Separating phases of allopolyploid evolution with resynthesized and natural Capsella bursa-pastoris
Figures
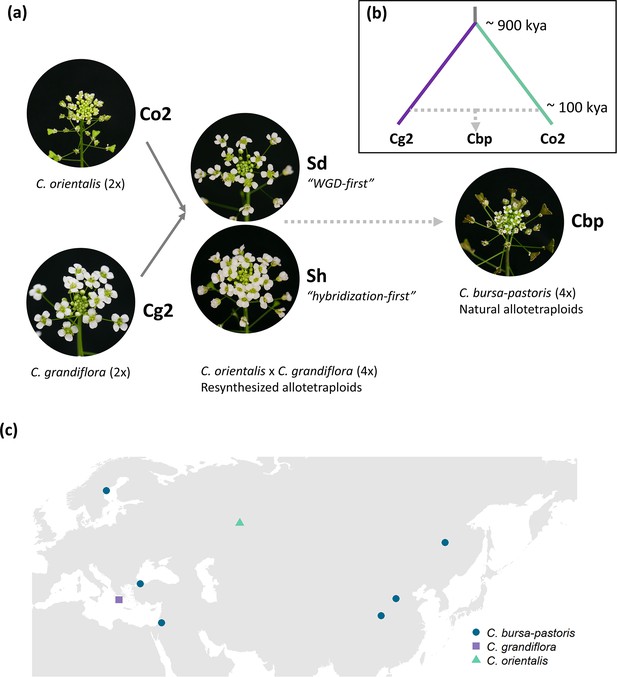
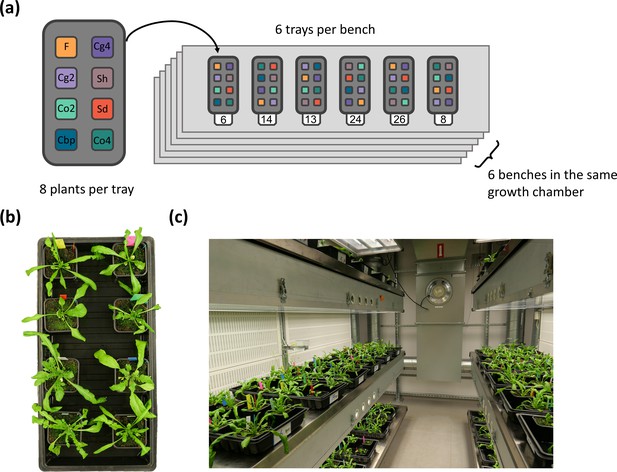
Plant material used in the present study.
(a) Five groups of Capsella plants. Diploid species (Co2 and Cg2 groups) and the second generation of resynthesized allotetraploids (Sd and Sh groups) were from Duan et al., 2023. Samples of natural allotetraploids, C. bursa-pastoris, were added to the present study. (b) Phylogenetic relationship of the three natural species used in the present study, modified from Douglas et al., 2015; C. bursa-pastoris originated from the hybridization between the ancestral population of C. orientalis and the (C. grandiflora + C. rubella) lineage, and C. rubella were omitted from the figure; kya: thousand years ago. (c) Geographic origin of the Capsella samples.
-
Figure 1—source data 1
Capsella plants used in the present study.
- https://cdn.elifesciences.org/articles/88398/elife-88398-fig1-data1-v1.docx
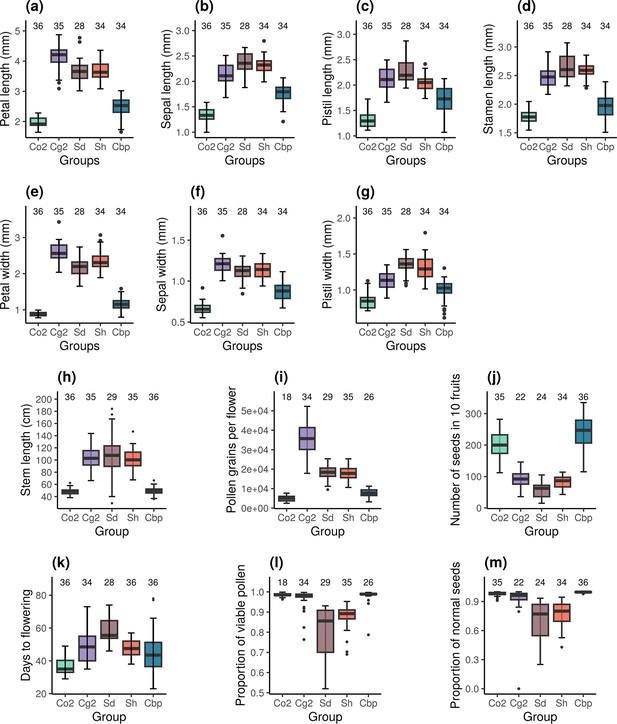
Phenotypic traits of the five Capsella groups.
