Sustained store-operated calcium entry utilizing activated chromatin state leads to instability in iTregs
Figures
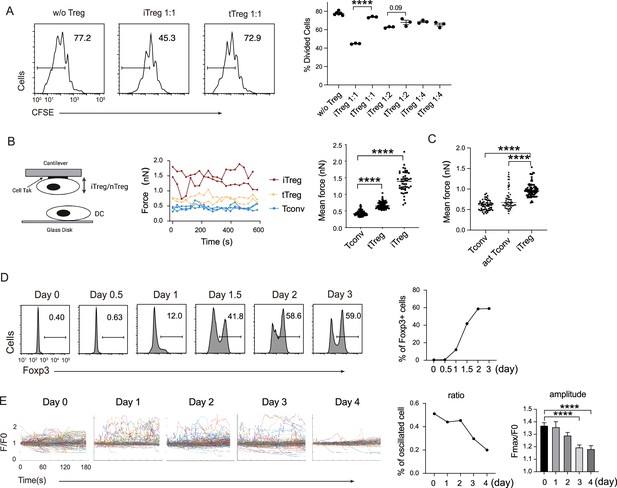
iTreg share similar suppressive mechanism to tTregs.
(A) Comparison of suppressive activity between tTreg and iTreg. CFSE (carboxy fluoroscein succinimidyl Eester) -labeled OT-II T cells were stimulated with OVA-pulsed DC, then Foxp3-GFP+ iTregs and tTregs were added to the culture to suppress the OT-II proliferation. After 4 days, CFSE dilution were analyzed. n = 3, N = 3. Left, representative histograms of CFSE in divided Tconvs. Right, graph for the percentage of divided Tconvs. (B) iTregs possessed stronger binding force to DCs than tTreg. A schematic diagram for AFM-SCFS (atomic force microscope-single cell force spectroscopy) assay setup (left). SCFS force readings for Tconv, tTreg, and iTreg adhering to DC2.4 cells, one line represents a pair of T–DC, every dot represents force reading from each contact. Mean force of Tconv, tTreg, and iTregs adhering to DC2.4 cells. (C) iTregs showed increased binding force compared with activated Tconvs. Mean force of Tconv, activated Tconv, and iTreg adhering to DC2.4 cells. n > 45, N = 3. (D) Precise expression of Foxp3 was assessed during iTreg induction. Naive Tconvs were stimulated with anti-CD3 and anti-CD28, in the presence of TGF-β and IL-2 to induce iTregs. Cells were harvested at the indicated time and Foxp3 expression was analyzed by intracellular staining. (E) Basal Ca2+ oscillation was assessed during iTreg induction. Naive Tconvs were stimulated with anti-CD3 and anti-CD28, in the presence of TGF-β and IL-2 to induce iTregs. Cells were harvested at the indicated time and loaded with Fluo-4 AM, and Fluo-4 fluorescence over time were recorded with confocal microscope. The change of intracellular free Ca2+ concentration over time were shown as F/F0. The ratio of oscillated cells and standard deviation of F/F0 were calculated. n > 150, N = 3. Here, ****p < 0.0001, by Student’s t-est.
-
Figure 1—source data 1
The suppression activity, AFM force of iTreg and nTreg, and calcium oscillation during iTreg induction.
- https://cdn.elifesciences.org/articles/88874/elife-88874-fig1-data1-v1.xlsx
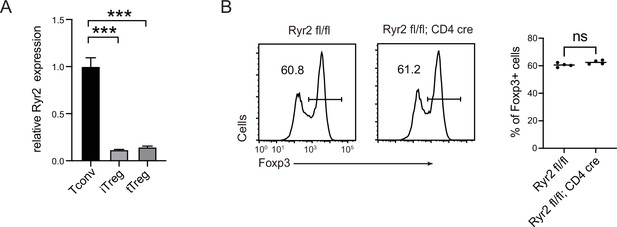
Expression of Ryr2 in iTreg and the effect of Ryr2 on iTreg induction.
(A) QPCR for Ryr2 mRNA expression in iTregs, tTregs, and Tconvs. n = 3, N = 4. (B) The effect of Ryr2 expression on iTreg induction. Naive Tconv cells were sorted from Ryr2 fl/fl; CD4 cre mice and control Ryr2 fl/fl mice, then cells were differentiated into iTregs for 4 days. The percentages of Foxp3+ cells were analyzed by intracellular staining after 4-day induction. Left, representative histograms of induced iTregs. Right, graph for the percentage of Foxp3+ cells in all CD4+ cells. n = 3, N = 3. Here, ***p < 0.001, by Student’s t-test.
