Breaking enhancers to gain insights into developmental defects
Figures
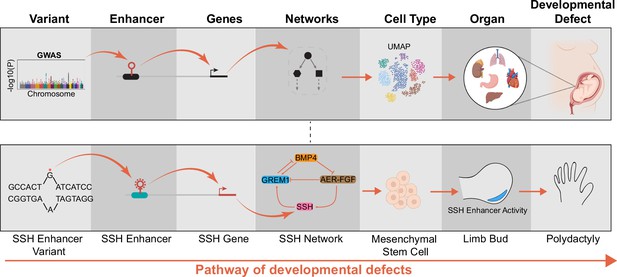
Pathway of how enhancer variants cause developmental defects.
The functional characterization of enhancer variants is a multi-step process linking genotypes to molecular phenotypes (target genes and networks), cellular phenotypes (cell state, morphology), and organismal phenotypes (developmental defect) (top row). A genetic variant of a Sonic Hedgehog enhancer driving polydactyly is one well-characterized example (bottom row). However, the role of most cis-regulatory elements in developmental disease remains unclear.
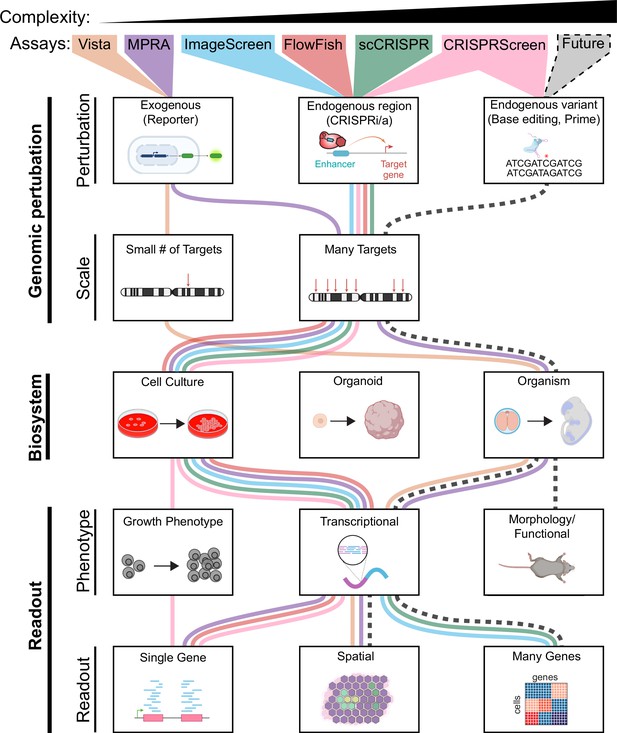
Features of enhancer perturbation studies.
Studies of enhancer variants in developmental disease have been carried out using diverse genomic perturbation methods, biosystems, and readouts. Each of these three layers can be organized into increasing levels of complexity with increasing biomedical relevance. We envision that future technologies (black dashed line) will enable analyses of higher complexity.
Tables
Summary of genomic approaches to characterize enhancers and variants.
| Approach | Application | Pros | Cons | Example studies |
|---|---|---|---|---|
| VISTA | Measures in vivo enhancer reporter activity in transgenic mice. | in vivo and spatial readouts of enhancer activity | single time point (E11.5), low throughput | Pennacchio et al., 2006; Visel et al., 2007 |
| Massively parallel reporter assays (MPRA), Self-transcribing active regulatory region (STARR)-Seq | Measures the activity of enhancer sequences and variants with high throughput reporter assays. Single-cell MPRA gives readouts on cell-specific enhancer activity. | very high throughput, variant-level activity | lacks endogenous genomic context, readouts can depend on the design of reporter constructs | Inoue et al., 2019; Kircher et al., 2019; Arnold et al., 2013; Zhao et al., 2023; Lalanne et al., 2022 |
| CRISPR screen | Measures endogenous activity of enhancer by perturbating sequences with CRISPR/Cas9, using sgRNA dropout as a phenotypic readout. | high throughput, endogenous genomic context | requires a selectable phenotype, the readout is only sgRNA abundance | Sanjana et al., 2016; Korkmaz et al., 2016 |
| CRISPRi FlowFISH, HCR FlowFISH | Measures endogenous enhancer activity on the expression of candidate genes with high sensitivity. | medium throughput, endogenous genomic context, sensitive transcriptional readout | only a small number of genes can be measured in each experiment | Fulco et al., 2016; Reilly et al., 2021 |
| Single-cell CRISPRi screen | Measures endogenous enhancer activity on transcriptome-wide phenotypes. | high throughput, endogenous genomic context, transcriptome-wide readout | low sensitivity for lowly expressed genes or enhancers with modest effects; expensive | Genga et al., 2019; Armendariz et al., 2022 |
| Base editing screen | Measures endogenous activity of enhancer variants after high throughput base editing. | variant-level perturbations more relevant to disease modeling, endogenous genomic context | some base substitutions incompatible with current base editors, limited editing window restricts sgRNA design; modest effect sizes | Martin-Rufino et al., 2023; Chen et al., 2022 |
| merFISH, seqFISH, osmFISH | Measures spatial RNA expression with high sensitivity. | spatial context, sensitive readout of many transcripts | existing screens are low throughput, expensive, specialized equipment | Xie et al., 2017; Eng et al., 2019; Codeluppi et al., 2018 |
| Imaging screen (Cell Painting, optical) | Measures morphological phenotypes after perturbation. | spatial readout, morphological phenotypes of multiple cellular components | enhancer perturbations may not cause morphological phenotypes; lacks gene expression readout; limited cell type compatibility | Bray et al., 2016 |





