Novel regulators of islet function identified from genetic variation in mouse islet Ca2+ oscillations
Figures
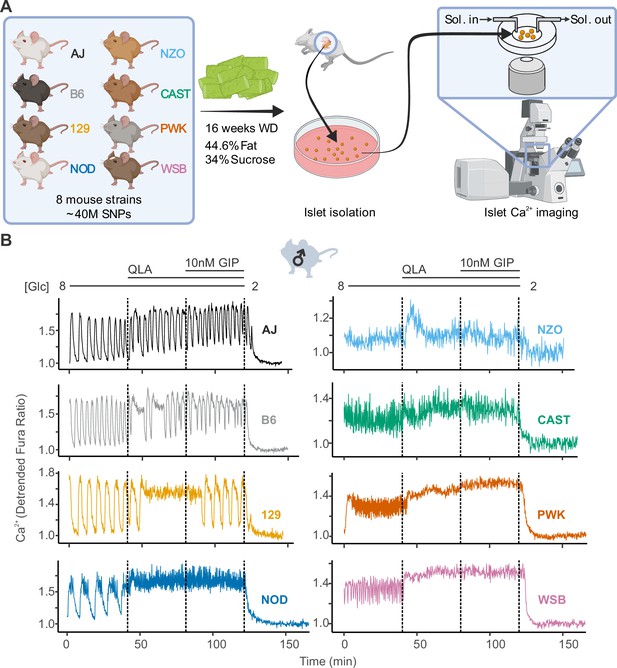
High diversity in Ca2+ oscillations across eight genetically distinct mouse strains.
(A) Male and female mice from eight strains (A/J; C57BL/6J (B6); 129S1/SvImJ (129); NOD/ShiLtJ (NOD); NZO/HILtJ (NZO); CAST/EiJ (CAST); PWK/PhJ (PWK); and WSB/EiJ (WSB)) were placed on a Western diet (WD) for 16 weeks before their islets were isolated. The islets were then imaged on a confocal microscope using Fura Red dye under conditions of 8 mM glucose (8G); 8G + 2 mM L-glutamine, 0.5 mM L-leucine, and 1.25 mM L-alanine (8G/QLA); 8G/QLA + 10 nM glucose-dependent insulinotropic polypeptide (8G/QLA/GIP); and 2 mM glucose. (B) Representative Ca2+ traces for male mice (n = 3–8 mice per strain, and 15–83 islets per mouse), with the transitions between solution conditions indicated by dashed lines. Abbreviations: ‘[Glc]’ = ‘concentration of glucose in mM’; ‘Sol.’ = ‘solution’; ‘SNPs’ = ‘single-nucleotide polymorphisms’.

The high diversity in Ca2+ oscillation in males is also observed in female mice.
(A) Representative Ca2+ traces for female mice (n = 3–7 mice per strain, and 11–94 islets per mouse) exhibit a high degree of variability across the eight strains. Abbreviations: ‘[Glc]’ = ‘concentration of glucose in mM’. (B) Representative Ca2+ trace for non-diabetic NZO female (left panel, fasting plasma glucose 69 mg/dl) and a diabetic NZO female (right trace, fasting plasma glucose >300 mg/dl). Related to Figure 1.
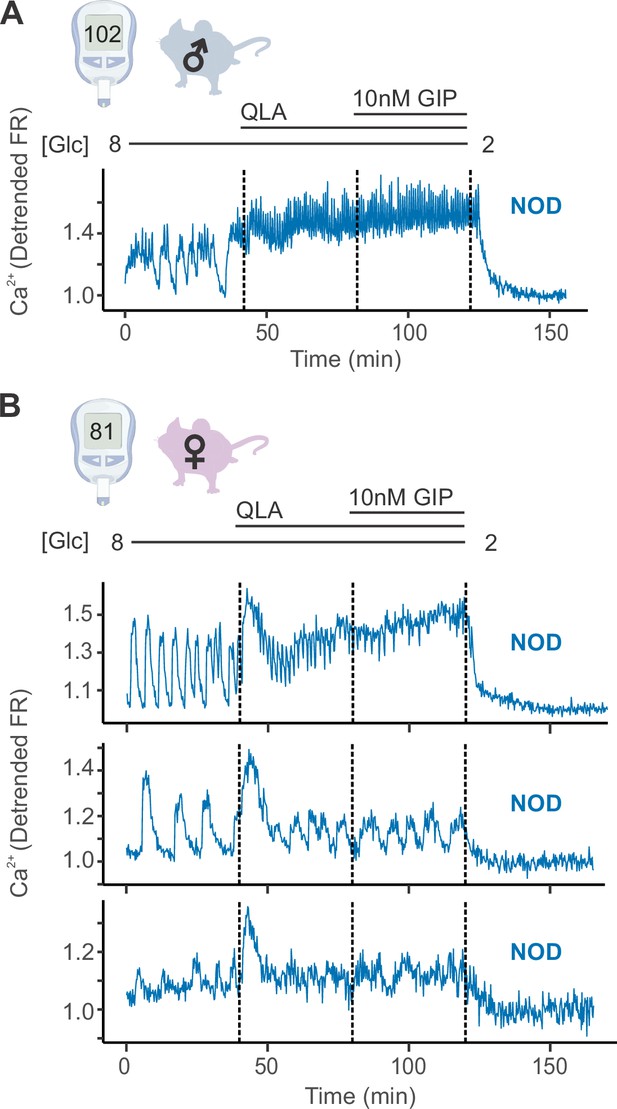
Diverse responses in non-diabetic NOD females’ islets.
