Loss of the extracellular matrix protein Perlecan disrupts axonal and synaptic stability during Drosophila development
Figures
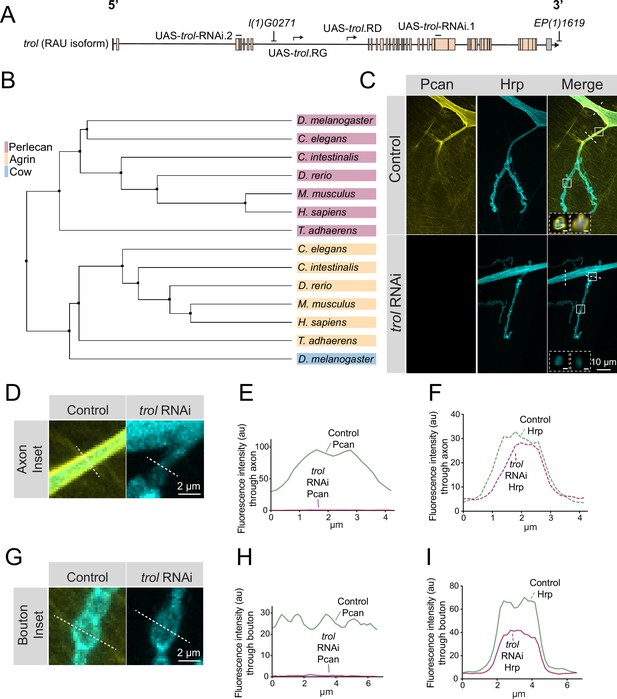
Perlecan conservation and localization within the Drosophila PNS.
(A) Diagram of the trol-RAU isoform with exons (boxes) and introns (lines) indicated. Sequence locations targeted by two UAS-trol RNAi lines and start sites for two overexpression constructs (UAS-trol.RD and UAS-trol.RG) used in this study are noted. The location of P-elements l(1)G0271 and EP(1)1619 previously mobilized to generate the trolnull deletion allele (Voigt et al., 2002) is also shown. (B) Phylogenetic tree of Perlecan, Agrin and Carrier of Wingless (Cow) from the indicated species generated using BLOSUM62 average distance. (C) Representative images of muscle 4 NMJs stained for Perlecan (Pcan) and Hrp in control (trolGFP, UAS-trol-RNAi.1/+;+;+) or Perlecan knockdown (trolGFP, UAS-trol-RNAi.1/+;+;tub-Gal4/+) larvae. White boxes denote location of insets depicted in D and G. Dashed white lines denote location of insets depicted in lower left corner. Left inset displays orthogonal section through axon bundles, showing Perlecan signal in the neural lamella (scale bar 2 μm). Right inset displays orthogonal section through individual axon, showing Perlecan signal in the neural lamella (scale bar 1 μm). (D, G) Magnified view of control and trol RNAi axons (D) and boutons (G), highlighting loss of Perlecan following RNAi knockdown. Dashed lines show representative sites for line scanning quantification for panels E-F, H-I. (E–F, H–I) Line scan profiles of Perlecan (E, H) or Hrp (F, I) mean fluorescent intensity through axons (E, F) or synaptic boutons (H, I) at muscle 4 in segment A2 (control: 14 axons from 8 larvae; trol RNAi: 19 axons from 11 larvae; control: 14 NMJs from 8 larvae; trol RNAi: 22 NMJs from 11 larvae). Control measurements are denoted in green and trol RNAi in magenta.
-
Figure 1—source data 1
Raw Values and Statistics for Figure 1 Perlecan Localization.
- https://cdn.elifesciences.org/articles/88273/elife-88273-fig1-data1-v1.xlsx
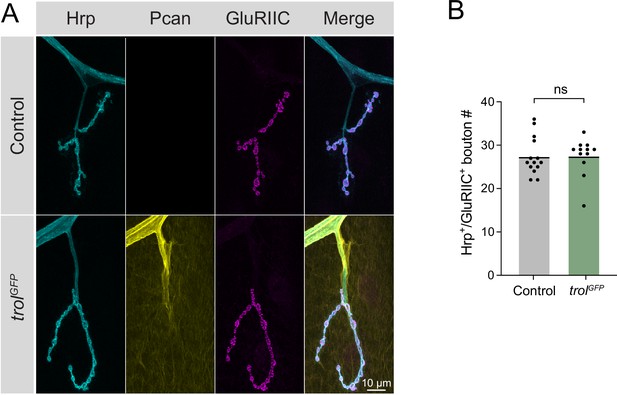
Endogenous TrolGFP strain produces a functional Perlecan protein.
(A) Representative images of muscle 4 NMJs from segment A4 in control (CS) and TrolGFP (trolGFP/y;+;+) larvae stained for Hrp, Perlecan (Pcan) and the glutamate receptor subunit GluRIIC. Merged images are shown on the right. (B) Quantification of Hrp+/GluRIIC+ Ib synaptic bouton number in control (27.4±1.2, 14 NMJs from 7 larvae) and TrolGFP (27.5±1.3, 12 NMJs from 6 larvae, p=0.9654) larvae. Each point represents the number of boutons at one muscle 4 NMJ in segment A4. The mean is depicted by the solid black line.
-
Figure 1—figure supplement 1—source data 1
Raw Values and Statistics for Figure 1—figure supplement 1 on Bouton Number.
