A new pipeline SPICE identifies novel JUN-IKZF1 composite elements
Figures
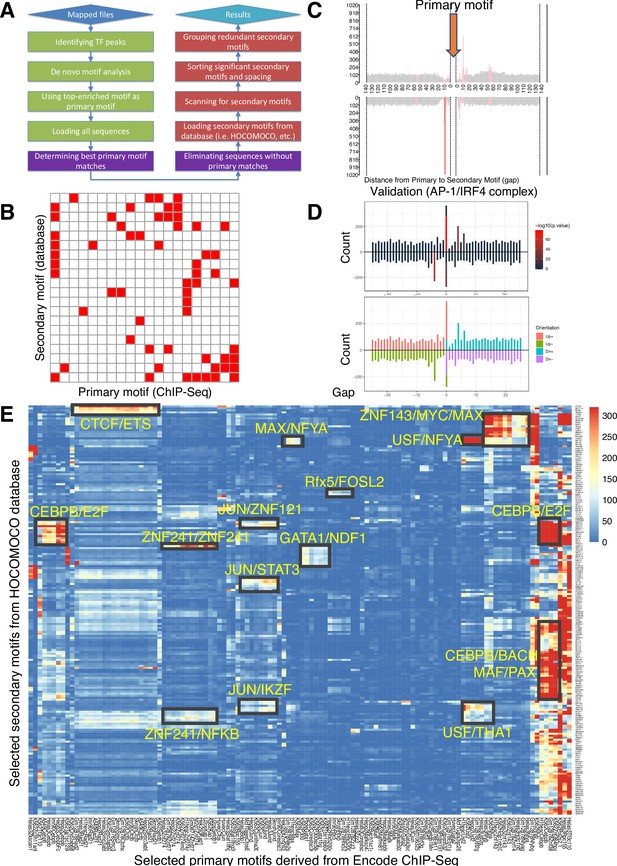
Spacing Preference Identification of Composite Elements (SPICE) predicts known and novel composite elements and spacing preferences.
(A) Schematic step-by-step flowchart of the SPICE pipeline to predict transcription factor composite elements and their spacing preferences (see Methods for details). (B) Schematic of a heatmap depicting hypothetically enriched significant motif pairs (shown as solid red boxes). This is an interaction matrix of primary and secondary transcription factor motifs. It depicts the potential composite elements that can be predicted when a Chromatin immunoprecipitation coupled with DNA sequencing (ChIP-seq)-derived primary transcription factor motif region is compared with a database of secondary transcription factor motifs, based on the distribution of spacing between primary and secondary motifs. The red boxes indicate composite elements based on the primary and secondary motifs on the x- and y-axis, respectively. (C) Schematic bar graphs showing the spacing distribution of secondary motifs centered on a given primary motif, with bars highlighted in red, indicating the most preferred spacing. (D) SPICE successfully predicts and validates known composite elements. Shown is the validation of SPICE’s ability to rediscover the known AP-1/IRF4 composite element (AICE). (E) Potential transcription factor composite elements identified from SPICE. Heatmap of the significant motif pairs, with at least one pair of motifs exhibiting an E-value <1e–10. This 118×205 spatial interaction matrix using Transcription Factor Binding Sites (TFBS) from the ENCODE project represents a subset of the primary and secondary motifs shown in the interaction matrix shown in Figure 1—figure supplement 2. The x-axis represents the primary motifs derived from ChIP-seq libraries, and the y-axis indicates the transcription factor motifs from Homo sapiens Comprehensive Model Collection (HOCOMOCO) database. Potential novel combinatory complexes are highlighted in black boxes and yellow font (e.g. CTCF/ETS composite elements and JUN/IKZF composite elements).
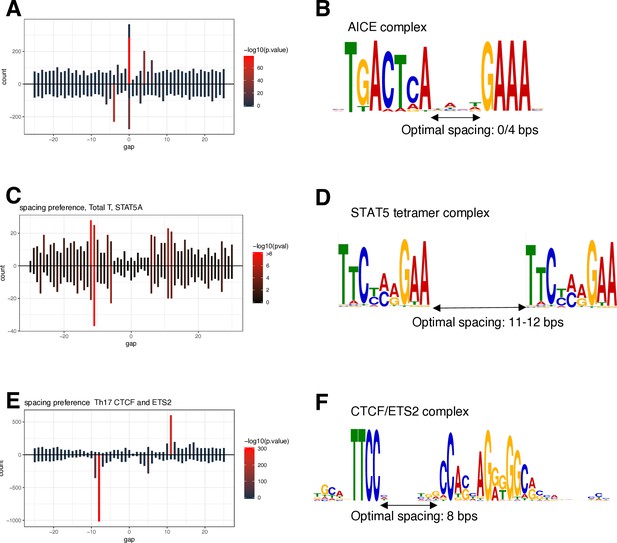
Spacing Preference Identification of Composite Elements (SPICE) successfully predicts AICE, STAT5 tetramers, and CTCF/ETS complexes.