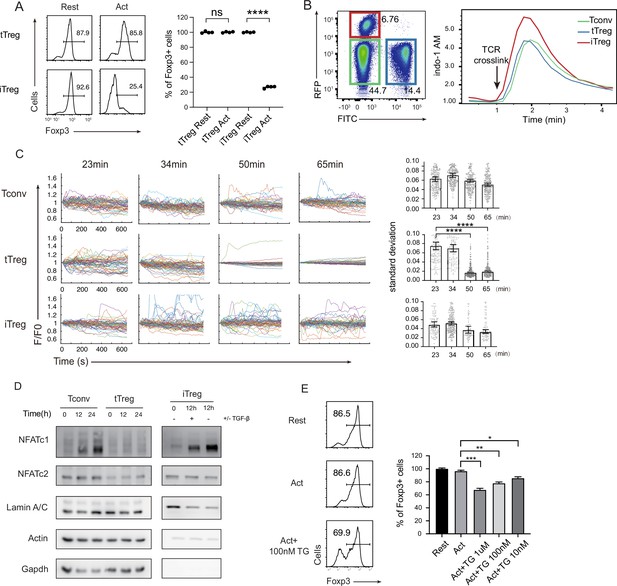
Diminished store-operated calcium entry (SOCE) signal and NFAT translocation in tTregs, but not in iTregs.
(A) Comparison of Treg stability between tTregs and iTregs. iTregs and tTregs were sorted and restimulated with anti-CD3 and anti-CD28 antibodies. Cells were harvested after 2-day restimulation and Foxp3 expression was analyzed by intracellular staining. The percentages of Foxp3+ cells were monitored by FACS (fluorescence-activated cell sorting). Left, representative histograms of restimulated iTregs. Right, graph for the percentage of Foxp3+ cells in all CD4+ cells. n = 4, N = 5. (B) Early SOCE signal was measured in iTreg, tTreg, and Tconv by flow cytometry. Sorted Foxp3-RFP+ iTreg and Foxp3-GFP+ tTreg and double negative Tconv cells were mixed and loaded with Indo-1 AM, then stained with biotin anti-CD3 and biotin anti-CD28 for 1 hr, the baseline fluorescence was recorded for 1 min, and then TCR crosslink was perform by the addition of streptavidin. Left, the gate of three mixed cells. Right, Indo-1 AM ratio of these cells upon TCR crosslink. N = 3. (C) Long-term SOCE were truncated in tTreg, but sustained in iTreg. Tconv, tTreg, and iTreg cells were loaded with Fluo-4 AM and activated by anti-CD3 and anti-CD28 in confocal dish. Fluorescence was recorded in the indicated time after stimulation with the interval of 10 s. Left, the F/F0 of mean fluorescence intensities were calculated and presented. Right, graph for standard deviation of fluorescence in these cells. n > 50, N = 3. (D) NFAT accumulate much in nucleus of iTreg, but not in tTreg. Tconv, tTreg, and iTreg cells were stimulated by anti-CD3 and anti-CD28, after the indicated times, cells were lysed and the cytoplasmic/nuclear components were separated. The cytoplasmic and nuclear NFATc1 and NFATc2 were analyzed by western blot. Actin and GAPDH were used as loading control of cytoplasmic proteins, and LaminA/C as nuclear. N = 4. (E) Forcibly sustained calcium signal destabilizes tTreg. Foxp3-RFP+ tTregs were stimulated by anti-CD3 and anti-CD28. After 1 hr, various concentrations TG were added in the culture medium. Cells were collected after 24-hr stimulation and Foxp3 expression was analyzed by intracellular staining. Left, representative histograms of treated tTregs. Right, graph for the percentage of Foxp3+ cells in all tTregs. n = 3, N = 3. Here, *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, by Student’s t-test.
-
Figure 2—source data 1
Treg stability, early store-operated calcium entry (SOCE) signal, long-term SOCE signal in iTreg and nTreg.
- https://cdn.elifesciences.org/articles/88874/elife-88874-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Original imaging data of long-term store-operated calcium entry (SOCE) signal in Tconv, nTreg, and iTreg, related to Figure 2C.
- https://cdn.elifesciences.org/articles/88874/elife-88874-fig2-data2-v1.zip
-
Figure 2—source data 3
Original western data of NFAT nuclear translocation in Tconv, nTreg, and iTreg, related to Figure 2D.
- https://cdn.elifesciences.org/articles/88874/elife-88874-fig2-data3-v1.zip
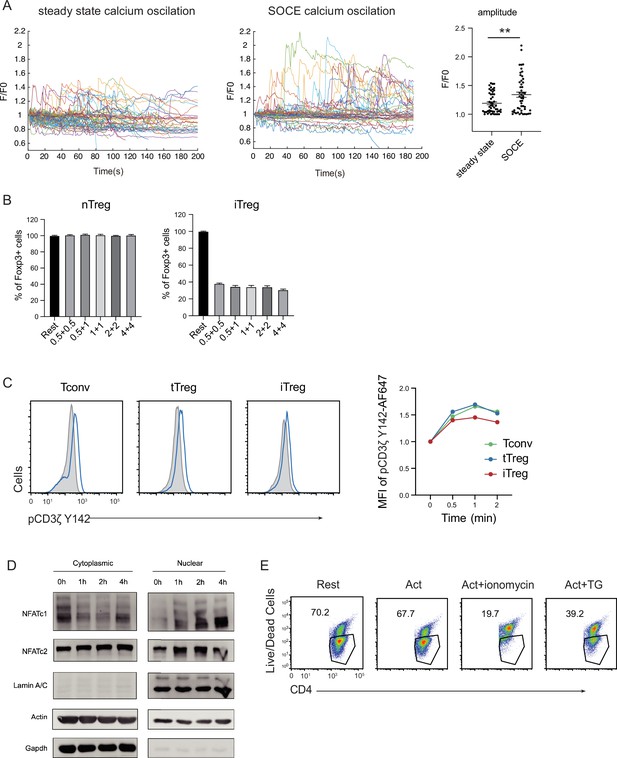
TCR responsiveness and NFAT translocation in tTregs and iTregs.