Co2: diploid C. orientalis; Cg2: diploid C. grandiflora; Sd: whole-genome-duplication-first resynthesized allotetraploids; Sh: hybridization-first resynthesized allotetraploids; Cbp: natural allotetraploid C. bursa-pastoris. The measured traits were (a) petal length, (b) sepal length, (c) pistil length, (d) stamen length, (e) petal width, (f) sepal length, (g) pistil width, (h) length of the longest stem, (i) number of pollen grains per flower, (j) number of seeds in ten fruits, (k) number of days from germination to the opening of the first flower, (l) proportion of viable pollen grains, and (m) proportion of normal seeds in ten fruits. Sample sizes are shown above the groups.
-
Figure 2—source data 1
Effects of plant group and positions (tray ID) on phenotypes.
- https://cdn.elifesciences.org/articles/88398/elife-88398-fig2-data1-v1.docx
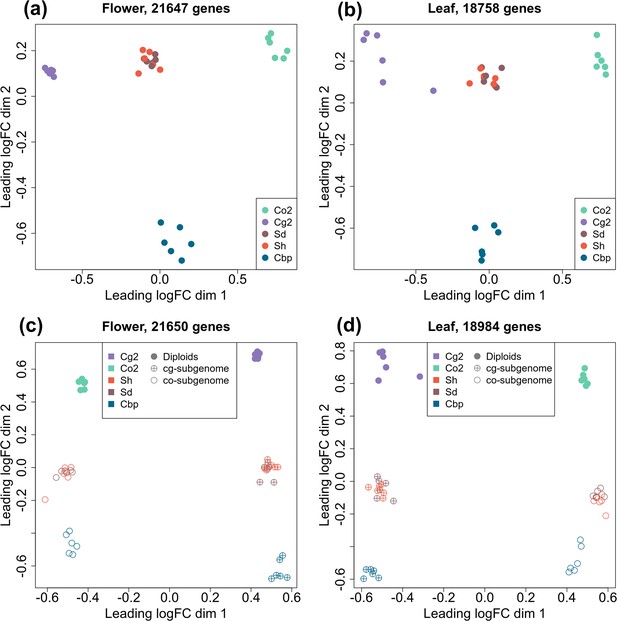
Multi-dimensional scaling (MDS) analyses of gene or homoeolog expression in two tissues.
Down-sampled gene expressions were used to compare gene expression patterns of the five plant groups in either flowers (a) or leaves (b). The five groups of Capsella plants are: diploid parental species C. grandiflora (Cg2) and C. orientalis (Co2), hybridization-first (Sh) and whole-genome-duplication-first (Sd) resynthesized allotetraploids, and natural allotetraploid C. bursa-pastoris (Cbp). Separated homoeolog expressions in allotetraploids were then compared with rescaled gene expression of diploid groups in both flowers (c) and leaves (d). All the MDS analyses used genes with count-per-million (CPM) >1 in at least two samples, and expression levels were normalized with the trimmed mean of M-values (TMM) method.
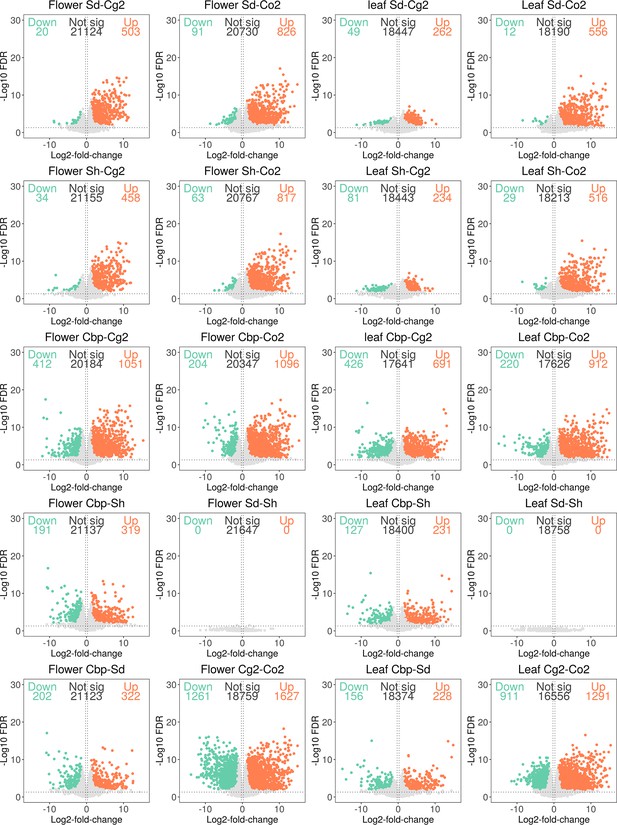
Differentially expressed genes (DEGs) in pair-wise contrasts among the five Capsella plant groups in flowers and leaves.