(A) Representative Ca2+ traces for a male NOD mouse, for which islets closely resemble the trace pattern shown. (B) Example traces from islets of a single non-diabetic NOD female mouse. The pattern observed, where some of the mouse’s islets appeared similar to the NOD male islets (top panel), some appeared similar to the diabetic NZO islets (bottom panel), and some presented with an intermediate phenotype (oscillations present in all stimulatory conditions, but with a more pronounced 8/QLA initial peak and diminished amplitudes; middle panel), was consistently observed for all the NOD females. Abbreviations: ‘[Glc]’ = ‘concentration of glucose in mM’. Related to Figure 1.
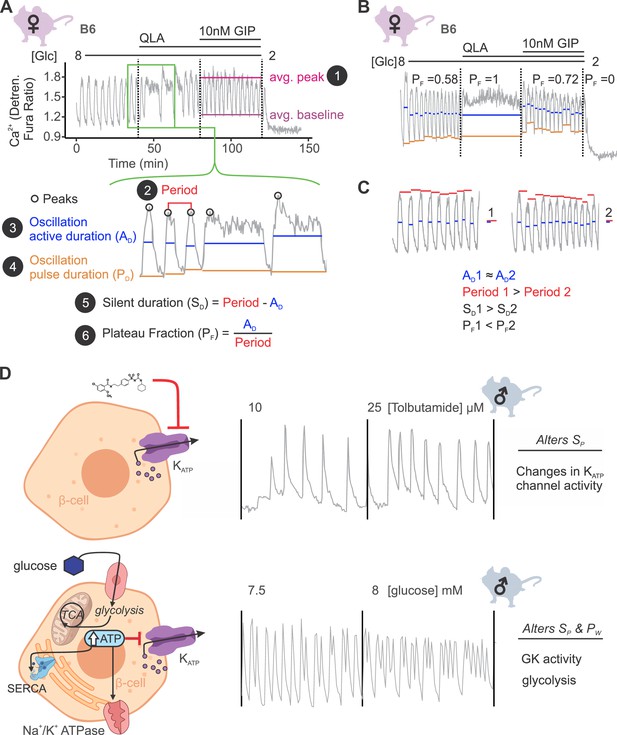
Ca2+ wave breakdown reveals mechanisms underlying Ca2+ responses.
(A) An example B6 female Ca2+ wave, showing that the islet oscillations can change in their average peak (1) and average baseline in response to different nutrients. Additionally, shifts in wave shape (green box) can be broken down into changes in time between peaks (period, 2), the time in the active phase (active duration, AD, 3), and the length of the oscillation (pulse duration, PD, 4). From these, the time inactive between oscillations (silent duration, SD, 5), and the relative time in the active phase, or plateau fraction (PF, 6), can be calculated. Each parameter can be changed by different underlying mechanisms. (B) For islets that plateaued, as in the example islet in 8G/QLA, they were assigned a plateau fraction of one and a period of zero. For islets that ceased to oscillate, such as the example islet in 2 mM glucose, they were assigned a plateau fraction of zero and a period of the time of measurement (40 min). (C) For trace 1 (left), which has a longer period (red bars) than trace 2 (right), but the same active duration (blue bars), the silent duration is greater and consequently the PF is shorter, in contrast to the trace in (A) where the PF increases between 8G and 8G/QLA are largely due to increases in AD. (D) Changes in specific Ca2+ wave parameters can reflect different mechanisms in β-cells. For example, changing KATP activity pharmacologically (upper panels) predominantly increases PF by altering SD, whereas increasing glucose concentrations by elevating glucose or activating GK cause significant alterations in both AD and SD to increase PF. Abbreviations: ‘[Glc]’ = ‘concentration of glucose in mM’; ‘GK’ = ‘glucokinase’.

Example of spectral density breakdown for Ca2+ traces.
(A) For the trace segment shown, the wave can be broken down into individual periodic waves (e.g. sine waves) of discrete amplitudes that, when added together, reproduce the trace. The frequency of each of these waves and its relative contribution to the overall trace signal is shown in (B), with frequency on the x axis and the spectral density, or the strength of each component signal, on the y axis. This was computed using scripts in R. The strongest two frequencies, denoted 1st and 2nd, respectively, are indicated. Related to Figure 2.
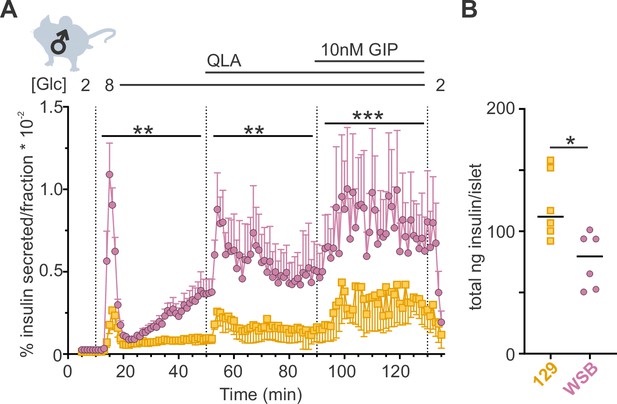
WSB mice secrete significantly more insulin than 129 mice.
(A) Insulin secretion was measured for perifused islets from WSB (n = 6, magenta circles) and 129 (n = 5, yellow squares) male mice in 2 mM glucose, 8G, 8G/QLA, and 8G/QLA/GIP. Transitions between solutions are indicated by dotted lines and the conditions for each are indicated above the graph. ‘[Glc]’ denotes the concentration of glucose in mM. Data are shown as a percentage of total islet insulin (mean ± standard error of the mean [SEM]). (B) Average total insulin per islet for the WSB and 129 males used in (A) with one exception: islets from one of the 129 mice were excluded from perifusion analysis due to technical issues with perifusion system on the day those animals’ islets were perifused. Dots represent individual values, and the mean is denoted by the black line. For (A), asterisks denote strain effect for the area-under-the-curve of the section determined by two-way analysis of variance (ANOVA), mixed effects model; **p < 0.01, ***p < 0.001. For (B), asterisk denotes p < 0.05 from Student’s t-test with Welch’s correction.