- https://cdn.elifesciences.org/articles/88273/elife-88273-fig1-figsupp1-data1-v1.xlsx
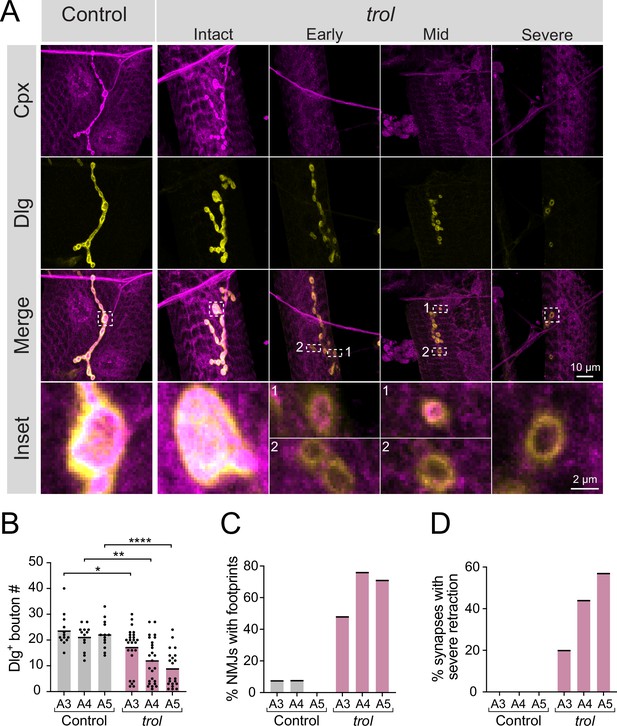
Synaptic retraction in trolnull motoneurons.
(A) Representative images of muscle 4 NMJs in control (trolnull/+;+;+, left panel) and trolnull (trolnull/y;+;+) larvae stained for Cpx (magenta) and Dlg (yellow). Example trol NMJs of increasing retraction severity are shown in the right panels. Highlighted areas in the merged image are shown as insets at the bottom. For mild and moderately retracted NMJs, inset 1 displays a bouton with pre- and postsynaptic material still co-localized while inset 2 highlights a synaptic footprint with only postsynaptic material remaining. (B) Quantification of Dlg +Ib bouton number from control and trol muscle 4 NMJs for abdominal segments A3-A5. Each point represents the number of boutons at one NMJ, with mean bouton number indicated with the solid black line. Quantification of bouton number: control A3: 23.5±1.8, 13 NMJs from 7 larvae; trol A3: 17.2±1.6, 25 NMJs from 13 larvae, p<0.05; control A4: 21.1±1.3, 13 NMJs from 7 larvae; trol A4: 12.0±1.7, 25 NMJs from 13 larvae, p<0.01; control A5: 22.1±1.5, 13 NMJs from 7 larvae; trol A5: 8.9±1.6, 21 NMJs from 13 larvae, p<0.0001. (C) Percentage of control or trol NMJs with one or more postsynaptic footprints (Dlg bouton lacking Cpx) for segments A3-A5 from the dataset in B. (D) Percentage of control or trol NMJs with severe retraction for segments A3-A5 from the dataset in B.
-
Figure 2—source data 1
Raw Values and Statistics for Figure 2 on Synapse Number and Retraction.
- https://cdn.elifesciences.org/articles/88273/elife-88273-fig2-data1-v1.xlsx

Synapse retraction occurs in trolnull mutants over deficiency and in trol RNAi knockdown larvae.
(A) Representative images of muscle 4 NMJs in Df(1)ED411/+ controls and Df(1)ED411/trolnull larvae stained for the presynaptic AZ protein Brp (yellow), glutamate receptor GluRIIC (magenta) and anti-Hrp (cyan, only shown in inset). The merged image is shown below with location of insets boxed. Insets show a control bouton and an intact and retracting bouton from trolDf/trolnull NMJs. (B) Quantification of Hrp+/GluRIIC +Ib bouton number from control and trolDf/trolnull muscle 4 NMJs in segments A3 and A4. Each point represents the number of boutons at one NMJ, with mean bouton number indicated with the solid black line. Quantification of bouton number: control: 20.4±0.7, 14 NMJs from 4 larvae; trolDf/trolnull: 15.1±1.4, 20 NMJs from 5 larvae, p<0.005. (C) Percentage of control or trolDf/trolnull NMJs with one or more postsynaptic footprints from the dataset in B. (D) Representative images of muscle NMJs in RNAi (+;UAS-trol-RNAi.2/+;+) and Gal4 driver (+;+;tubulin-Gal4/+) controls and trol knockdown (+;UAS-trol-RNAi.2/+;tubulin-Gal4/+) larvae stained stained for Brp (yellow), GluRIIC (magenta), and anti-Hrp (cyan, only shown in inset). The merged image is shown below with location of the insets boxed. Insets show control or intact, moderate, or severe retraction boutons in trol RNAi knockdown larvae. (E) Quantification of Hrp+/GluRIIC +Ib bouton number from controls and trol RNAi knockdown muscle 4 NMJs in segments A3-A5. Each point represents the number of boutons at one NMJ, with mean bouton number indicated with the solid black line. Quantification of bouton number: RNAi control A3: 26.8±1.6, 12 NMJs from 6 larvae; Gal4 driver control A3: 27.9±1.6, 12 NMJs from 6 larvae; tub >RNAi A3: 9.5±2.3, 12 NMJs from 6 larvae, RNAi versus driver controls p=0.99, RNAi control and tub >RNAi p<0.0001; RNAi control A4: 27.3±1.8, 12 NMJs from 6 larvae; Gal4 control A4: 25.2±0.6, 11 NMJs from 6 larvae; tub >RNAi A4: 6.3±2.2, 12 NMJs from 6 larvae, RNAi versus driver controls p=0.95, RNAi control and tub >RNAi p<0.0001; RNAi control A5: 26.6±1.2, 12 NMJs from 6 larvae; Gal4 control A5: 31.6±2.3, 12 NMJs from 6 larvae; tub >RNAi A5: 4.7±1.6, 12 NMJs from 6 larvae, RNAi versus driver controls p=0.25, RNAi control and tub >RNAi p<0.0001. (F) Percentage of control or trol RNAi knockdown NMJs with one or more postsynaptic footprints for segments A3-A5 from the dataset in E.
-
Figure 2—figure supplement 1—source data 1
Raw Values and Statistics for Figure 2—figure supplement 1 on Trol Mutant and RNAi Retraction.