(A) Calcium oscillation were recorded in steady state and after anti-CD3 and anti-CD28 activation. Tconv cells were loaded with Fluo-4 AM, and left in steady state without anti-CD3 and anti-CD28 stimulation, or treated with anti-CD3 and anti-CD28 stimulation to record store-operated calcium entry (SOCE) signal. Fluorescence was recorded with the interval of 10 s. Left, the F/F0 of mean fluorescence intensities were calculated and presented, Right, graph for standard deviation of fluorescence in these cells. n > 50, N = 3. (B) tTreg and iTreg cells were sorted and restimulated with various concentrations of anti-CD3 and anti-CD28 antibodies. The percentage of Foxp3+ cells was analyzed by intracellular staining after 2-day restimulation. n = 3, N = 3. (C) Early TCR signal of pCD3ζ in tTreg and iTreg after TCR crosslinking. Sorted iTreg, tTreg, and Tconv cells were mixed and stained with biotin anti-CD3 and biotin anti-CD28 for 1 hr. TCR crosslink was perform by the addition of streptavidin. Cells were fixed at the indicated times and stained with pCD3ζ Y142 antibody. Left, representative flow overlays for pCD3ζ Y142 in rest and activated cells. Right, graph for the MFI changes in activated cells after indicated time. (D) Foxp3-RFP+ iTreg were stimulated by anti-CD3 and anti-CD28, after the indicated times, cells were collected and the cytoplasmic/nuclear component were separated. The cytoplasmic and nuclear NFATc1 and NFATc2 were analyzed by western blot. Actin and GAPDH were used as loading control of cytoplasmic protein, and Lamin A/C as nuclear. N = 3. (E) Foxp3-RFP+ tTregs were stimulated by anti-CD3 and anti-CD28, after 1 hr ionomycin and TG were added in the culture medium. After 24 hr of stimulation, the percentages of live cells in CD4+ cells were monitored. N = 3. Here, **p < 0.01, by Student’s t-test.
-
Figure 2—figure supplement 1—source data 1
Original western data of NFAT nuclear translocation upon early activation in Tconv, nTreg, and iTreg, related to Figure 2—figure supplement 1D.
- https://cdn.elifesciences.org/articles/88874/elife-88874-fig2-figsupp1-data1-v1.zip
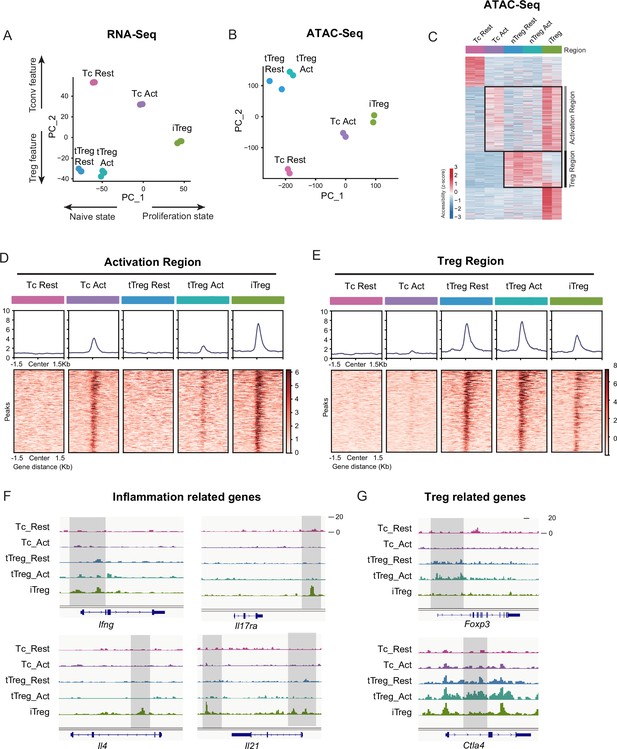
iTregs display highly open chromatin state at the activation and differentiation-related genes.