SPICE successfully predicted previously identified composite elements, including AICE (AP-1/IRF4 composite elements), based on BATF, JUN, and IRF4 ChIP-seq data in TCR pre-activated mouse T cells (A, B), STAT5 tetramer complex formation based on STAT5 ChIP-seq in TCR pre-activated and IL-2-treated T cells (C, D), and CTCF/ETS composite elements based on available ETS2 ChIP-seq data (E, F).
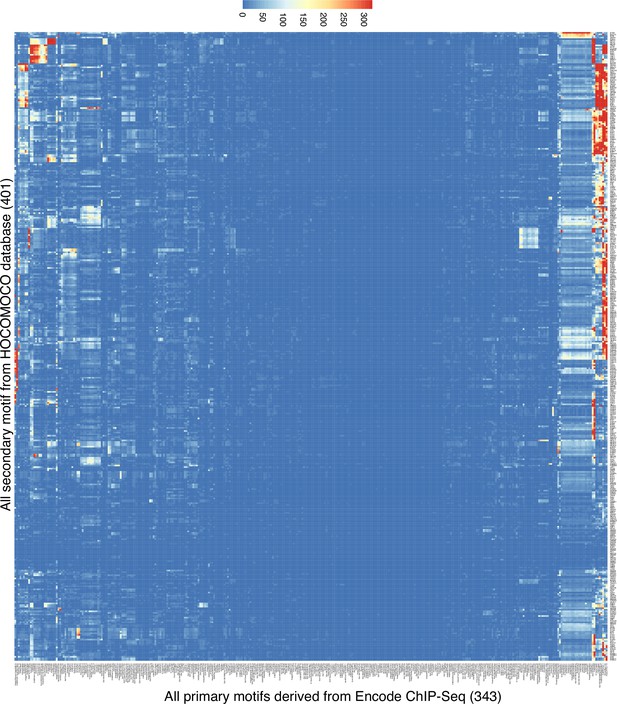
Spacing Preference Identification of Composite Elements (SPICE) predicts transcription factor composite elements using the Encode Chromatin immunoprecipitation coupled with DNA sequencing (ChIP-seq) data.
Heatmap of the motif interaction matrix of ChIP-seq libraries from Transcription Factor Binding Sites (TFBSs) from the ENCODE project. The x-axis represents the primary motifs derived from 343 Encode ChIP-seq libraries, and the y-axis indicates the 401 motifs from the Homo sapiens Comprehensive Model Collection (HOCOMOCO) database (these are also in Supplementary file 2). The color scale represents the -log10 transformed E-value, which is the lowest p-value of any spacing of the secondary motif times the number of secondary motifs.
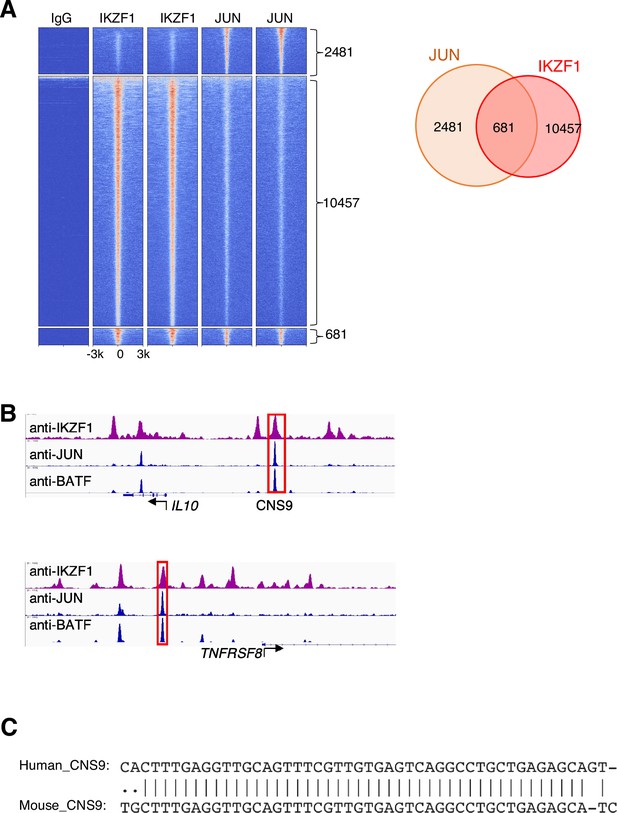
JUN-IKZF composite elements in GM12878 cells.