Average Ca2+ for the stimulatory conditions.
Average Ca2+ (detrended Fura Red ratio) for 8 mM glucose (8G, left panel), 8G with 1.25 mM L-alanine, 2 mM L-glutamine, and 0.5 mM L-leucine (8G/QLA, middle panel), and 8G/QLA with 10 nM GIP (8G/QLA/GIP, right panel) are shown for each strain/sex. n = 3–7 mice per strain. Related to Figures 3 and 4.
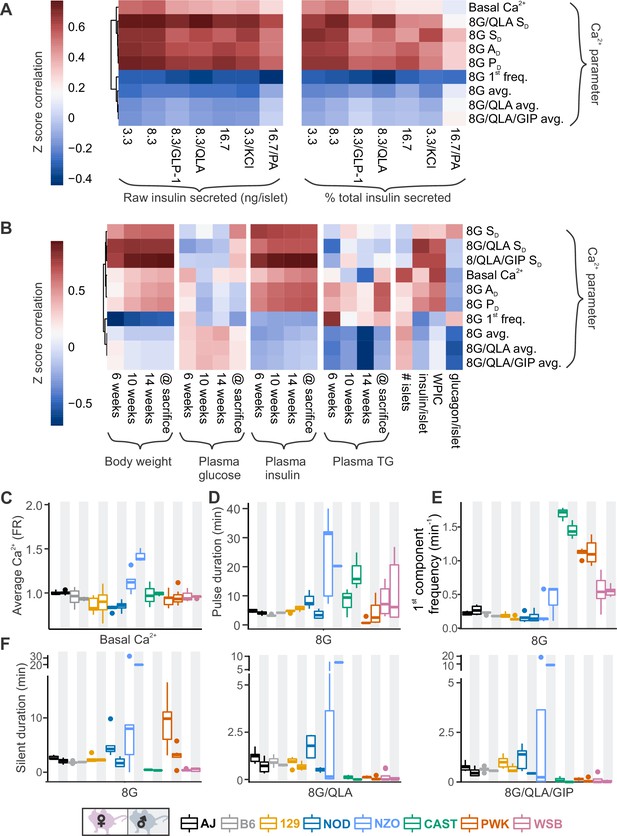
Comparing sex and strain patterns for Ca2+ metrics, insulin secretion, and clinical traits nominates Ca2+ metrics of interest.
(A) The Z-score correlation coefficient was calculated for Ca2+ parameters and raw insulin secreted and % total insulin secreted. Insulin measurements were previously collected for seven different secretagogues (16.7 mM glucose + 0.5 mM palmitic acid (16.7G/PA); 3.3 mM glucose + 50 mM KCl (3.3G/KCl); 16.7 mM glucose (16.7G); 8.3 mM glucose + 1.25 mM L-alanine, 2 mM L-glutamine, and 0.5 mM L-leucine (8.3G/QLA); 8.3 mM glucose + 100 nM GLP-1 (8.3G/GLP-1); 8.3 mM glucose (8.3G); and 3.3 mM glucose (3.3G)) (Mitok et al., 2018). (B) Correlation of the Ca2+ parameters to the clinical measurements in the founder mice which include (1) plasma insulin, triglycerides (TG), and glucose at 6, 10, and 14 weeks as well as at time of sacrifice; (2) number of islets; (3) whole-pancreas insulin content (WPIC); and (5) islet content for insulin and glucagon. For (A) and (B), the Ca2+ parameters shown here include average Ca2+ in 2 mM glucose (basal Ca2+); average Ca2+ in 8 mM glucose (8G avg.); average Ca2+ in 8 mM glucose + 1.25 mM L-alanine, 2 mM L-glutamine, and 0.5 mM L-leucine (8G/QLA avg); average Ca2+ in 8 mM glucose + QLA + 10 nM GIP (8G/QLA/GIP avg.); pulse duration in 8 mM glucose (8G PD); active duration in 8G (8G AD); silent duration in 8G (8G SD), 8G/QLA (8G/QLA/SD), and 8G/QLA/GIP (8G/QLA/GIP SD); and 1st component frequency in 8 mM glucose (8G 1st freq.). Other parameters analyzed are indicated in Figure 4—figure supplement 2 and Figure 4—figure supplement 3. (B–E) Sex and strain variability for (C) average Ca2+ determined by the Fura-ratio (FR) in 2 mM glucose, (D) pulse duration of oscillations in 8G, (E) 1st component frequency in 8G, and (F) silent duration of oscillations in 8G, 8G/QLA, and 8G/QLA/GIP.
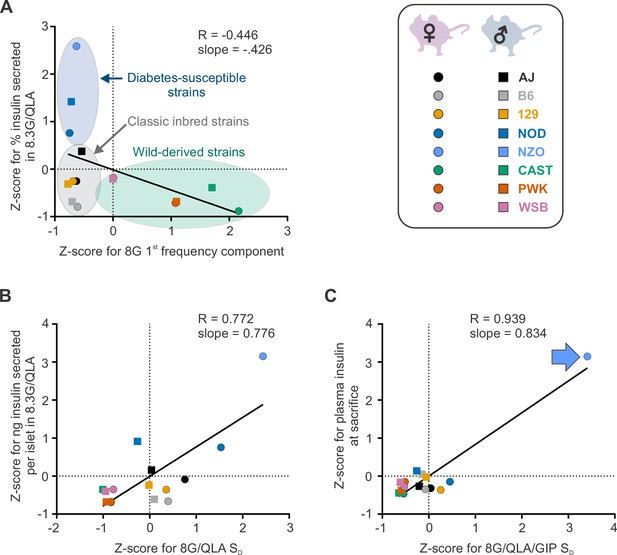
Differential strain and sex effects in correlations between traits.