- https://cdn.elifesciences.org/articles/88273/elife-88273-fig2-figsupp1-data1-v1.xlsx
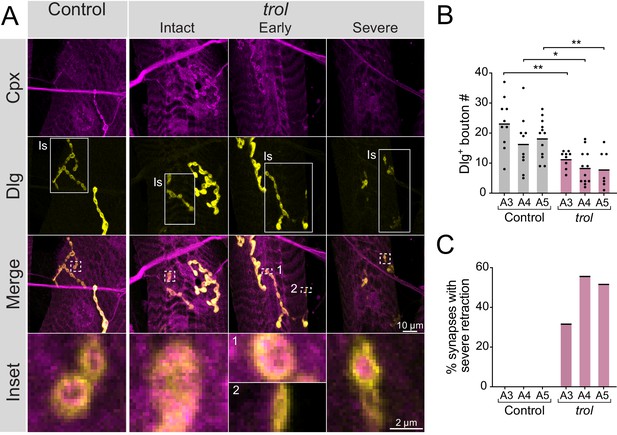
trolnull Is synapses retract.
(A) Representative images of Is muscle 4 NMJs in control (trolnull/+;+;+) and trolnull (trolnull/y;+;+) larvae stained for Cpx (magenta) and Dlg (yellow). Example trol NMJs that are intact or in the early or severe stages of retraction are shown in the right panels. White boxes in the 2nd row of images highlight Is innervation, with the remaining Dlg staining coming from the co-innervating Ib MN. Highlighted areas in the merged image are shown as insets in the bottom panel. For the NMJ in early stages of retraction, inset 1 displays a bouton containing both pre- and postsynaptic material, while inset 2 highlights a synaptic footprint with only postsynaptic material remaining. For the severely retracted NMJ, inset displays a bouton with pre- and postsynaptic material remaining (top) and one with only postsynaptic material (bottom). (B) Quantification of Dlg +Is bouton number from control and trolnull muscle 4 NMJs for segments A3-A5. Each point represents the number of boutons at one NMJ, with mean bouton number indicated with the solid black line. Quantification of bouton number: control A3: 23.3±2.7, 10 NMJs from 7 larvae; trol A3: 11.4±1.0, 6 NMJs from 13 larvae, p<0.001; control A4: 16.5±2.9, 10 NMJs from 7 larvae; trol A4: 8.5±1.6, 12 NMJs from 13 larvae, p<0.03; control A5: 18.3±1.9, 12 NMJs from 7 larvae; trol A5: 8.0±2.3, 7 NMJs from 13 larvae, p<0.01. (C) Percentage of control or trolnull Is NMJs with severe retraction for segments A3-A5 from the dataset in B.
-
Figure 2—figure supplement 2—source data 1
Raw Values and Statistics for Figure 2—figure supplement 2 on Segmental Retraction.
- https://cdn.elifesciences.org/articles/88273/elife-88273-fig2-figsupp2-data1-v1.xlsx
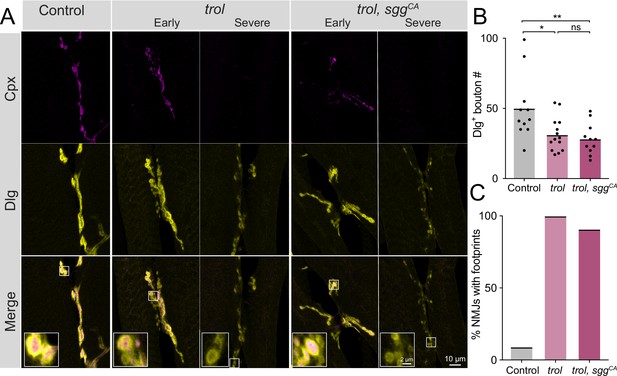
Synaptic retraction in trol mutants is not prevented by blocking presynaptic Wg signaling.
(A) Representative images of muscle 6/7 NMJs at segment A4 in control (+;UAS-sggS9A/+;+, left panel), trolnull (trolnull,vGlut-Gal4/y;+;+, middle panels) and trolnull, sggCA (trolnull,vGlut-Gal4/y;UAS-sggS9A/+;+, right panels) larvae stained for Cpx (magenta) and Dlg (yellow). Example NMJs that are in early or severe stages of retraction are shown for both trol genotypes. Highlighted areas in the merge are shown as insets in the bottom panel with either intact (early) or retracting (severe) boutons. (B) Quantification of Dlg +Ib and Is bouton number from control, trolnull and trolnull, sggCA muscle 6/7 NMJs for abdominal segment A4. Each point represents the number of boutons at one NMJ, with mean bouton number indicated with the solid black line. Quantification of bouton number: control: 50.1±7.0, 11 NMJs from 6 larvae; trol: 31.3±3.2, 14 NMJs from 7 larvae, p<0.05 compared to control; trol, sggCA: 28.2±3.4, 11 NMJs from 6 larvae; p<0.01 compared to control, p=0.8814 compared to trol. (C) Percentage of control, trol, or trol, sggCA NMJs with one or more postsynaptic footprints for the dataset in B.
-
Figure 3—source data 1
Raw Values and Statistics for Figure 3 on Perlecan - Wingless Pathway Interactions.
- https://cdn.elifesciences.org/articles/88273/elife-88273-fig3-data1-v1.xlsx
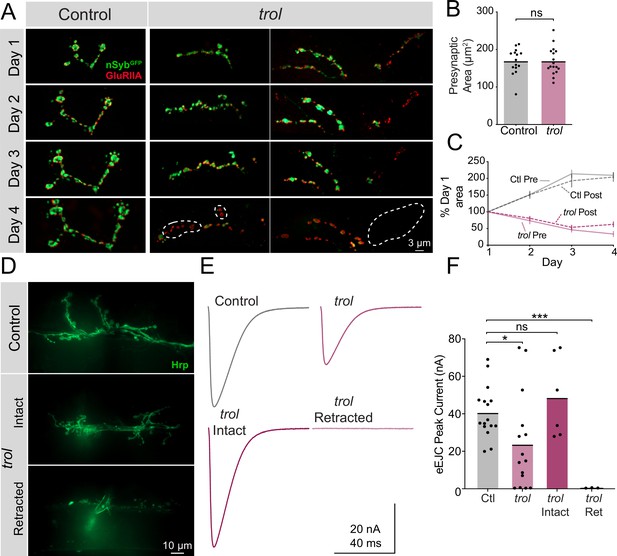
Trol mutant NMJs form normally but are not maintained over development.