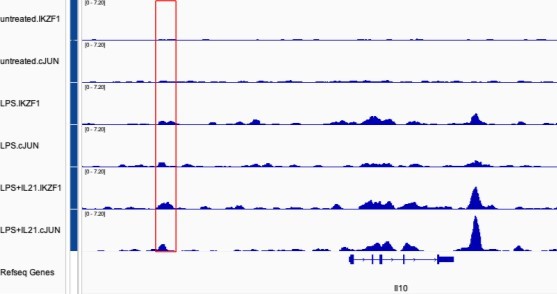
(A) Heatmap of genome-wide Chromatin immunoprecipitation coupled with DNA sequencing (ChIP-seq) binding intensities in a±3 kb genomic region centered on summits of combined IKZF1 and JUN peaks in GM12878 cells from the ENCODE project; two biological replicate experiments are shown in the heatmap. (B) Co-localization of IKZF1, JUN, and BATF peaks at the IL10 and TNFRSF8 genomic loci. A conserved cis-regulatory element in the human IL10 and mouse Il10 loci, cis-regulatory element (CNS9), is highlighted in the red box. (C) Sequence alignment reveals CNS9 region is highly conserved in human and mouse.
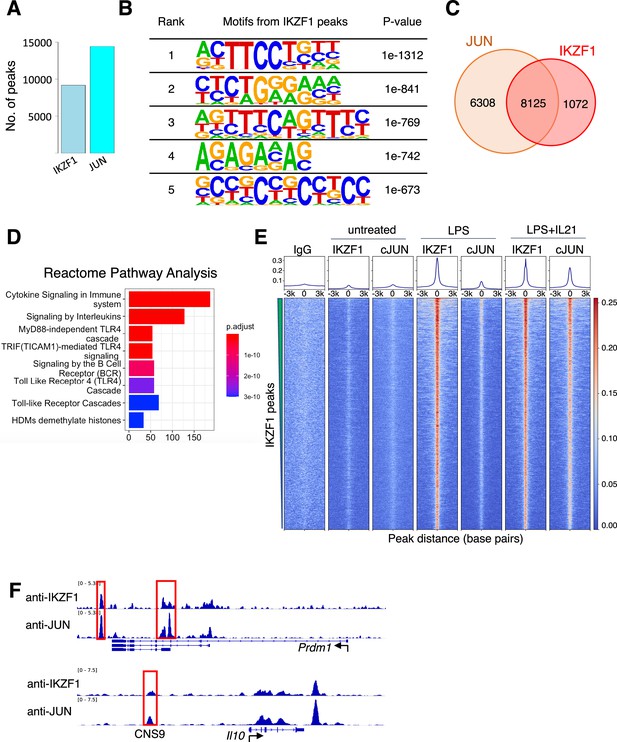
JUN co-localizes with IKZF1.
(A) Number of Chromatin immunoprecipitation coupled with DNA sequencing (ChIP-seq) peaks identified for IKZF1 and JUN in experiments from primary mouse B cells that were treated with LPS + IL-21. (B) De novo motif discovery (HOMER) from IKZF1 peaks in primary mouse B cells treated with LPS + IL-21. The most significantly enriched motifs and their corresponding p-values are shown. (C) Venn diagram showing that most (88.3%) IKZF1 peaks co-localize with JUN. (D) Reactome pathways analysis from the IKZF1-JUN co-localized peaks reveal that cytokine-related pathways were most enriched, including ‘Cytokine Signaling in Immune System,’ ‘Signaling by Interleukins,’ and several TLR4-related pathways/cascades. (D) Heatmap showing that IKZF1 binding was potently activated by LPS or LPS + IL-21 treatment and that IKZF1 binds in proximity to JUN. Shown are normalized ChIP-seq signals ± 3 kb, centered on IKZF1 peak summits. (E) IGV browser file shows IKZF1 and JUN bound to the Prdm1 and Il10 genes. The red boxes indicate known regulatory elements. The position of a known upstream cis-regulatory element (CNS9) that controls Il10 expression is indicated.