(A) Principal component analysis (PCA) visualization of transcriptional profiles of Tconvs, tTregs, and iTregs with or without TCR stimulation. Color indicates cell types. (B) PCA visualization of chromatin accessibility profiles of different cell types. Color indicates cell type. (C) Heatmap showing the chromatin accessibility of cell type specifically accessible peaks. As shown, two major groups of genes were labeled on right. (D) Line plots (top) and heatmaps (bottom) of activation regions in Tconvs, iTregs, and tTregs. Activation regions were determined by a threshold of adjusted p < 0.05 calculated by DESeq2. (E) Line plots (top) and heatmaps (bottom) of Treg regions in Tconvs, iTregs, and tTregs. Treg regions were determined by a threshold of adjusted p < 0.05 calculated by DESeq2. (F) Genomic track showing the chromatin accessibility of Ifng, Il4, Il17ra, and Il21. (G) Genomic track showing the chromatin accessibility of Foxp3 and Ctla4.
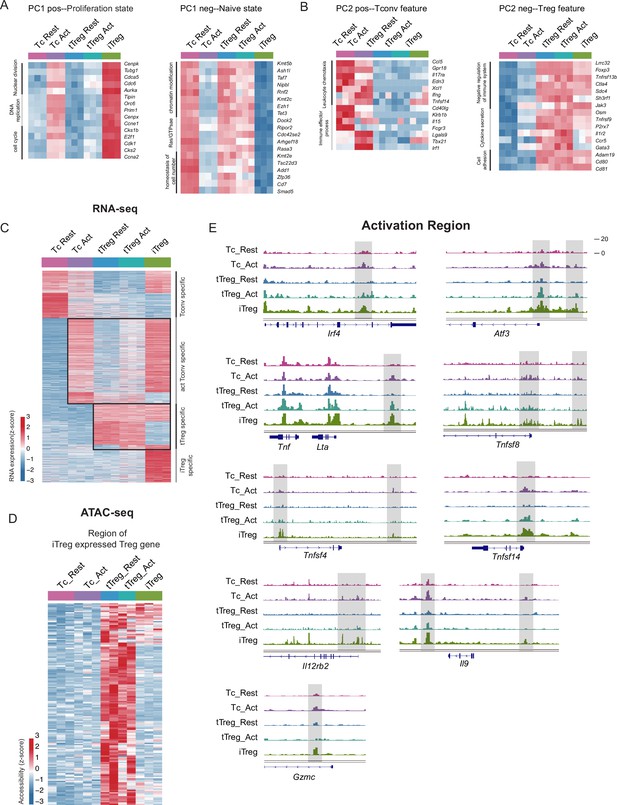
iTreg has highest proliferation state and partial Treg feature.
(A) Left, heatmap showing the top genes that positively contribute to PC1. Right, heatmap showing the top genes that negatively contribute to PC1. As shown, genes contributing positively to PC1 mainly belong to proliferation such as nuclear division, DNA replication, and cell cycle, while the negatively associated ones mainly belong to chromatin state regulations such as histone/DNA modification. (B) Left, heatmap showing the top genes that positively contribute to PC2. Right, heatmap showing the top genes that negatively contribute to PC2. As shown, the main contributors to PC2 axis were the signature genes of Tconvs and Tregs, such as Ifng, Il17ra, T-bet in the positive direction, Foxp3, Ctla4, Tnfrsf9 in the negative direction. (C) Heatmap showing the expression of cell type specifically expressed genes in RNA-seq data. As shown, four major groups of genes were identified, based on the significant higher expression in specific cell types. Different group of genes were labeled. (D) Heatmap showing the chromatin accessibility of tTreg specifically accessible peaks in tTreg specifically expressed genes. (E) Genomic track showing chromatin accessibility of Irf4, Atf3, Lta, Tnfsf8, Tnfsf4, Tnsfsf14, Il12rb2, Il9, Gzmc in activated Tconv and iTreg.
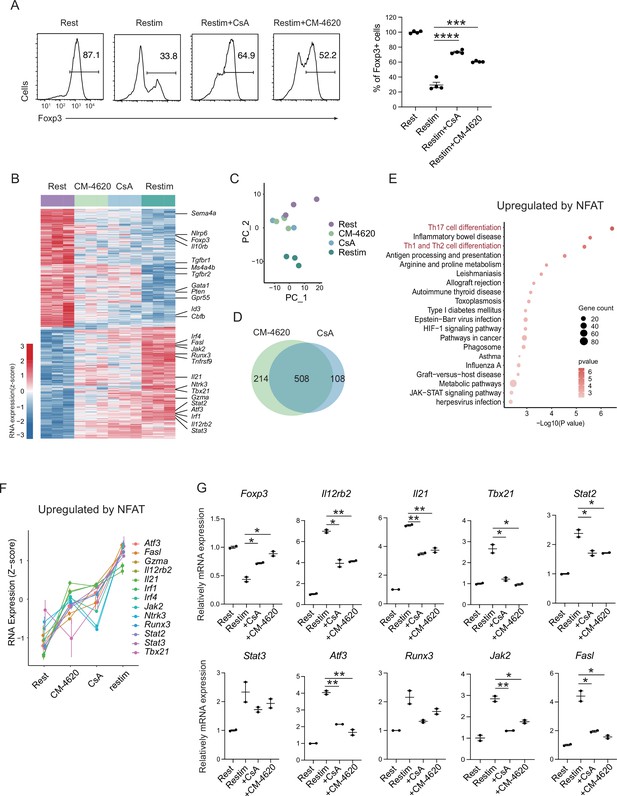
Store-operated calcium entry (SOCE) signaling and NFAT can disrupt iTreg stability.