(A) Representative NMJ images during serial intravital imaging of muscle 26 in control (trolnull/+;+;nSybGFP/GluRIIARFP, left) and trolnull (trolnull/y;+;nSybGFP/GluRIIARFP, right) larvae over 4 days starting at the 2nd instar stage. nSybGFP is shown in green and GluRIIARFP in red. Dashed lines highlight areas where presynaptic nSyb is missing while postsynaptic GluRIIA remains (retraction footprints). Severe NMJ retraction is seen in both example trolnull NMJs by day 4, while control NMJs continue to grow. (B) Quantification of muscle 26 presynaptic NMJ area in control and trolnull 2nd instar larvae at the beginning of serial intravital imaging sessions. No differences in NMJ area are present during this stage of early development. Quantification of NMJ area: control: 168.5±8.974, 15 NMJs from 4 larvae; trol: 168.5±8.855, 17 NMJs from 4 larvae, p=0.9991. NMJs from multiple abdominal segments were imaged. (C) Percent change in NMJ area during each day of imaging for control and trolnull larvae. Both presynaptic and postsynaptic area continue to increase in controls, while synaptic area is lost in trolnull larvae. (D) Representative images of muscle 6/7 NMJs stained with anti-Hrp following TEVC physiology in control (trolnull/+;+;+) and trolnull (trolnull/y;+;+) 3rd instar larvae. Examples of intact (middle) and retracted (bottom) NMJs are shown. (E) Average eEJC traces in 0.3 mM Ca2+ saline in control and all trolnull NMJs combined (top), together with average traces from NMJs of only intact or retracted trolnull NMJs (bottom). (F) Quantification of average eEJC peak amplitude (nA) per NMJ in segments A3 and A4 for the indicated genotypes. Intact and retracted trol NMJs were determined post-hoc blinded following anti-Hrp staining and paired with their corresponding eEJC data. Quantification of EJC amplitude: control: 40.8±3.5 nA, 16 NMJs from 7 larvae; trol combined: 23.8±6.6, 15 NMJs from 6 larvae, p<0.05 compared to control; trol intact: 48.8±8.9 nA, 6 NMJs from 4 larvae, p=0.3189 compared to control; trol retracted: 0.5±0.1, 3 NMJs from 2 larvae, p<0.0001 compared to control.
-
Figure 4—source data 1
Raw Values and Statistics for Figure 4 on Trol Serial Imaging and Physiology.
- https://cdn.elifesciences.org/articles/88273/elife-88273-fig4-data1-v1.xlsx
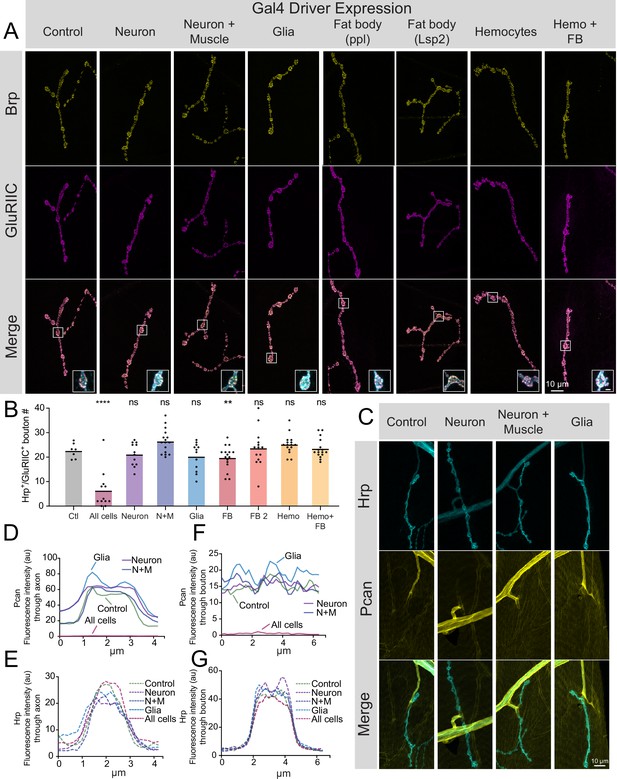
Perlecan acts in non-cell autonomous fashion to control synaptic maintenance.