The correlations between Z-scores for the calcium trait (x axis) and the other indicated trait (y axis) are shown for the eight strains (key, upper right). Males are indicated as boxes (□), females as circles (○) in each plot, where the color indicates the strain. (A) The correlation between the Z-scores for the 1st frequency component in 8 mM glucose (8G) and the percent insulin secreted in 8.3 mM glucose with 1.25 mM L-alanine, 2 mM L-glutamine, and 0.5 mM L-leucine (8.3G/QLA) reveal clustering of strains into the disease susceptible strains (blue ellipse), classic inbred strains (gray ellipse), and wild-derived strains (green ellipse). (B) The correlation between the Z-scores for silent duration (SD) in 8G/QLA and the ng of insulin secreted per islet in 8.3G/QLA show less of a separation between the three groups. (C) Some correlations, such as that between Z-scores for the SD in 8 mM glucose with 1.25 mM L-alanine, 2 mM L-glutamine, 0.5 mM L-leucine, and 10 nM GIP (8G/QLA/GIP) and the plasma insulin level at sacrifice show strong effects of single strains (e.g. NZO, indicated by blue arrow). In each plot, the Pearson’s R value and slope are indicated. Related to Figure 4.
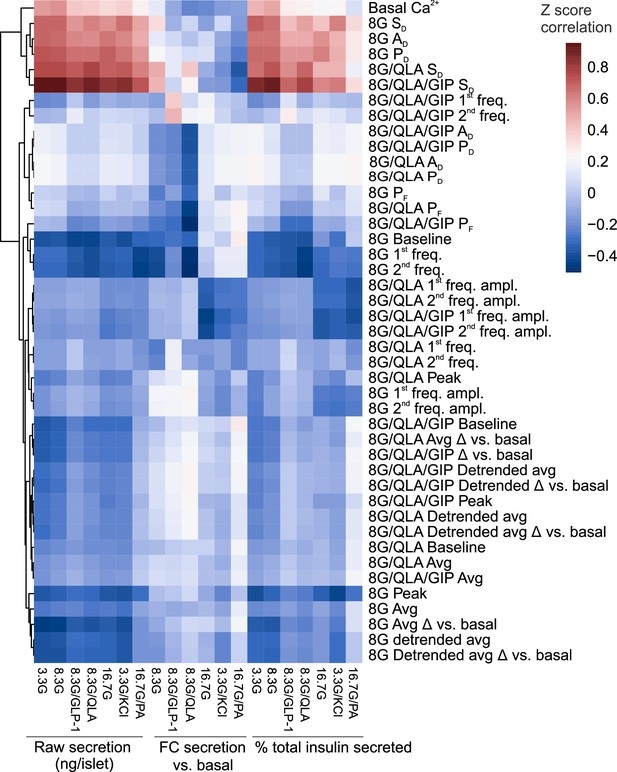
Correlation reveals specific Ca2+ parameters highly associated with insulin secretion.
Heatmap displaying the correlation coefficients between the Z-scores for Ca2+ wave metrics and the Z-scores for raw insulin secreted, fold-change (FC) over basal insulin secreted, and % of total islet insulin secreted. Insulin measurements were previously collected (Mitok et al., 2018) for seven different secretagogues (16.7 mM glucose + 0.5 mM palmitic acid (16.7G/PA); 3.3 mM glucose + 50 mM KCl (3.3G/KCl); 16.7 mM glucose (16.7G); 8.3 mM glucose + 1.25 mM L-alanine, 2 mM L-glutamine, and 0.5 mM L-leucine (8.3G/QLA); 8.3 mM glucose + 100 nM GLP-1 (8.3G/GLP-1); 8.3 mM glucose (8.3G); and 3.3 mM glucose (3.3G)). Perifusion conditions included 8 mM glucose (8G); 8 mM glucose + 1.25 mM L-alanine, 2 mM L-glutamine, and 0.5 mM L-leucine (8G/QLA); 8 mM glucose + QLA + 10 nM GIP (8G/QLA/GIP). Unsupervised clustering of the Ca2+ wave parameters revealed several parameters highly correlated to multiple insulin secretion conditions. Parameters included: average Ca2+ in 2 mM glucose (basal Ca2+), in 8 mM glucose (8G avg.), in 8G/QLA (8G/QLA avg), and in 8G/QLA/GIP (8G/QLA/GIP avg.); average detrended Ca2+ in 8 mM glucose (8G detr. avg.), in 8G/QLA (8G/QLA detr. avg), and in 8G/QLA/GIP (8G/QLA/GIP detr. avg.); average change in Ca2+ vs. basal in 8 mM glucose (8G avg. Δ vs. 2G), in 8G/QLA (8G/QLA avg. Δ vs. 2G), and in 8G/QLA/GIP (8G/QLA/GIP avg. Δ vs. 2G); change in detrended average Ca2+ vs. basal in 8 mM glucose (8G detr. Δ vs. 2G), in 8/QLA (8G/QLA detr. Δ vs. 2G), and in 8G/QLA/GIP (8G/QLA/GIP detr. Δ vs. 2G); average oscillation peak Ca2+ in 8G (8G peak), in 8G/QLA (8G/QLA peak), and in 8G/QLA/GIP (8G/QLA/GIP peak); average oscillation baseline Ca2+ in 8G (8G baseline), in 8G/QLA (8G/QLA baseline), and in 8G/QLA/GIP (8G/QLA/GIP baseline); pulse duration in 8G (8G PD), in 8G/QLA (8G/QLA PD), and in 8G/QLA/GIP (8G/QLA/GIP PD); active duration in 8 mM glucose (8G AD), in 8G/QLA (8G/QLA AD), and in 8G/QLA/GIP (8G/QLA/GIP AD); silent duration in 8 mM glucose (8G SD), in 8G/QLA (8G/QLA SD), and in 8G/QLA/GIP (8G/QLA/GIP SD); plateau fraction in 8 mM glucose (8G PF), in 8G/QLA (8G/QLA PF), and in 8G/QLA/GIP (8G/QLA/GIP PF); spectral density 1st component frequency in 8 mM glucose (8G 1st freq.), in 8G/QLA (8G/QLA 1st freq.), and in 8G/QLA/GIP (8G/QLA/GIP 1st freq.); spectral density 2nd component frequency in 8 mM glucose (8G 2nd freq.), in 8G/QLA (8G/QLA 2nd freq.), and in 8G/QLA/GIP (8G/QLA/GIP 2nd freq.); contribution of the 1st component to the Ca2+ waveform for 8 mM glucose (8G 1st freq. amp.), for 8G/QLA (8G/QLA 1st freq. amp.), and for 8G/QLA/GIP (8G/QLA/GIP 1st freq. amp.); and contribution of the 2nd component to the Ca2+ waveform for 8 mM glucose (8G 2nd freq. amp.), for 8G/QLA (8G/QLA 2nd freq. amp.), and for 8G/QLA/GIP (8G/QLA/GIP 2nd freq. amp.). Related to Figure 4.