Genes with expression (CPM >1) in at least two samples were used for the differential expression (DE) analysis. DE analysis was conducted with the R package edgeR (version 3.28.1; Robinson et al., 2010), using TMM normalized unphased gene expression levels. Genes with fold-change >2 and false discovery rate <0.05 were considered significant DEGs.
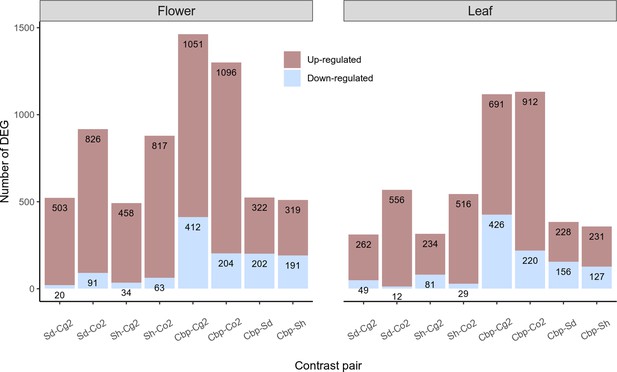
Summary of differential expression analyses of allotetraploid groups.
The number of up-regulated and down-regulated differentially expressed genes (DEGs) in each group pair were annotated on plots. DE analysis was performed on 21,937 genes in flowers and 18,999 genes in leaves, with a threshold of FC >2 and FDR <0.05. Gene expression levels were normalized with the trimmed mean of M-values (TMM) method.

Additive and non-additive expression in allotetraploid groups.
Sd: whole-genome-duplication-first resynthesized allotetraploids; Sh: hybridization-first resynthesized allotetraploids; Cbp: natural allotetraploid C. bursa-pastoris. (a) Number of genes that showed additive expression (ADD, including partial expression level dominance [ELD]), complete ELD, and transgressive expression (TRE) in each allotetraploid group. (b) Genes with complete ELD or TRE were further classified by whether they were up- or down-regulated in allotetraploids, and whether the expression level in allotetraploids was similar to C. grandiflora (Cg-ELD) or C. orientalis (Co-ELD). (c) Venn diagram of genes with complete ELD of the three allotetraploid groups in flowers, separated by directions of ELD.
-
Figure 4—source data 1
Additive and non-additive gene expression in allotetraploid groups.
- https://cdn.elifesciences.org/articles/88398/elife-88398-fig4-data1-v1.docx
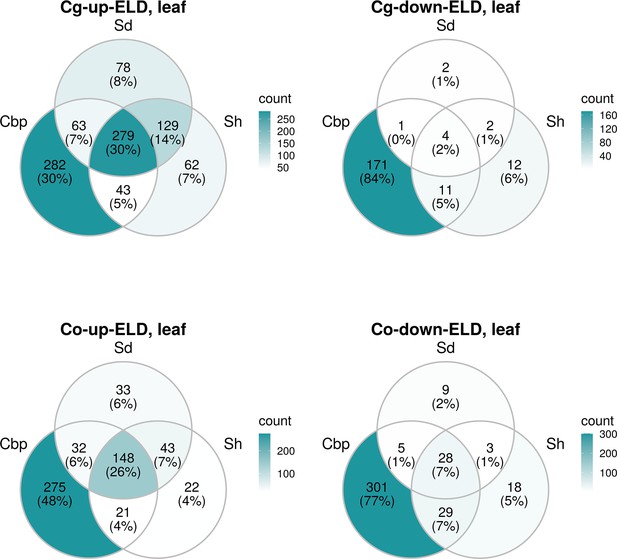
Genes showed expression level dominance (ELD) of the three allotetraploid groups in leaves.
Venn diagram of genes showed ELD of the three allotetraploid groups in leaves, separated by directions of ELD.