(A) Representative images of larval muscle 4 NMJs at segment A4 stained for Brp (yellow), GluRIIC (magenta) and anti-Hrp (cyan, only shown in inset) in control (+;UAS-trol-RNAi.2/+;+) and trol RNAi knockdown in the indicated cell types (UAS-trol-RNAi.2 (one copy) driven by elavC155, elavC155 and mef2-Gal4, repo-Gal4, ppl-Gal4, Lsp2-Gal4, Hml-Gal4, and c564-Gal4 (one copy)). The merged image is shown below with location of the insets highlighting single boutons. Inset scale bar is 2 μm. (B) Quantification of Hrp+/GluRIIC +positive Ib bouton number at muscle 4 NMJs in segment A4 in controls and following cell-type specific trol RNAi knockdown. Each knockdown was analyzed in separate experiments with both UAS only and Gal4 only controls. Significance was calculated for each experimental comparison, but a single control that represents the average bouton number of every experiment is plotted for ease of visualization. Quantification of trol knockdown with tubulin-Gal4 at A4 (from Figure 2—figure supplement 1) is included for comparison. Unlike pan-cellular RNAi, cell-type specific RNAi does not induce synaptic retraction. Quantification of bouton number: neuron UAS only control: 23.25±1.399, 12 NMJs from 6 larvae; neuron Gal4 only control: 23.75±1.226, 12 NMJs from 6 larvae; elavC155 >UAS-trol-RNAi.2: 21.17±1.359, 12 NMJs from 6 larvae; p=0.4424 compared to UAS control; p=0.2994 compared to Gal4 control; neuron and muscle UAS only control: 21.21±1.130, 14 NMJs from 7 larvae; neuron and muscle Gal4 only control: 23.43±1.189, 14 NMJs from 7 larvae; elavC155, mef2 >UAS-trol-RNAi.2: 26.44±1.248, 16 NMJs from 8 larvae; p=0.0066 (<0.01) compared to UAS control; p=0.1437 compared to Gal4 control; glia UAS only control: 26.83±1.375, 12 NMJs from 6 larvae; glia Gal4 only control: 23.79±1.407, 14 NMJs from 7 larvae; repo >UAS-trol-RNAi.2: 20.25±1.643, 12 NMJs from 6 larvae; p=0.0078 (<0.01) compared to UAS control; p=0.1666 compared to Gal4 control; fat body (ppl) UAS only control: 25.57±1.312, 14 NMJs from 7 larvae; fat body (ppl) Gal4 only control: 26.21±0.7644, 14 NMJs from 7 larvae; ppl >UAS-trol-RNAi.2: 19.75±1.216, 16 NMJs from 8 larvae; p<0.01 compared to UAS and Gal4 controls (0.0014 compared to UAS; 0.0004 compared to Gal4); fat body 2 (Lsp2) UAS only control: 19.08±1.412, 13 NMJs from 7 larvae; fat body 2 (Lsp2) Gal4 only control: 25.07±0.7593, 14 NMJs from 7 larvae; Lsp2 >UAS-trol-RNAi.2: 23.64±2.053, 14 NMJs from 7 larvae; p=0.0731 compared to UAS control; p=0.7249 compared to Gal4 control; hemocyte UAS only control: 18.93±0.8285, 14 NMJs from 7 larvae; hemocyte Gal4 only control: 26±1.441, 14 NMJs from 7 larvae; Hml >UAS-trol-RNAi.2: 25.19±0.9841, 16 NMJs from 8 larvae; p=0.0005 (<0.001) compared to UAS control; p=0.8242 compared to Gal4 control; hemocyte and fat body UAS only control: 23.14±1.181, 14 NMJs from 7 larvae; hemocyte and fat body Gal4 only control: 24.25±0.8972, 12 NMJs from 6 larvae; c564 >UAS-trol-RNAi.2: 23.44±0.9999, 16 NMJs from 8 larvae; p=0.9705 compared to UAS control; p=0.8149 compared to Gal4 control. (C) Representative images of muscle 4 NMJs stained for Perlecan and Hrp in control (TrolGFP,UAS-trol-RNAi.1) or trol RNAi knockdown by elavC155, elavC155 and mef2-Gal4, or repo-Gal4 (one copy of UAS and Gal4 constructs). (D–G) Line scanning profiles of Perlecan (D,F) or Hrp (E,G) mean fluorescent intensity through axons (D,E) or synaptic boutons (F,G) at muscle 4 in segment A4. Measurements are color-coded as indicated: control (green), trol RNAi in neurons (magenta), neurons and muscles (blue), or glia (light blue). No reduction in Perlecan around nerves or on the muscle surface surrounding boutons was observed compared with tubulin-Gal4 knockdown (Figure 1F–I, replicated here (‘All cells’) for comparison).
-
Figure 5—source data 1
Raw Values and Statistics for Figure 5 on Cell-type Specfic Trol Knockdown.
- https://cdn.elifesciences.org/articles/88273/elife-88273-fig5-data1-v1.xlsx

Overexpression of Perlecan in trolnull motoneurons does not rescue synaptic retraction phenotypes.
(A) Representative images of muscle 4 NMJs in larval segments A4 for controls (+;+;UAS-trol.RG/+, 1st panel; +;+;UAS-trol.RD/+, 4th panel), trolnull larvae expressing Gal4 only (trolnull, vGlut-Gal4/y;+;+, 2nd and 5th panels) and trolnull larvae expressing UAS-trol.RG or UAS-trol.RD with vGlut-Gal4 (trolnull, vGlut-Gal4/y;+;UAS-trol.RG/+, 3rd panel; trolnull, vGlut-Gal4/y;+;UAS-trol.RD/+, 6th panel). Larvae were stained for Cpx (magenta) and Dlg (yellow), with the merged image below. In this set of panels, retracting NMJs had brightness enhanced to facilitate visualization of dim synaptic material. (B) Quantification of Dlg +Ib bouton number from control, trol (trolnull, vGlut-Gal4), or trol rescues (trolnull, vGlut-Gal4 driving UAS-trolRG or trolRD) at muscle 4 NMJs from segment A4. Each point represents the number of boutons at one NMJ, with mean bouton number indicated with the solid black line. Quantification of bouton number: RG experiments - UAS-trolRG: 28.4±2.0, 12 NMJs from 6 larvae; trolnull, vGlut-Gal4: 5.5±1.7, 16 NMJs from 8 larvae, p<0.0001 compared to control; trol, vGlut >UAS-trolRG rescues: 8.1±1.7, 14 NMJs from 7 larvae; p<0.0001 compared to control, p=0.54 compared to trolnull, vGlut-Gal4; RD experiments - UAS-trolRD: 22.0±1.4, 12 NMJs from 6 larvae; trolnull, vGlut-Gal4: 8.5±1.7, 10 NMJs from 5 larvae, p<0.0001 compared to control; trol, vGlut >UAS-trolRD rescues: 4.3±0.5, 15 NMJs from 8 larvae; p<0.0001 compared to control, p=0.043 (<0.05) compared to trolnull, vGlut-Gal4. (C) Percentage of NMJs with one or more postsynaptic footprints for segments A4 from the genotypes in panel B.