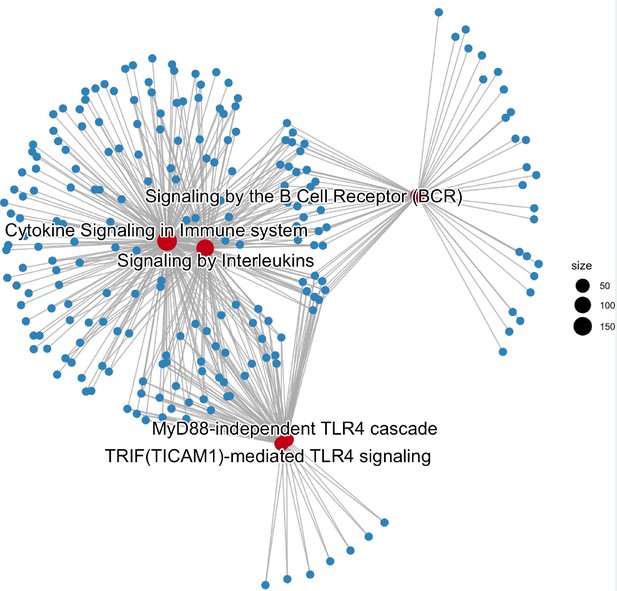
Gene-concept network depicts the linkages of genes and biological concepts of the top five Reactome Pathways that were enriched in IKZF1-JUN shared peaks.
The highlighted red dots represent the top 5 enriched pathways and the dot size indicates the number of genes involved in each pathway.
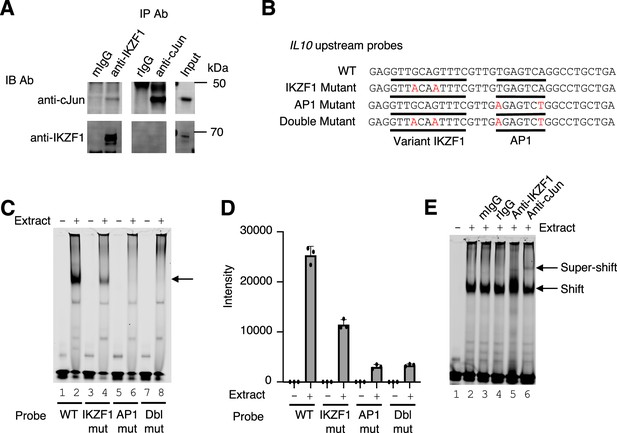
Cooperative binding of IKZF1 and JUN at an IL10 upstream site, cis-regulatory element (CNS9).
(A) Co-immunoprecipitation reveals IKZF1 physically associates with JUN. Nuclear protein lysates from 25×106 cells were used for each IP reaction. Protein extracts (input) were immunoprecipitated with antibodies to IKZF1, JUN, or normal IgG (mouse and rabbit) and resolved by SDS-PAGE. Western blotting was then performed with antibodies to IKZF1 or JUN. (B) Wild-type and mutant probes at the IL10 CNS9 upstream site; variant IKZF1 and AP-1 motifs are underlined, with mutant nucleotides colored in red. (C) Representive EMSA with IL10 upstream probes (wild-type, or IKZF1, AP-1, or IKZF1/AP-1 mutants; see panel B) and nuclear extracts from MINO cells. 100 nM of IR700-labeled probes were used per EMSA reaction. 5 μg of MINO nuclear extract was used in lanes 2, 4, 6, and 8; no nuclear extract was added in control lanes 1, 3, 5, and 7. (D) Average intensities from three independent EMSA experiments (one of which is shown in panel C). Band intensities were calculated using Image Studio software (LI-COR). (E) EMSA super-shifting was performed with 5 μg of nuclear extracts from MINO cells and mouse IgG, rabbit IgG, anti-IKZF1, and anti-JUN. Nuclear lysates were pre-incubated for 20 min on ice with 1 μg of the indicated antibodies prior to the addition of the probe. EMSAs were performed at least three times.