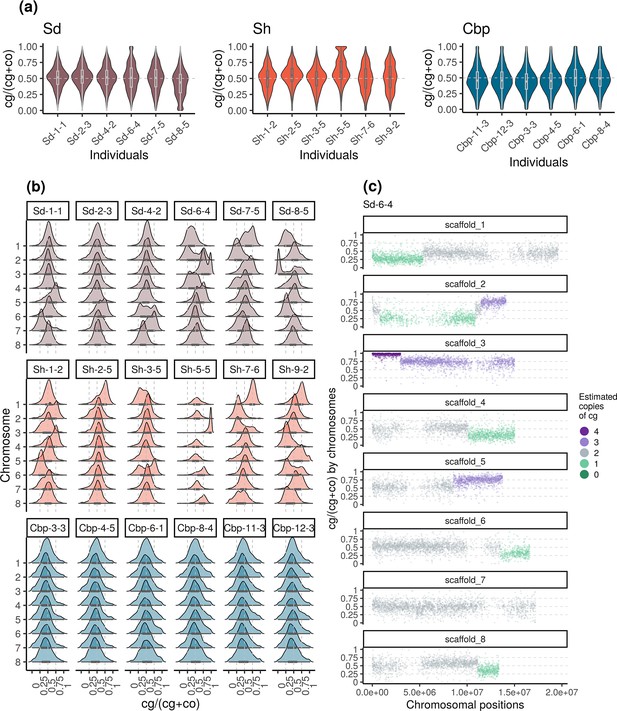
Variation of homoeolog expression bias (HEB) of the three allotetraploid groups in flowers.
Sd: whole-genome-duplication-first resynthesized allotetraploids; Sh: hybridization-first resynthesized allotetraploids; Cbp: natural allotetraploid C. bursa-pastoris. Gene-wise HEB was calculated as the expression level of cg-homoeolog divided by the total expression level of both cg- and co- homoeologs (cg/(cg +co)). For each individual, HEB was calculated for 18,255 genes, which had count-per-million>1 in all flower samples. The distribution of HEB was shown by (a) individuals and (b) chromosomes. (c) HEB was also plotted along chromosome positions to show the sudden change of average HEB between genomic blocks, taking individual Sd-6–4 as an example. The number of cg-homoeologs at each gene estimated by the five-state Hidden Markov Model (HMM) was indicated by five colors. Dark green, light green, grey, light purple, and dark purple represent (0, 1, 2, 3, 4) cg-homoeologs and (4, 3, 2, 1, 0) co-homoeologs, respectively.
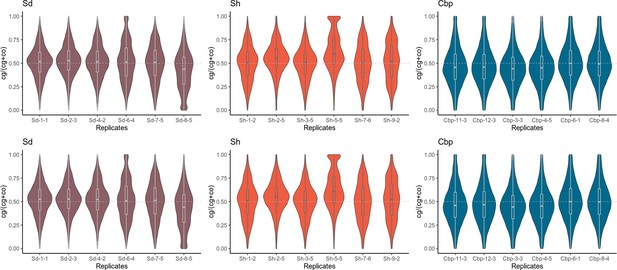
Distribution of gene homoeolog expression bias (HEB) by individuals.
Distribution of gene HEB of each allotetraploid individual in flowers (upper panel) and leaves (lower panel). For each gene, HEB was estimated as the expression of C. grandiflora-derived homoeolog divided by the total expression of both C. grandiflora- and C. orientalis-derived homoeolog (cg/(cg +co)). HEBs were calculated for 18,255 genes in flowers and 15,581 genes in leaves.
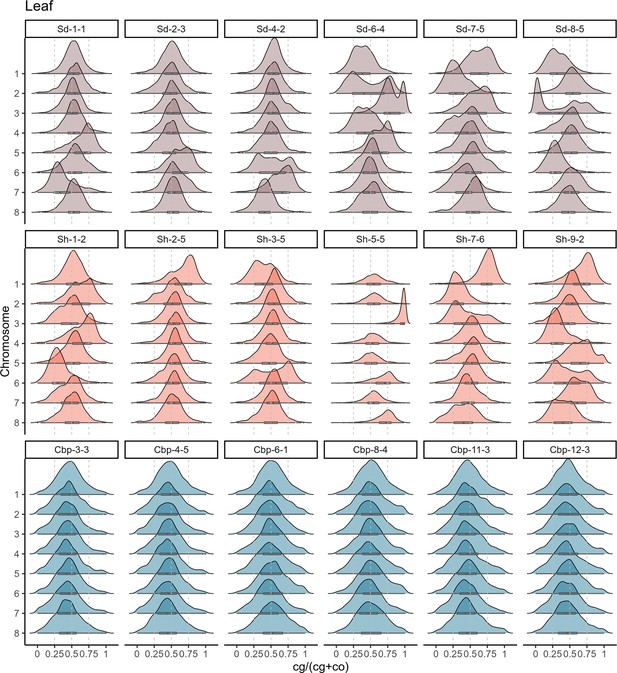
Distribution of gene homoeolog expression bias (HEB) by chromosomes in leaves.
The distribution of gene HEB of resynthesized and natural Capsella allotetraploids was showed by eight major chromosomes in leaves.