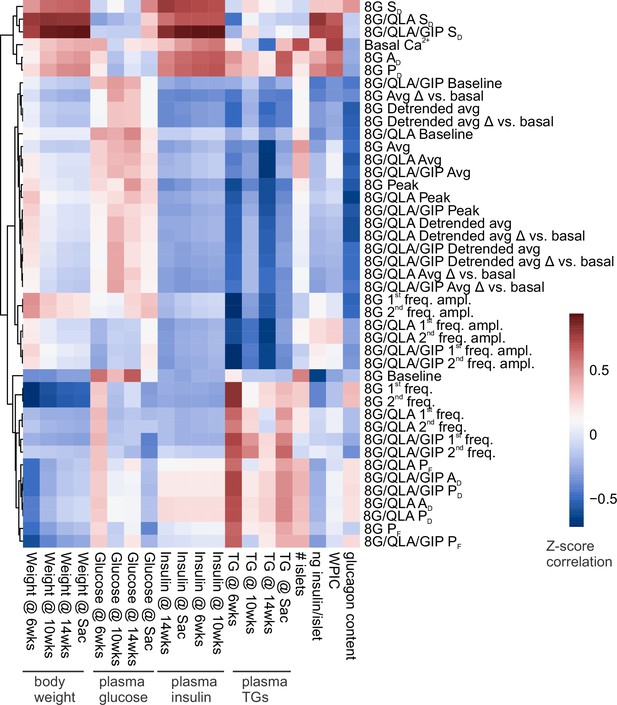
Correlation reveals specific Ca2+ parameters highly associated with in vivo traits.
Heatmap displaying the correlation coefficients between the Z-scores for Ca2+ wave metrics and the Z-scores for mouse in vivo metrics previously collected (Mitok et al., 2018) for the same strains and sexes. These traits included plasma insulin, glucose, and triglycerides (TGs) (measured at 6, 10, and 14 weeks of age as well as at sacrifice) as well as whole body weights at those time points. Whole-pancreas insulin content (WPIC), islet glucagon, islet number, and islet insulin content were also determined. Ca2+ perifusion conditions included 8 mM glucose (8G); 8 mM glucose + 1.25 mM L-alanine, 2 mM L-glutamine, and 0.5 mM L-leucine (8G/QLA); 8 mM glucose + QLA + 10 nM GIP (8G/QLA/GIP). Unsupervised clustering of the Ca2+ wave parameters revealed several parameters highly correlated to multiple in vivo traits. Parameters included: average Ca2+ in 2 mM glucose (basal Ca2+), in 8 mM glucose (8G), in 8G/QLA (8G/QLA), and in 8G/QLA/GIP (8G/QLA/GIP); average detrended Ca2+ in 8 mM glucose (8G detr.), in 8G/QLA (8G/QLA detr.), and in 8G/QLA/GIP (8G/QLA/GIP detr.); average change in Ca2+ vs. basal in 8 mM glucose (8G Δ vs. basal), in 8G/QLA (8G/QLA Δ vs. basal), and in 8G/QLA/GIP (8G/QLA/GIP Δ vs. basal); change in detrended average Ca2+ vs. basal in 8 mM glucose (8G detr. Δ vs. basal), in 8/QLA (8G/QLA detr. Δ vs. basal), and in 8G/QLA/GIP (8G/QLA/GIP detr. Δ vs. basal); average oscillation peak Ca2+ in 8G (8G peak), in 8G/QLA (8G/QLA peak), and in 8G/QLA/GIP (8G/QLA/GIP peak); average oscillation baseline Ca2+ in 8G (8G baseline), in 8G/QLA (8G/QLA baseline), and in 8G/QLA/GIP (8G/QLA/GIP baseline); pulse duration in 8G (8G PD), in 8G/QLA (8G/QLA PD), and in 8G/QLA/GIP (8G/QLA/GIP PD); active duration in 8 mM glucose (8G AD), in 8G/QLA (8G/QLA AD), and in 8G/QLA/GIP (8G/QLA/GIP AD); silent duration in 8 mM glucose (8G SD), in 8G/QLA (8G/QLA SD), and in 8G/QLA/GIP (8G/QLA/GIP SD); plateau fraction in 8 mM glucose (8G PF), in 8G/QLA (8G/QLA PF), and in 8G/QLA/GIP (8G/QLA/GIP PF); spectral density 1st component frequency in 8 mM glucose (8G 1st freq.), in 8G/QLA (8G/QLA 1st freq.), and in 8G/QLA/GIP (8G/QLA/GIP 1st freq.); spectral density 2nd component frequency in 8 mM glucose (8G 2nd freq.), in 8G/QLA (8G/QLA 2nd freq.), and in 8G/QLA/GIP (8G/QLA/GIP 2nd freq.); contribution of the 1st component to the Ca2+ waveform for 8 mM glucose (8G 1st freq. amp.), for 8G/QLA (8G/QLA 1st freq. amp.), and for 8G/QLA/GIP (8G/QLA/GIP 1st freq. amp.); and contribution of the 2nd component to the Ca2+ waveform for 8 mM glucose (8G 2nd freq. amp.), for 8G/QLA (8G/QLA 2nd freq. amp.), and for 8G/QLA/GIP (8G/QLA/GIP 2nd freq. amp.). Related to Figure 4.