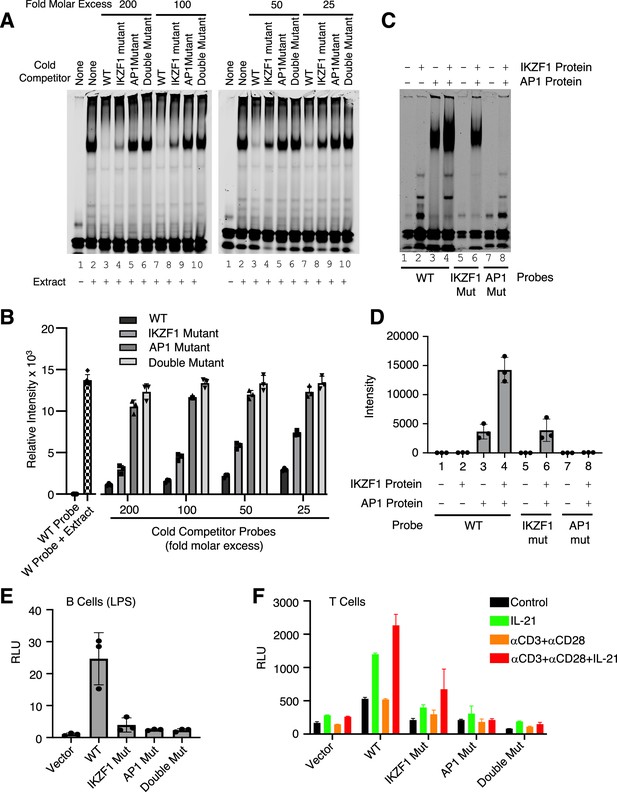
Both IKZF1 and JUN are critical for cooperative binding and transcriptional activation.
(A, B) Cooperative binding of IKZF1 and AP1. EMSA was performed with a wild-type (WT) IL10 probe corresponding to cis-regulatory element (CNS9) and nuclear extracts from MINO cells stimulated with LPS + IL-21. Un-labeled ‘cold’ wild-type or mutant double-stranded oligonucleotides (25, 50, 100, or 200-fold molar excess relative to the IR700-labeled probes) were added to 5 μg nuclear extracts (prepared from MINO cells treated with LPS + IL-21) prior to the addition of 100 nM of WT IR700-labeled probe. Shown are a representative EMSA (A) and the summary of relative band intensities from three independent experiments (B). Relative intensities were calculated using Image Studio software (LI-COR). (C, D) EMSA using human recombinant AP-1 or IKZF1 with IL10 CNS9 upstream probes (wild-type, or IKZF1/AP-1 mutants). 300 ng of recombinant AP1 protein (150 ng of each cJUN and cFOS) and 1 μg of recombinant IKZF1 were used as indicated. Band intensities from three independent experiments are shown in panel D. (E) WT or mutant reporter constructs were transfected via electroporation into mouse primary B cells pre-stimulated with LPS for 24 hr. After electroporation, cells were treated with LPS for 24 hr before dual luciferase activity was measured (n=3; mean ± S.D). (F) WT or mutant reporter constructs were transfected into mouse primary pre-activated CD8+ T cells and the cells were then treated as indicated for 24 hr (n=3; mean ± S.D). RLU, relative light units.
Additional files
-
Supplementary file 1
Spacing Preference Identification of Composite Elements (SPICE) can predict possible tetramers for STAT family member proteins.
SPICE is capable of identifying canonical STAT binding motifs, also known as interferon gamma-activated sequence (GAS) motifs, and then predicts STAT tetramer formation and the optimal spacing for tetramer formation for STAT1, STAT3, STAT4, STAT5A, and STAT5B, whereas STAT2 was predicted to not form tetramers. The various STAT family members were activated in the indicated cell types by the indicated cytokine, Chromatin immunoprecipitation coupled with DNA sequencing (ChIP-seq)-derived canonical GAS motifs, percentage of GAS motifs, likelihood of tetramer formation, and predicted optimal spacing were determined.
- https://cdn.elifesciences.org/articles/88833/elife-88833-supp1-v1.pdf
-
Supplementary file 2
The spatial interaction matrix of primary motifs from Chromatin immunoprecipitation coupled with DNA sequencing (ChIP-seq) libraries and secondary motifs from the Homo sapiens Comprehensive Model Collection database (HOCOMOCO) database.
Each row represents one of the 401 motifs from the HOCOMOCO database, and each column is derived motifs from 343 TFBS ChIP-seq libraries. Each cell has an E-value indicating the significance of motif pairs, which is the lowest p-value of any spacing of the secondary motif times the number of secondary motifs. The value of 1 indicates there is no interaction between motif pairs.
- https://cdn.elifesciences.org/articles/88833/elife-88833-supp2-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/88833/elife-88833-mdarchecklist1-v1.docx