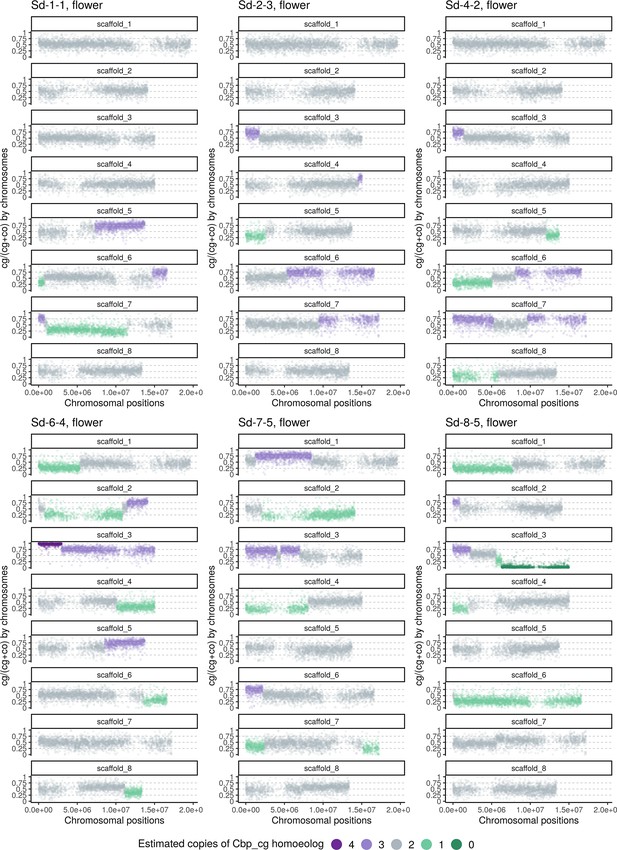
Homoeolog expression bias along chromosome positions in the inflorescence sample of the Sd group (‘Whole-genome-duplication-first’” resynthesized Capsella allotetraploids).
The number of cg-homoeologs at each gene estimated by the five-state Hidden Markov Model (HMM) was indicated by five colors. Dark green, light green, grey, light purple, and dark purple represent (0, 1, 2, 3, 4) cg-homoeologs and (4, 3, 2, 1, 0) co-homoeologs, respectively.
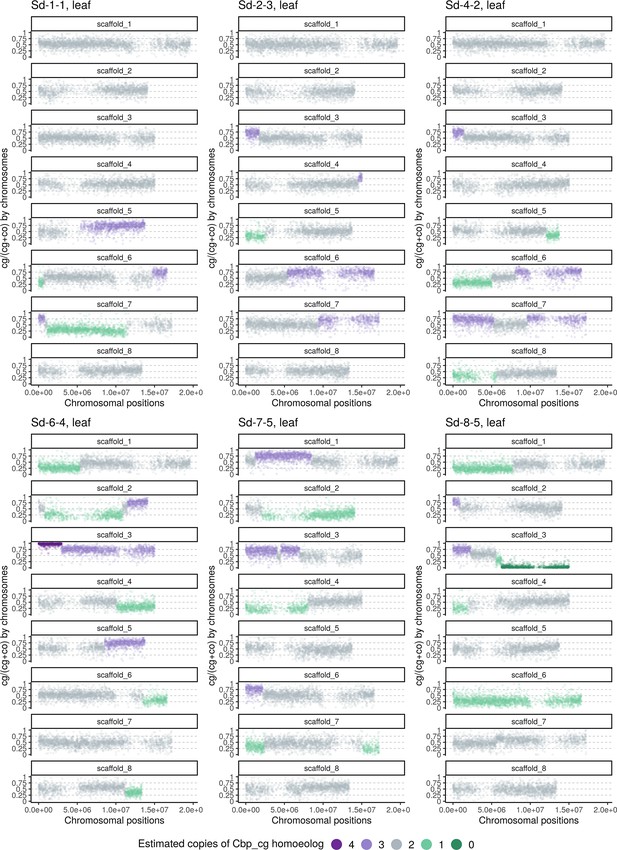
Homoeolog expression bias along chromosome positions in the leaf sample of the Sd group (‘Whole-genome-duplication-first’ resynthesized Capsella allotetraploids).
The number of cg-homoeologs at each gene estimated by the five-state Hidden Markov Model (HMM) was indicated by five colors. Dark green, light green, grey, light purple, and dark purple represent (0, 1, 2, 3, 4) cg-homoeologs and (4, 3, 2, 1, 0) co-homoeologs, respectively.