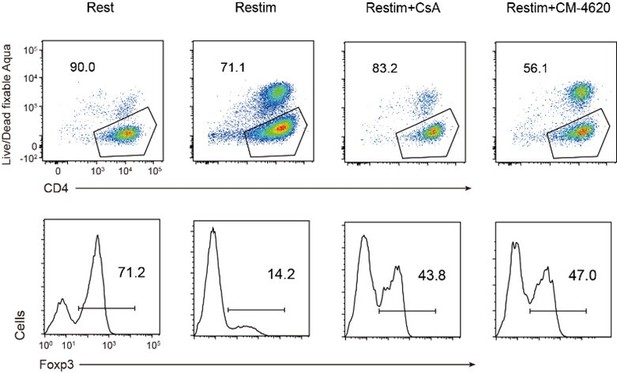
(A) Impact of calcium signal and NFAT on iTreg stability. Sorted Foxp3-GFP+ iTregs were rested for 1 day, then restimulated by anti-CD3 and CD28 in the presence of cyclosporine A (CsA) and CM-4620. Percentages of Foxp3+ cells were analyzed by intracellular staining after 2-day restimulation. Left, representative histograms of CsA and CM-4620-treated iTregs. Right, graph for the percentages of CsA and CM-4620-treated Foxp3+ cells in all CD4+ cells. n = 4, N = 8. (B) Heatmap showing the decreased changes for restimulated iTregs with CM-4620 or CsA. Typical genes were highlighted. (C) Principal component analysis (PCA) visualization of transcriptional profiles of iTregs at different states. Color indicates cell states. (D) Venn plot showing the overlap of DEGs rescued by adding CM-4620 or CsA. (E) Biological terms enriched by the significant upregulated genes after iTreg restimulation rescued by inhibiting calcium or NFAT. (F) Representative genes upregulated after iTreg restimulation rescued by inhibiting calcium or NFAT. (G) QPCR of Foxp3 and Th-differentiated gene expression in the resting, restimulated and CsA/CM-4620-treated iTregs. Here, *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, by Student’s t-test.
-
Figure 4—source data 1
iTreg stability, QPCR of related gene expression upon activation after CsA and CM-4620 blockade.
- https://cdn.elifesciences.org/articles/88874/elife-88874-fig4-data1-v1.xlsx

Store-operated calcium entry (SOCE) signaling and NFAT cause instability and downregulate Treg-related gene.
(A) Stability of iTreg treated with CM-4620, cyclosporine A (CsA), and BTP-2 upon restimulation. iTregs were restimulated by anti-CD3 and CD28 in the presence of CsA, CM-4620, and BTP-2. Percentages of Foxp3+ cells were analyzed by intracellular staining after 2-day restimulation. Left, representative histograms. Right, graph for the percentages of Foxp3. n = 3, N = 3. (B) Stability of ionomycin and CsA-treated iTreg. iTreg were treated with ionomycin and CsA, percentages of Foxp3+ cells were analyzed by intracellular staining after 1-day restimulation. n = 2, N = 3. (C) Stability of NF-κb and c-Jun/c-Fos inhibitor-treated iTreg. Sorted Foxp3-GFP+iTregs were rested for 1 day, then restimulated by anti-CD3 and CD28 in the presence of indicated inhibitors. Percentages of Foxp3+ cells were analyzed by intracellular staining after 2-day restimulation. n = 3. (D) Representative genes downregulated after iTreg restimulation rescued by inhibiting calcium or NFAT. (E) Biological terms enriched by the significant downregulated genes after iTreg restimulation rescued by inhibiting calcium or NFAT. Here, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, by Student’s t-test.
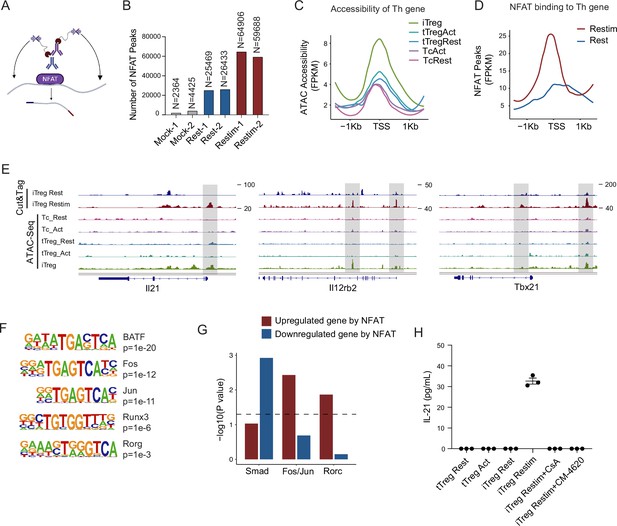
NFAT disrupt iTreg stability by upregulating prime-opened TH genes.