-
Figure 5—figure supplement 1—source data 1
Raw values and statistics for Figure 5—figure supplement 1 on cell-type specific trol rescue.
- https://cdn.elifesciences.org/articles/88273/elife-88273-fig5-figsupp1-data1-v1.xlsx
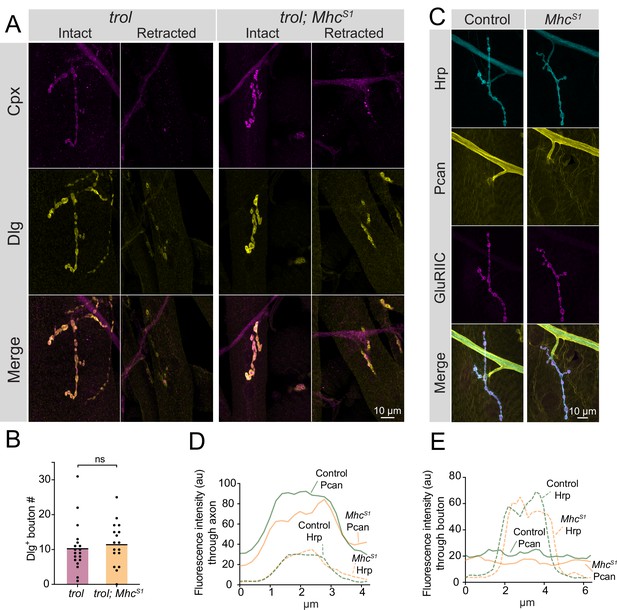
Enhanced muscle contraction does not exacerbate synaptic retraction in trol mutants.
(A) Representative images of muscle 4 NMJs at segment A3 in trol (trolnull/y;+;+) and trol; MhcS1 (trolnull/y;MhcS1/+;+) larvae stained for Cpx (magenta) and Dlg (yellow). Examples of intact and retracted NMJs are shown for both genotypes. Brightness for images of retracted NMJs was enhanced to show residual synaptic material. (B) Quantification of Dlg +Ib bouton number from trol and trol; MhcS1 muscle 4 NMJs at segment A3. Each point represents the number of boutons at one NMJ, with mean bouton number indicated with the solid black line. Quantification of bouton number: trol: 10.5±1.6, 19 NMJs from 10 larvae; trol; MhcS1: 11.7±1.5, 19 NMJs from 10 larvae, p=0.597. (C) Representative images of muscle 4 NMJs stained for Perlecan, Hrp and GluRIIC in control (trolGFP/y;+;+) or MhcS1 (trolGFP/y;MhcS1/+;+.) larvae. (D,E) Line scanning profiles of Perlecan and Hrp fluorescent intensity through axons (D) or synaptic boutons (E) at muscle 4 in segment A4. For both axons and boutons, line profiles from 12 control NMJs from 6 larvae and 10 MhcS1 NMJs from 5 larvae were averaged. Control measurements are denoted in green and MhcS1 in orange.
-
Figure 6—source data 1
Raw Values and Statistics for Figure 6 on Trol - Mhc Interactions.
- https://cdn.elifesciences.org/articles/88273/elife-88273-fig6-data1-v1.xlsx
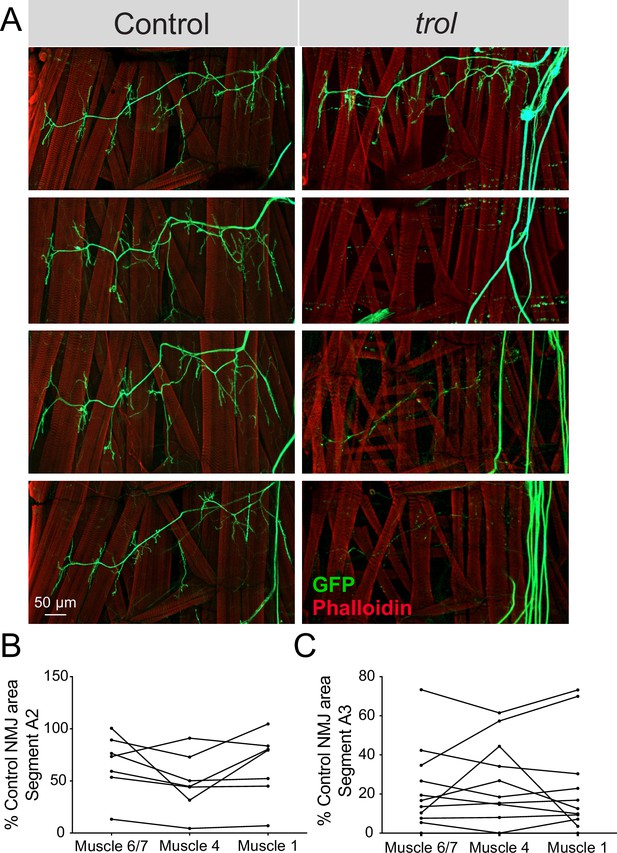
NMJ loss occurs in a temporally coordinated manner across abdominal hemisegments in trol mutants.
(A) Representative images of larval hemisegments in four control (trolnull,vGlut-Gal4/+;+;UAS-10xGFP/+, left panels) or trolnull (trolnull,vGlut-Gal4/y;+;UAS-10xGFP/+, right panels) larvae expressing 10X-GFP in motoneurons (green) and stained for Phalloidin to label muscle Actin (red). (B) Area of listed NMJs (muscle 6/7, muscle 4, muscle 1) in trol segment A2 as a percentage of mean control NMJ area. Percent area is largely consistent across NMJs along the hemisegment, indicating synapses retract or are maintained together with their hemisegment. (C) Area of listed NMJs (muscle 6/7, muscle 4, muscle 1) in trol segment A3 as a percentage of mean control NMJ area.