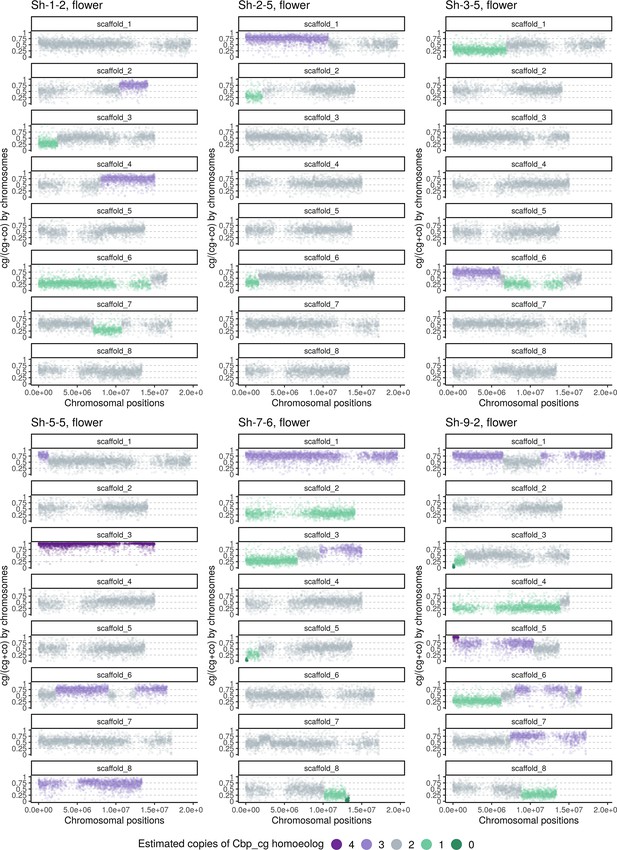
Homoeolog expression bias along chromosome positions in the inflorescence sample of the Sh group (‘hybridization-first’ resynthesized Capsella allotetraploids).
The number of cg-homoeologs at each gene estimated by the five-state Hidden Markov Model (HMM) was indicated by five colors. Dark green, light green, grey, light purple, and dark purple represent (0, 1, 2, 3, 4) cg-homoeologs and (4, 3, 2, 1, 0) co-homoeologs, respectively.
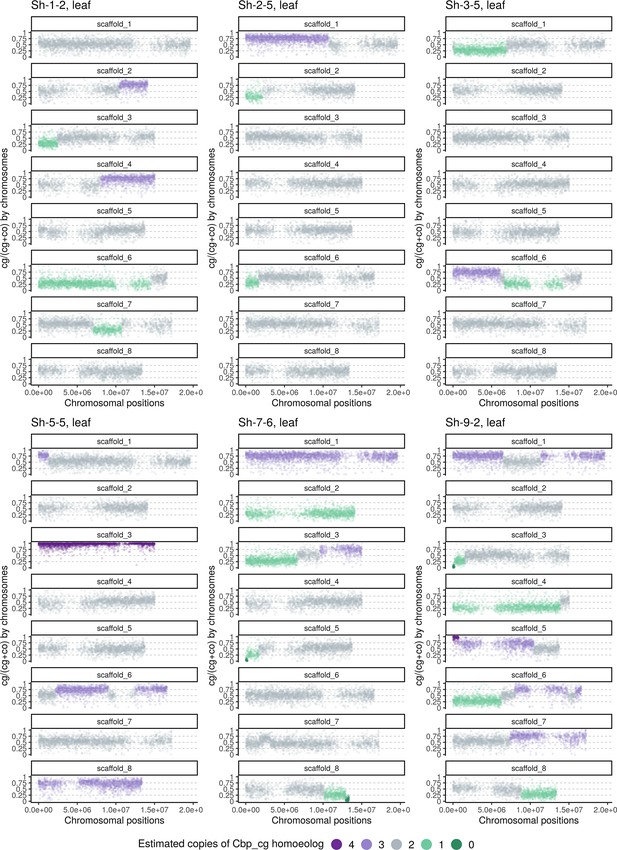
Homoeolog expression bias along chromosome positions in the leaf sample of the Sh group (‘hybridization-first’ resynthesized Capsella allotetraploids).
The number of cg-homoeologs at each gene estimated by the five-state Hidden Markov Model (HMM) was indicated by five colors. Dark green, light green, grey, light purple, and dark purple represent (0, 1, 2, 3, 4) cg-homoeologs and (4, 3, 2, 1, 0) co-homoeologs, respectively.