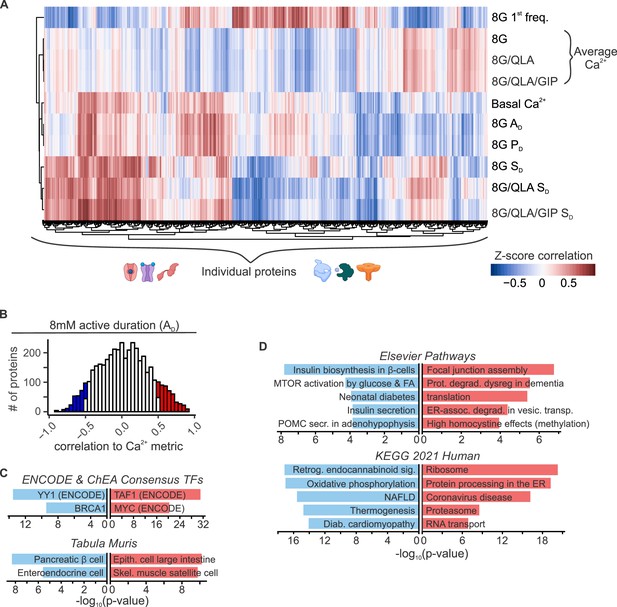
Islet proteins show correlation architecture to specific Ca2+ parameters.
(A) Unsupervised clustering of correlation coefficients between protein abundance Z-scores and Z-scores for the Ca2+ parameters indicated. Islet proteins show differential correlation values to basal Ca2+, excitatory Ca2+ (detrended average values for 8G, 8/QLA, and 8/QLA/GIP), active duration and pulse duration in 8G (8G PD and AD), and silent durations (SD) in 8G, 8G/QLA, and 8G/QLA/GIP. Correlation coefficients for other parameters are indicated in Figure 5—figure supplement 1. (B) Histograms representing the number of proteins that are correlated (red) and anticorrelated (blue) to 8G AD. ENCODE & CHEA Consensus transcription factor motif database and Tabula Muris tissue single-cell RNA-seq signature database (C) as well as pathway enrichments for the Elsevier Pathway database and KEGG 2021 Human pathway database (D) (−log10(p-values)), for the highly correlated (red) and anticorrelated (blue) proteins to 8 AD metric. Databases were queried using Enrichr (Chen et al., 2013; Kuleshov et al., 2016).

Correlation reveals proteins highly associated with specific Ca2+ parameters.
Heatmap displaying unsupervised clustering of the correlation coefficients between the Z-scores for Ca2+ wave metrics and the Z-scores for normalized islet protein abundance. Islet proteins were quantified previously (Mitok et al., 2018). Perifusion conditions included 8 mM glucose (8G); 8G mM glucose + 1.25 mM L-alanine, 2 mM L-glutamine, and 0.5 mM L-leucine (8G/QLA); 8 mM glucose + QLA + 10 nM GIP (8G/QLA/GIP). Unsupervised clustering of the Ca2+ wave parameters revealed several parameters highly correlated to multiple insulin secretion conditions. Parameters included: average Ca2+ in 2 mM glucose (basal Ca2+), in 8 mM glucose (8G avg.), in 8G/QLA (8G/QLA avg), and in 8G/QLA/GIP (8G/QLA/GIP avg.); average detrended Ca2+ in 8 mM glucose (8G detr. avg.), in 8G/QLA (8G/QLA detr. avg), and in 8G/QLA/GIP (8G/QLA/GIP detr. avg.); average change in Ca2+ vs. basal in 8 mM glucose (8G avg. Δ vs. 2G), in 8G/QLA (8G/QLA avg. Δ vs. 2G), and in 8G/QLA/GIP (8G/QLA/GIP avg. Δ vs. 2G); change in detrended average Ca2+ vs. basal in 8 mM glucose (8G detr. Δ vs. 2G), in 8G/QLA (8G/QLA detr. Δ vs. 2G), and in 8G/QLA/GIP (8G/QLA/GIP detr. Δ vs. 2G); average oscillation peak Ca2+ in 8G (8G peak), in 8G/QLA (8G/QLA peak), and in 8G/QLA/GIP (8G/QLA/GIP peak); average oscillation baseline Ca2+ in 8G (8G baseline), in 8G/QLA (8G/QLA baseline), and in 8G/QLA/GIP (8G/QLA/GIP baseline); pulse duration in 8G (8G PD), in 8G/QLA (8G/QLA PD), and in 8G/QLA/GIP (8G/QLA/GIP PD); active duration in 8G (8G AD), in 8G/QLA (8G/QLA AD), and in 8G/QLA/GIP (8G/QLA/GIP AD); silent duration in 8 mM glucose (8G SD), in 8G/QLA (8G/QLA SD), and in 8G/QLA/GIP (8G/QLA/GIP SD); plateau fraction in 8 mM glucose (8G PF), in 8G/QLA (8G/QLA PF), and in 8G/QLA/GIP (8G/QLA/GIP PF); spectral density 1st component frequency in 8 mM glucose (8G 1st freq.), in 8G/QLA (8G/QLA 1st freq.), and in 8G/QLA/GIP (8G/QLA/GIP 1st freq.); spectral density 2nd component frequency in 8 mM glucose (8G 2nd freq.), in 8G/QLA (8G/QLA 2nd freq.), and in 8G/QLA/GIP (8G/QLA/GIP 2nd freq.); contribution of the 1st component to the Ca2+ waveform for 8 mM glucose (8G 1st freq. amp.), for 8G/QLA (8G/QLA 1st freq. amp.), and for 8G/QLA/GIP (8G/QLA/GIP 1st freq. amp.); and contribution of the 2nd component to the Ca2+ waveform for 8 mM glucose (8G 2nd freq. amp.), for 8G/QLA (8G/QLA 2nd freq. amp.), and for 8G/QLA/GIP (8G/QLA/GIP 2nd freq. amp.). Related to Figure 5.