-
Figure 7—source data 1
Raw Values and Statistics for Figure 7 on Hemi-segment Coordinated NMJ Retraction.
- https://cdn.elifesciences.org/articles/88273/elife-88273-fig7-data1-v1.xlsx
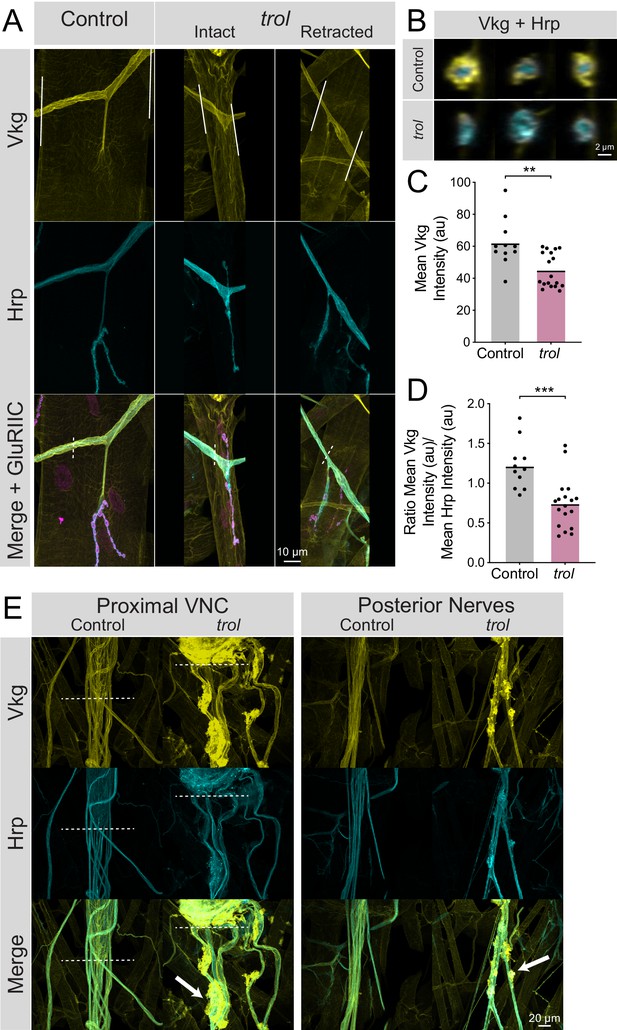
Loss of Perlecan disrupts the neural lamella.
(A) Representative images of muscle 4 NMJs in control (trolnull/+;vkgGFP/+;+) and trolnull (trolnull/y;vkgGFP/+;+) larvae stained for Vkg (yellow), Hrp (cyan), and GluRIIC (magenta). Images of intact and retracted trol NMJs are shown. White lines in Vkg panels indicate borders for quantification of axon bundle fluorescence in panels C and D. (B) Representative cross-sections of control (top) and trol (bottom) axon bundles with Vkg in yellow and Hrp in cyan. (C) Quantification of mean Vkg fluorescence in the axon bundle crossing over muscle 4. Each point represents one axonal segment measurement. (Control: 7 larvae; n=11; 62.00±4.476; trol: 9 larvae; n=18; 45.04±2.549; P=0.0014). (D) Quantification of the ratio of mean Vkg fluorescence divided by mean Hrp fluorescence in the axon bundle crossing over muscle 4. Each point represents the ratio for one axonal segment measurement. (Control: 7 larvae; n=11; 1.219±0.08998; trol: 9 larvae; n=18; 0.7402±0.07467, p<0.001). (E) Representative images of axon bundles stained for Vkg (yellow) and Hrp (cyan) exiting the proximal VNC or those located more posteriorly. Control nerve bundles are on the left, with trol nerve bundles on the right. White dashed lines indicate the posterior tip of the VNC. White arrows note areas of Vkg accumulation and protrusions from the neural lamella.
-
Figure 8—source data 1
Raw Values and Statistics for Figure 8 on Vkg Alterations in Neural Lamella.
- https://cdn.elifesciences.org/articles/88273/elife-88273-fig8-data1-v1.xlsx
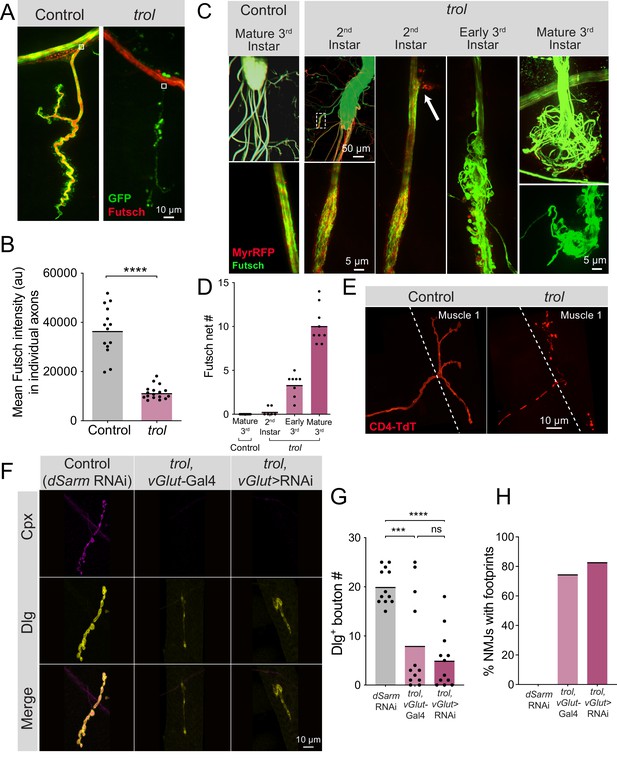
Fragmentation of the microtubule cytoskeleton and axons in trol mutants.