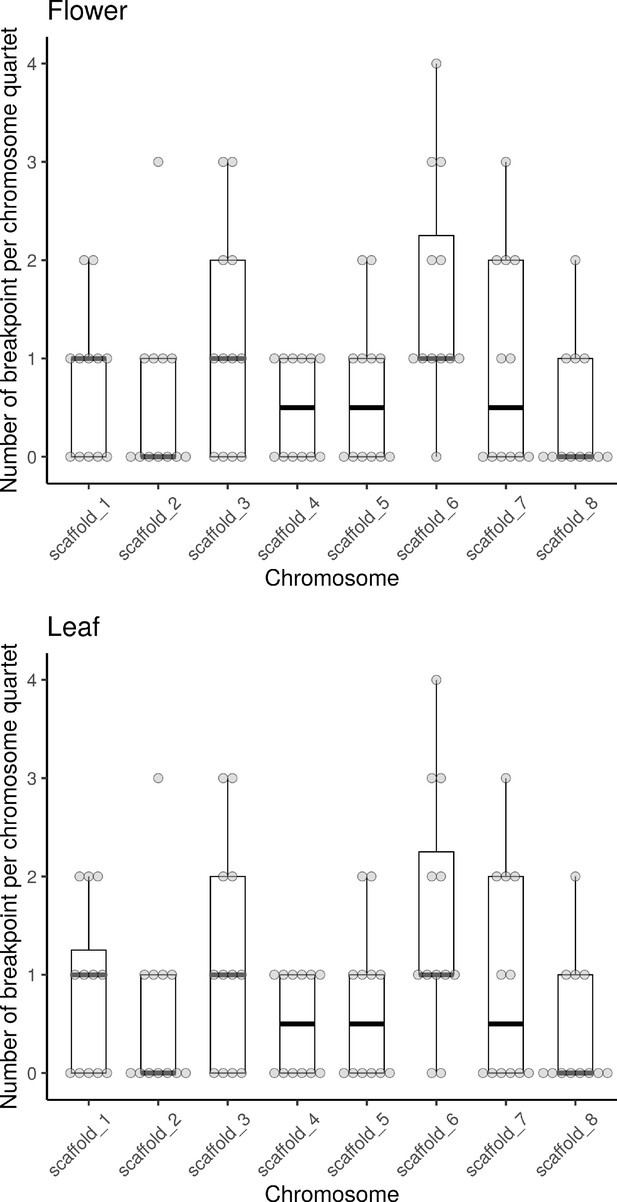
Estimated number of breakpoints per chromosome quartet in resynthesized allotetraploids.
The number of breakpoints between chromosome segments with distinct average homoeologous expression bias (HEB) per chromosome was estimated with a five-state Hidden Markov Model for inflorescence and leaf sample of the 12 resynthesized allotetraploid individuals (groups Sd and Sh). Each dot represents the estimation of one chromosome in one individual. Quartiles and median were shown by boxes and thicker bars.

Homoeolog expression bias (cg/(cg +co)) along chromosomes of natural allotetraploid C. bursa-pastoris in flowers, taking four pairs of chromosome quartets with typical patterns as an example.
The number of cg-homoeologs estimated by the five-state Hidden Markov Model was indicated by five colors. The two chromosome quartets in the same row are from the two individuals of the same major genetic cluster of natural C. bursa-pastoris (Kryvokhyzha et al., 2016), showing that some estimated segments with an unbalanced number of cg- and co-homoeologs were shared between the individuals from the same genetic cluster.

Homoeolog expression bias along chromosome positions in the inflorescence sample of the Cbp group (natural Capsella bursa-pastoris).
The number of cg-homoeologs at each gene estimated by the five-state Hidden Markov Model (HMM) was indicated by five colors. Dark green, light green, grey, light purple, and dark purple represent (0, 1, 2, 3, 4) cg-homoeologs and (4, 3, 2, 1, 0) co-homoeologs, respectively.

Homoeolog expression bias along chromosome positions in the leaf samples of the Cbp group (natural Capsella bursa-pastoris).
The number of cg-homoeologs at each gene estimated by the five-state Hidden Markov Model (HMM) was indicated by five colors. Dark green, light green, grey, light purple, and dark purple represent (0, 1, 2, 3, 4) cg-homoeologs and (4, 3, 2, 1, 0) co-homoeologs, respectively.

Loss of homoeolog expression in resynthesized (Sd and Sh) and natural allotetraploids (Cbp).
The number of genes with the expression loss of C. orientalis-homoeolog (a,c) or C. grandiflora-homoeolog (b,d) per individual was compared among the three groups of allotetraploids in flowers (a,b) or leaves (c,d). Homoeologous genes that had obvious expression (count per million >5) in all individuals of the corresponding diploid species but almost no expression (count per million <0.5) in one allotetraploid individual were considered cases of homoeolog expression loss.

Relationships between homoeolog expression change and non-additive gene expression in flowers.
(a) Expected patterns of homoeolog expression change in each scenario that may explain the cause of expression level dominance (ELD) in resynthesized allotetraploids. (b) Observed homoeolog expression change among genes with ELD in resynthesized allotetraploids (Sd and Sh). (c) Observed homoeolog expression change among genes with Cbp-specific non-additive expression.
-
Figure 8—source data 1
Expression level fold-change (log2FC) of homoeologs relative to the corresponding gene in diploid groups among genes with expression level dominance (ELD) in flowers or leaves.