(A) Model of CUT&Tag experiments to capture the binding sites of NFATc1. (B) The number of NFAT Cut&Tag peaks in mock control, resting iTregs, and restimulated iTregs. (C) Normalized counts of ATAC-seq reads in resting and activated Tconvs, resting and activated tTregs and iTregs, centered on the transcription start site (TSS) region of NFAT-upregulated TH genes. (D) Normalized counts of NFAT Cut&Tag reads in resting and restimulated iTreg, centered on the TSS region NFAT-upregulated TH genes. (E) Genome track visualization of NFAT-binding profiles and chromatin accessibility profiles in typical genes Il21, Il12rb2, and Tbx21. (F) Motif enriched in peaks with higher NFAT Cut&Tag signals in restimulated iTreg versus resting iTreg. List of five representative motifs ranked based on the p-values. The enrichment was performed by using HOMER. (G) The enrichment of motif occurrence for typical NFAT cofactors Smad, Fos/Jun, Rorc in the NFAT peaks in NFAT-upregulated or -downregulated genes after restimulation. p-value was from Fisher’s exact test. (H) IL-21 secretion in tTreg and iTreg upon activation. iTregs and tTregs were sorted and restimulated with anti-CD3 and anti-CD28 antibodies, in the presence of cyclosporine A (CsA) and CM-4620. Cell culture supernatants were harvested after 2-day restimulation and IL-21 secretion was analyzed by ELISA. n = 3.
-
Figure 5—source data 1
IL-21 secretion in tTreg and iTreg upon activation.
- https://cdn.elifesciences.org/articles/88874/elife-88874-fig5-data1-v1.xlsx
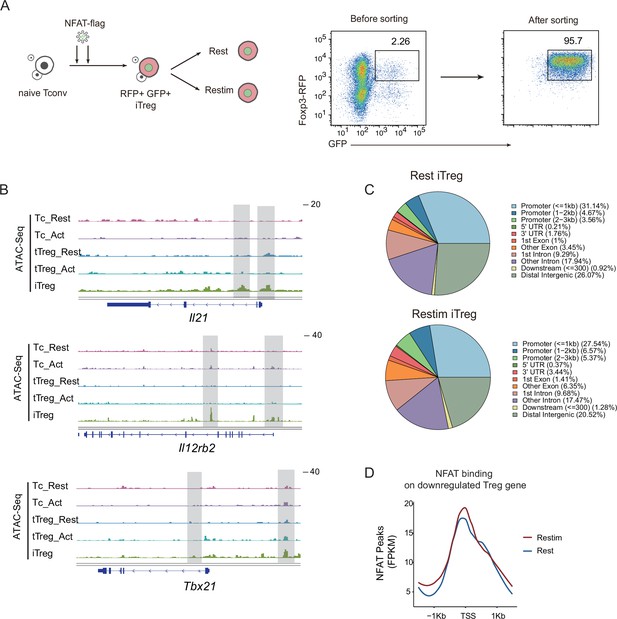
iTregs have highest accessibility in TH-associated genes.
(A) Sorted Foxp3-RFP+ naive Tconvs were induced into iTregs in the presence of IL-2 and TGF-β for 4 days, on the second and third days, iTreg cells were infected with retrovirus packaging with NFAT-flag plasmid. The infected Foxp3-RFP+ iTregs were sorted and restimulated by anti-CD3 and CD28 for 1 day. After 1-day stimulation, the cells were collected and performed Cut&Tag assay. (B) Genome track visualization of chromatin accessibility profiles in typical genes Il21, Il12rb2, and Tbx21. (C) The genomic distribution of Cut&Tag peaks for NFAT in rest iTregs and restimulated iTregs. (D) Normalized counts of NFAT Cut&Tag reads in resting and restimulated iTreg, centered on the transcription start site (TSS) region NFAT-downregulated Treg genes.