(A) Representative images of Ib motoneurons innervating muscle 4 in control (trolnull,vGlut-Gal4/+;+;UAS-10xGFP/+, left panel) or trol (trolnull,vGlut-Gal4/y;+;UAS-10xGFP/+, right panel) larvae expressing 10X-GFP in motoneurons (green) and stained for Futsch to label microtubules (red). White boxes indicate area of axon with Futsch fluorescence quantified in B. (B) Quantification of mean Futsch fluorescence in individual axons. Each point represents the average fluorescence in one axon. Quantification of mean fluorescence: control: 36725±2679, 14 NMJs from 6 larvae; trol: 11476±675.7, 17 NMJs from 7 larvae, p<0.0001. Multiple abdominal segments were imaged. (C) Representative images of VNCs and axon bundles stained for Futsch (green) and expressing MyrRFP (red) in control (trolnull,vGlut-Gal4/+;UAS-myrRFP/+;+) and trol (trolnull,vGlut-Gal4/y; UAS-myrRFP/+;+) larvae at the indicated stage. Mature 3rd instar control axon bundles have continuous tracks of microtubules within intact axons. trol axons show progressive disruption, with early swelling of the axon and mildly twisted microtubules (white dashed box depicts location of swollen axon and twisted microtubules shown in inset). The 2nd instar axon bundle image is replicated in the next panel to indicate an area of membrane material leaving the axon boundary (white arrow). By late 3rd instar, trol axons are severed and nets of tangled microtubules form balls at either end of the axon bundle breakage. (D) Quantification of the number of Futsch nets in mature 3rd instar control and trol larvae at multiple developmental stages. Each point indicates the number of Futsch nets in one larvae. (E) Representative images of MN1-Ib-Gal4 driving expression of UAS-CD4-TdTomato (red) to visualize single axons and synapses in control (trolnull/+;+;MN1-Ib-Gal4,UAS-CD4-TdTomato) and trol (trolnull/y;+;MN1-Ib-Gal4,UAS-CD4-TdTomato) 3rd instar larvae. (F) Representative images of larval muscle 4 NMJs stained for Cpx (magenta) and Dlg (yellow) in control (+;UAS-dSarm-RNAi/+;+), trol, vGlut-Gal4 (trolnull,vGlut-Gal4/y;+;+) and trolnull expressing dSarm RNAi (trolnull,vGlut-Gal4/y;UAS-dSarm-RNAi/+,+). Brightness for images of retracted NMJs was enhanced to show residual synaptic material. (G) Quantification of Dlg +Ib bouton number at muscle 4 NMJs in segment 4 in control, trol, vGlut-Gal4, and trolnull expressing dSarm-RNAi. Each point represents bouton number from one NMJ with the mean indicated by the solid black line. Quantification of bouton number: control: 20.08±1.048, 12 NMJs from 6 larvae; trol, vGlut-Gal4: 8.083±2.811, 12 NMJs from 7 larvae, p<0.001 compared to control; trolnull expressing dSarm-RNAi: 5.083±1.667, 12 NMJs from 6 larvae, p<0.0001 compared to control, P=0.5387 compared to trolnull. (H) Percentage of control, trol, vGlut-Gal4 or trolnull expressing dSarm-RNAi NMJs with footprints from the dataset in F.
-
Figure 9—source data 1
Raw Values and Statistics for Figure 9 on Microtubule Disruption in Trol Mutants.
- https://cdn.elifesciences.org/articles/88273/elife-88273-fig9-data1-v1.xlsx
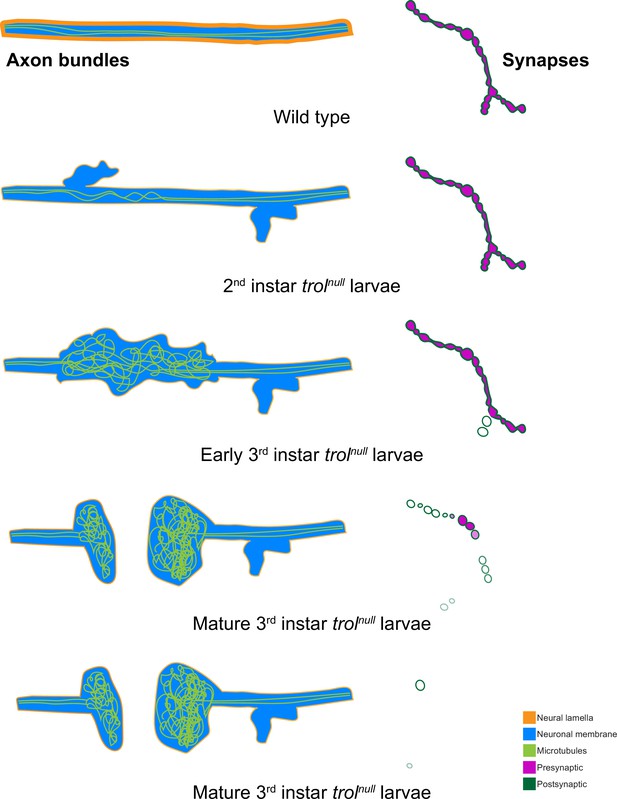
Model of progressive axonal and synaptic defects in the absence of Perlecan.
In control, axonal bundles (neuronal membrane in cyan) have a thick neural lamella (orange) and continuous, straight tracks of microtubules (light green). Synapses have apposed pre- (magenta) and postsynaptic (dark green) material. In 2nd instar trolnull larvae, microtubules appear twisted and small protrusions of neuronal membrane and a thinner neural lamella are found. In early 3rd instar larvae lacking Perlecan, microtubules are severely disorganized and neural lamella and axonal membrane protrusions are larger. Some presynaptic material is lost from synapses in the first stages of retraction. By the mature 3rd instar larval stage, axons have broken entirely, and tangled nets of microtubules are observed on both sides of the breakage. Synapses are retracting and continue to retract until few or no boutons remain.