- https://cdn.elifesciences.org/articles/88398/elife-88398-fig8-data1-v1.docx
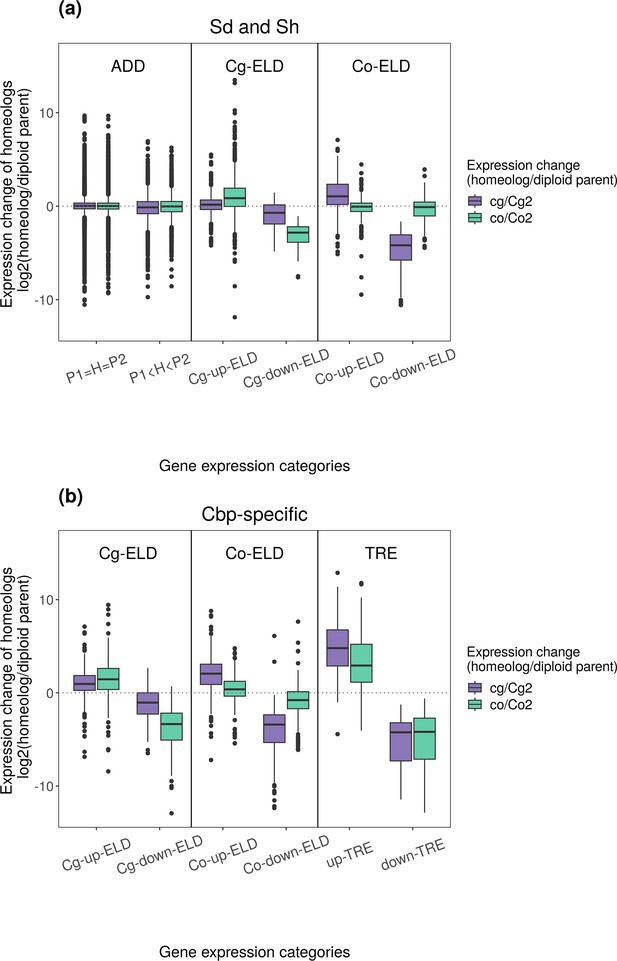
Relationships between homoeolog expression change and non-additive gene expression in leaves.
Gene (homoeolog) expression levels were normalized with the trimmed mean of M-values (TMM) method. (a) Homoeolog expression change among genes with ELD in resynthesized allotetraploids. (b) Homoeolog expression change among genes with Cbp-specific non-additive expression.
Tables
Classification of additive and non-additive gene expression pattern in allotetraploids.
| Group | Description | Classification criteria* |
|---|---|---|
| a | Additive expression with no parental differentiation | Cgi = xij = Coi |
| b | Partial ELD or additive expression with parental differentiation | (Cgi <xij < Coi) or (Coi <xij < Cgi) or (Cgi ≠ Coi and xij = Cgi and xij = Coi) |
| c | Up-regulated ELD toward Cg2 | xij = Cgi and xij >Coi |
| d | Down-regulated ELD toward Cg2 | xij = Cgi and xij <Coi |
| e | Up-regulated ELD toward Co2 | xij = Coi and xij >Cgi |
| f | Down-regulated ELD toward Co2 | xij = Coi and xij <Cgi |
| g | Up-regulated TRE with no parental differentiation | Cgi = Coi and xij >Cgi and xij >Coi |
| h | Up-regulated TRE with parental differentiation | Cgi ≠ Coi and xij >Cgi and xij >Coi |
| i | Down-regulated TRE with no parental differentiation | Cgi = Coi and xij <Cgi and xij <Coi |
| j | Down-regulated TRE with parental differentiation | Cgi ≠ Coi and xij <Cgi and xij <Coi |
-
*
Cgi: expression level of gene i in the Cg2 group; Coi: expression level of gene i in the Co2 group; xij: expression level of gene i in allotetraploid group j, and j∈(Sd, Sh, Cbp); ELD: expression level dominance; TRE: transgressive expression; The significance of differential expression between groups were determined by results of differential expression analysis on unphased gene expression, with a threshold of fold-change >2 and false discovery rate <0.05.
Additional files
-
Supplementary file 1
Tray ID of each Capsella plant and the position of eight plants within each tray.
- https://cdn.elifesciences.org/articles/88398/elife-88398-supp1-v1.csv
-
Supplementary file 2
RNA-sequencing information.
- https://cdn.elifesciences.org/articles/88398/elife-88398-supp2-v1.csv
-
Supplementary file 3
Gene ontology (GO) terms that were overrepresented in differentially expressed genes (FC >1.5 and FDR <0.05) between resynthesized and natural allopolyploids.
- https://cdn.elifesciences.org/articles/88398/elife-88398-supp3-v1.csv
-
MDAR checklist
- https://cdn.elifesciences.org/articles/88398/elife-88398-mdarchecklist1-v1.docx