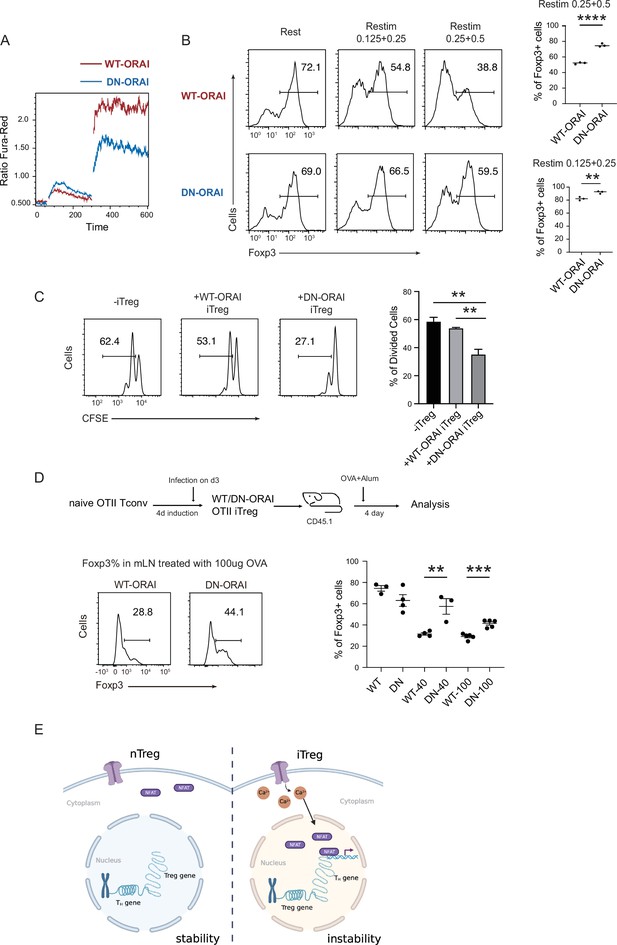
Manipulation of store-operated calcium entry (SOCE) can enhance iTreg stability.
(A) Manipulation of SOCE signal by dominant negative ORAI (DN-ORAI). SOCE was recorded in DN-ORAI iTreg cells loaded with Fura-Red by flow cytometry, TG was added after 1 min to induce ER depletion, 5 min later 2 mM calcium was added to induce calcium influx. N = 3. (B) Stability of iTreg was enhanced by DN-ORAI. iTregs were infected with WT-ORAI and DN-ORAI, and then restimulated by anti-CD3 and CD28 for 2 days. Percentages of Foxp3 were analyzed by intracellular staining. Left, representative histograms of Foxp3 expression in DN-ORAI iTregs. Right, graph for the percentages of Foxp3+ cells in all CD4+ cells. n = 3, N = 3. (C) DN-ORAI enhances iTreg suppressive capacity. CFSE-labeled OT-II T cells were stimulated with OVA-pulsed DC, Foxp3-GFP+ WT-ORAI/DN-ORAI iTregs were added to the culture to suppress the OT-II proliferation. After 40 hr, CFSE dilutions were analyzed. n = 3, N = 3. (D) Stability of DN-ORAI iTreg in vivo. WT-ORAI/DN-ORAI-GFP+-transfected CD45.2+ Foxp3-RFP+ OT-II iTregs were transferred i.v. into CD45.1 mice. Recipients were immunized with OVA323-339 in Alum adjuvant. On day 5, mLN and spleen were harvested and analyzed for Foxp3 expression by intracellular staining. Up, schematic representation of adoptive transfer experiment. Bottom right, histograms of Foxp3 expression in CD4+CD45.1+ cells in spleen and mLN; bottom left, graph for the percentages of Foxp3+ cells in all CD45.2+ cells in mLN. n = 3, N = 3. (E) Proposed model. The diminished calcium signal and closed chromatin structure in tTregs protect them from genetic and epigenetic disturbances, and the sustained calcium signal in iTregs cause NFAT aggregation in the nucleus, which makes use of a pre-opened gene loci to upregulate TH genes, thus resulting the instability of iTregs. Here, **p < 0.01; ***p < 0.001; ****p < 0.0001, by Student’s t-test.
-
Figure 6—source data 1
Store-operated calcium entry (SOCE) signal, iTreg stability in vitro and in vivo after over-expression DN-ORAI.
- https://cdn.elifesciences.org/articles/88874/elife-88874-fig6-data1-v1.xlsx
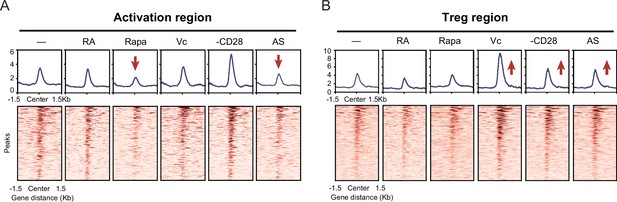
Different optimization of iTreg have various impact on activation and Treg regions.
(A) Line plots (top) and heatmaps (bottom) of activation regions in anti-CD3, anti-CD28, IL-2, and TGF-β-induced iTregs, treated with retinoic acid, rapamycin, vitamin C, removal of CD28, and AS2863619. (B) Line plots (top) and heatmaps (bottom) of Treg regions in anti-CD3, anti-CD28, IL-2, and TGF-β-induced iTregs, treated with retinoic acid, rapamycin, vitamin C, removal of CD28, and AS2863619.
