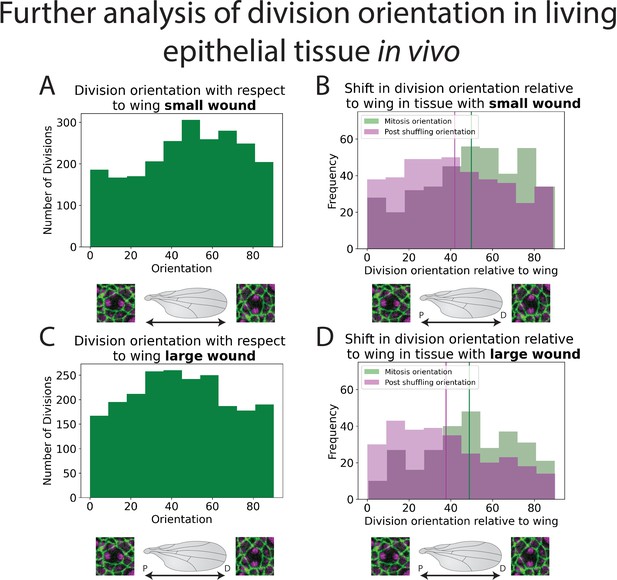
Further analysis of division orientation in living epithelial tissue.
(A) Distribution of the division orientations for small wounds. (B) Distribution of the division orientations with respect to the wing in small wounds (green), and the daughter cell orientation 20 min after dividing (magenta). (C) Distribution of the division orientations for large wounds. (D) Distribution of the division orientations with respect to the wing in large wounds (green), and the daughter cell orientation 20 min after dividing (magenta). Also see Figure 4.
Videos
Time-lapse imaging of the unwounded pupal epithelium over 3 hr.
Projected from a 3D stack using the stack focus algorithm with a radius of 5 pixels. Green indicates E-cadherin-GFP and magenta indicates Histone2Av-mRFP. The white circles show the divisions detected by the ‘U-NetCellDivision10’ and the white lines indicate the orientation of divisions determined by ‘U-NetOrientation’. Scale bar: 10 μm. Related to Figure 3.
Time-lapse imaging of a small wound in the pupal epithelium over 3 hr.
Projected from a 3D stack using the stack focus algorithm with a radius of 5 pixels. Green indicates E-cadherin-GFP and magenta indicates Histone2Av-mRFP. The white circles show the divisions detected by the ‘U-NetCellDivision10’ and the white lines indicate the orientation of divisions determined by ‘U-NetOrientation’. Scale bar: 10 μm. Related to Figure 3.
Time-lapse imaging of a large wound in the pupal epithelium over 3 hr.
Projected from a 3D stack using the stack focus algorithm with a radius of 5 pixels. Green indicates E-cadherin-GFP and magenta indicates Histone2Av-mRFP. The white circles show the divisions detected by the ‘U-NetCellDivision10’ and the white lines indicate the orientation of divisions determined by ‘U-NetOrientation’. Scale bar: 10 μm. Related to Figure 3.
Time-lapse imaging of a small wound in the pupal epithelium over 3 hr.
Projected from a 3D stack using the stack focus algorithm with a radius of 5 pixels. Greyscale background of epithelium with circles show the divisions detected by the ‘U-NetCellDivision10’, the lines indicate the orientation of divisions determined by ‘U-NetOrientation’ and the colour of labels display the orientations relative to wounds. Blue labelled divisions are orientated towards wounds, red away from wounds and white around 45°. The white dot is the centre of the wound and the closed wound site after closure. Scale bar: 10 μm. Related to Figure 3.
Time-lapse imaging of a large wound in the pupal epithelium over 3 hr.
Projected from a 3D stack using the stack focus algorithm with a radius of 5 pixels. Greyscale background of epithelium with circles show the divisions detected by the ‘U-NetCellDivision10’, the lines indicate the orientation of divisions determined by ‘U-NetOrientation’ and the colour of labels display the orientations relative to wounds. Blue labelled divisions are orientated towards wounds, red away from wounds and white around 45°. The white dot is the centre of the wound and the closed wound site after closure. Scale bar: 20 μm. Related to Figure 3.
Tables
Dice scores for the deep learning models.
| Model | True positives | False positive | False negative | Dice score |
|---|---|---|---|---|
| U-NetCellDivision3 | 797 | 216 | 310 | 0.752 |
| U-NetCellDivision10 | 1057 | 28 | 50 | 0.964 |
Dice scores for the segmentation methods.
| Segmentation | True positives | False positive | False negative | Dice score |
|---|---|---|---|---|
| Single focal plane + Tissue Analyzer | 8197 | 313 | 4317 | 0.780 |
| U-NetBoundary + Tissue Analyzer | 11,325 | 501 | 1189 | 0.931 |