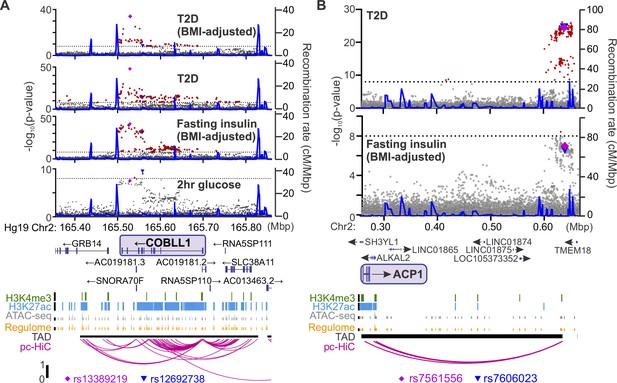
Identifying candidate protein targets by integrating human genome-wide association studies (GWAS).
(A) An example gene, COBLL1, orthologous to a gene coding for a protein identified as highly correlated to Ca2+ wave parameters in the founder mice. The recombination rate is indicated by the solid blue line. Significant single-nucleotide polymorphisms (SNPs; 8 < −log10(p), red) decorate the gene body for multiple glycemia-related parameters (in bold). Human islet chromatin data (Miguel-Escalada et al., 2019) for histone methylation (H3K4me3), histone acetylation (H3K27ac), ATAC-sequencing (ATAC-seq), and regulome score suggest active transcription of the gene within a topologically associated domain (TAD). Human islet promoter-capture HiC data (pc-HiC) (Miguel-Escalada et al., 2019) show contacts between the SNP-containing regions decorating the gene and its promoter. The highest SNP for 2 hr glucose (▼) and the other parameters (♦) are indicated. (B) Some orthologues did not show SNPs decorating the gene itself but did show looping to regions with SNPs for glycemic traits. The promoter of ACP1, for example, loops to a region within its topologically associated domain (black bar) with strong SNPs for type 2 diabetes risk and near-threshold SNPs for fasting insulin adjusted for body mass index (BMI). Some SNPs (▼, ♦) lie directly on the contact regions identified by HiC, whereas others lie immediately proximal to these contacts. For both panels, the significance of association (−log10 of the p-value) for the individual SNPs is on the left y axis and the recombination rate per megabasepair (Mbp) is on the right y axis. Chromosomal position in Mbp is aligned to Hg19. SNP data were provided by the Common Metabolic Diseases Knowledge Portal (https://hugeamp.org/).
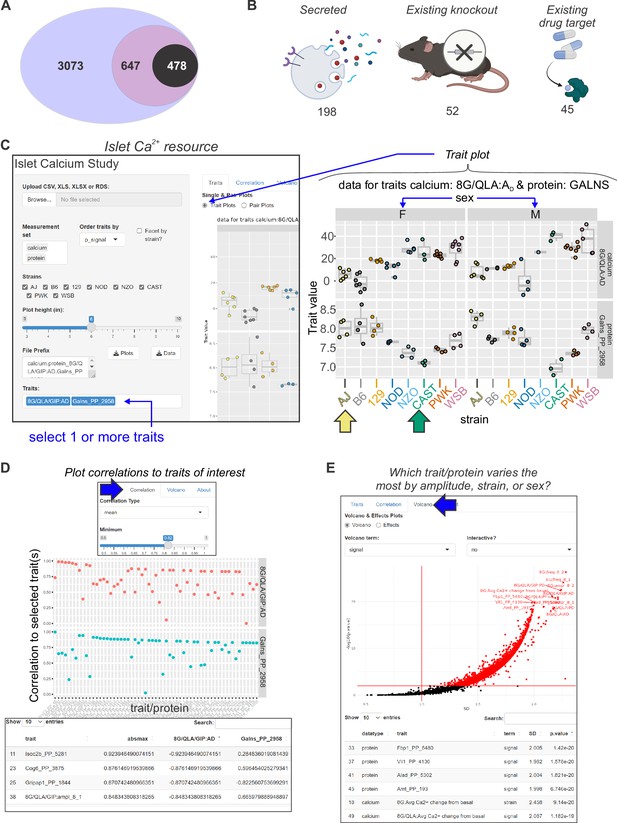
Mining Ca2+ data using a novel online resource .
(A) 3073 islet proteins significantly correlated to islet Ca2+ parameters of interest. Among the proteins, 647 had orthologues containing single-nucleotide polymorphisms (SNPs) for glycemic traits. Of these, 478 showed no results in our starting triage (see Methods) under any alias, suggesting they may be understudied in islet biology. (B) Of these 478 proteins, 198 were found to be secreted either as soluble proteins or in exosomes (Bateman et al., 2021; Thul et al., 2017; Uhlén et al., 2019; Navajas et al., 2022; Wang et al., 2013; Chen et al., 2019; Gonzales et al., 2009), 52 have existing knockout mice with annotated glycemia or pancreatic phenotypes (Groza et al., 2023; Blake et al., 2021), and 45 have existing compounds that target them (Stanford et al., 2021; Coker et al., 2019; Davies et al., 2015; Gaulton et al., 2017; Santos et al., 2017; Zhou et al., 2022). To make these data more accessible, we have developed an online resource that enables individuals to query the Ca2+ and proteomic data simultaneously. The user can select proteins and calcium traits (C) and display strain and sex distribution of these traits to determine the ideal backgrounds on which to test their traits or proteins of interest. In this example, GALNS is highly correlated to 8G/QLA/GIP AD, with the highest and lowest abundance strains for GALNS being AJ (yellow arrow) and CAST (green arrow), respectively. (D) The user can also query for the correlations between Ca2+ traits and proteins against one another or other traits of the same category. (E) The user can also see which of the traits or proteins has the largest change and most significant effects by sex, strain, or sex and strain.
Tables
Imaging medium formula.
Components are indicated by chemical abbreviation on the left and final concentration in mM is indicated in the right column.
| Component | Concentration (mM) |
|---|---|
| NaCl | 137 |
| KCl | 5.6 |
| MgCl2 | 1.2 |
| NaH2PO4·H2O | 0.5 |
| NaHCO3 | 4.2 |
| HEPES | 10 |
| CaCl2 | 2.6 |
Categories included in single-nucleotide polymorphism (SNP) queries.
These terms were considered as glycemia related and are categorized as such on the Common Metabolic Diseases Knowledge portal, which was queried for the relevant SNPs. Also included but not listed here were variations of these terms that were adjusted for body mass index (BMI).
| Fasting hormones | Glucose related | Tolerance test | Diabetes risk |
|---|---|---|---|
| Insulin | Fasting glucose | 2 hr glucose | T1D |
| Proinsulin | Random glucose | 2 hr insulin | T2D |
| C-peptide | Hba1c | 2 hr C-peptide | |
| Fasting glc–BMI interaction | Acute insulin response | ||
| Fasting ins–BMI interaction | SI-adjusted acute ins. Resp. | ||
| Gestational diabetes/altered fast glucose in pregnancy | AUC insulin | ||
| AUC insulin/AUC glucose | |||
| Corrected insulin response | |||
| HOMA-B | |||
| HOMA-IR | |||
| Ins. Secretion rate | |||
| Ins. Sensitivity | |||
| Incremental ins. @ 30 min OGTT | |||
| Insulin @ 30 min OGTT | |||
| Peak ins. response | |||
| Peak ins. Response adj SI |
Additional files
-
Supplementary file 1
Proteins correlated with Ca2+ parameters that have glycemic-related single-nucleotide polymorphisms (SNPs).
This includes protein IDs, gene names, gene IDs, and human orthologues for each of the proteins that correlate to one of the following metrics and have a glycemic-related SNP (see Table 2): basal Ca2+, 8G SD, 8G/QLA SD, 8G/QLA/GIP SD, 8G AD, 8G PD, and 8G 1st freq.
- https://cdn.elifesciences.org/articles/88189/elife-88189-supp1-v1.xlsx
-
Supplementary file 2
Proteins understudied in islet biology.
This table of proteins indicates the subset of proteins meeting our selection criteria (Supplementary file 1) that did not have any results in Pubmed, Google Scholar, or Google for any alias and the term ‘insulin secretion’, suggesting that they may be understudied in islet biology. The gene symbols for the mouse gene and human orthologue are indicated. The ‘Mouse?’ column indicates whether a knockout mouse with metabolic phenotypes exists (identified from sources in the subsequent ‘Source’ column). The ‘Drug?’ column similarly indicates whether any source (indicated in the following ‘Source’ column) shows existing compound(s) targeting the protein. Finally, the ‘Secreted?’ column indicates whether any source (indicated in the subsequent ‘Source’ column) shows an isoform of the protein to be secreted.
- https://cdn.elifesciences.org/articles/88189/elife-88189-supp2-v1.xlsx
-
Supplementary file 3
Enrichments for the highly correlated and anticorrelated proteins.
The Enrichr tool (Chen et al., 2013; Kuleshov et al., 2016) queries multiple databases for information regarding gene lists and queries can be stored for access later using hyperlinks. This Excel file contains five tabs. The ‘Key’ tab indicates the contents of the file. The ‘Uniprot_IDs_correlated’ and ‘Uniprot_IDs_anticorrelated’ tabs each, respectively, contain in their columns lists of the Uniprot IDs for peptides correlated (coefficient >0.5) or anticorrelated (coefficient <−0.5) to specific Ca2+ parameters listed in the row labeled ‘Traits’. These Uniprot IDs were queried to determine the gene names. The ‘Enrichr_correlated’ and ‘Enrichr_anticorrelated’ tabs each, respectively, contain these corresponding gene names for those proteins correlated (coefficient >0.5) or anticorrelated (coefficient <−0.5) to specific Ca2+ parameters listed in the row labeled ‘Traits’. In the row ‘Enrichr Link’ contains the Enrichr query hyperlinks for each protein list. For example, the Enrichr_correlated column B has the link https://maayanlab.cloud/Enrichr/enrich?dataset=affcd1912271cd603ec6e26304ac789e which is the Enrichr database search for the proteins listed in that column that correlate with the 1st frequency component in 8G. This table is available via the Zenodo repository (DOI 10.5281/zenodo.7776230).
- https://cdn.elifesciences.org/articles/88189/elife-88189-supp3-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/88189/elife-88189-mdarchecklist1-v1.docx