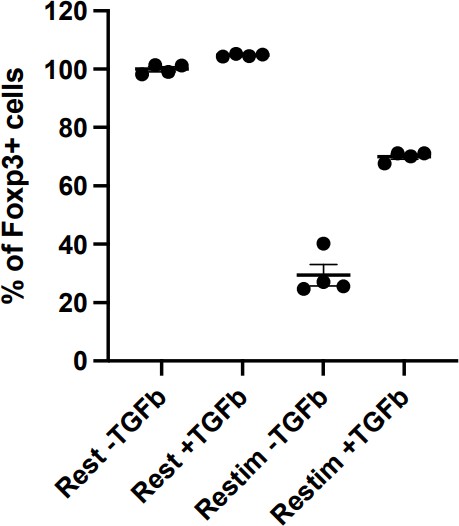
Restimulation with TGFb will persist iTreg inducing environment, resulting in less pronounced instability.
Sorted Foxp3-GFP+ iTregs were rested for 1d, and then rested or restimulated in the presence of TGF-β for 2 d. Percentages of Foxp3+ cells were analyzed by intracellular staining of Foxp3 after 2 d.
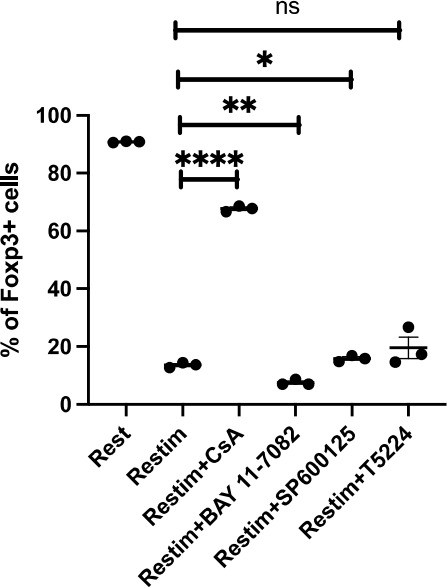
Comparing effects of NFAT, NF-kB and c-Jun/c-Fos inhibitors on iTreg instability.
Sorted Foxp3-GFP+ iTregs were rested for 1d, then restimulated by anti-CD3 and CD28 in the presence of listed inhibitors. Percentages of Foxp3+ cells were analyzed by intracellular staining after 2d restimulation.
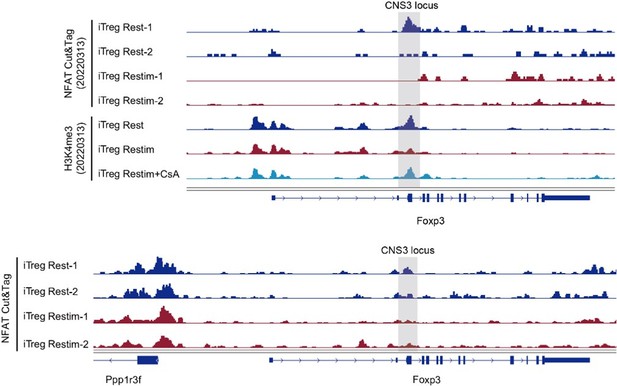
The NFAT binding and histone modification on Foxp3 gene locus.
Genome track visualization of NFAT binding profiles and H3K4me3 profiles in Foxp3 CNS3 locus in two batches of dataset.
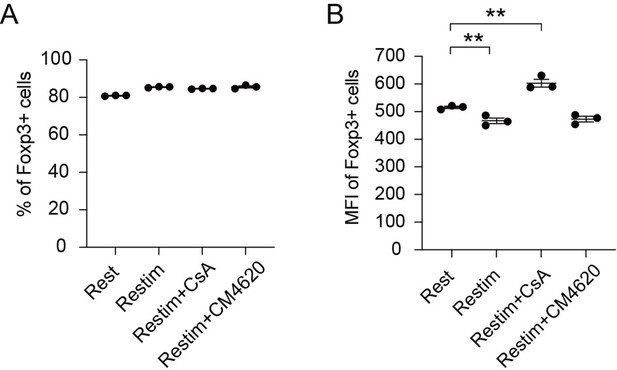
Effect of inhibiting NFAT and calcium on human iTreg stability.
Human naïve CD4 cells from PBMC were subjected to a two-week induction process to generate human iTreg. Subsequently, human iTreg were restimulated for 2 days with dynabeads followed by 2 days of rest in the prescence of CsA and CM-4620. Four days later, percentages of Foxp3+ cells and Foxp3 mean fluorescence intensity (MFI) were analyzed by intracellular staining.
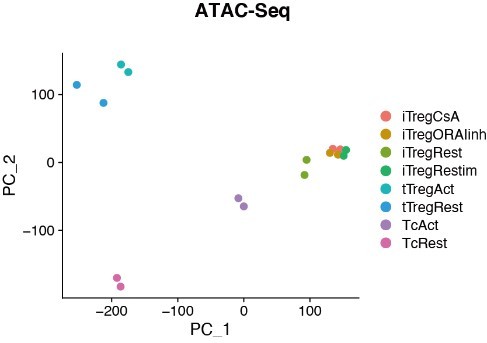
Chromatin accessibility of Rest, Retimulated, CsA/ORAIinh treated restimulated iTreg.
PCA visualization of chromatin accessibility profiles of different cell types. Color indicates cell type.
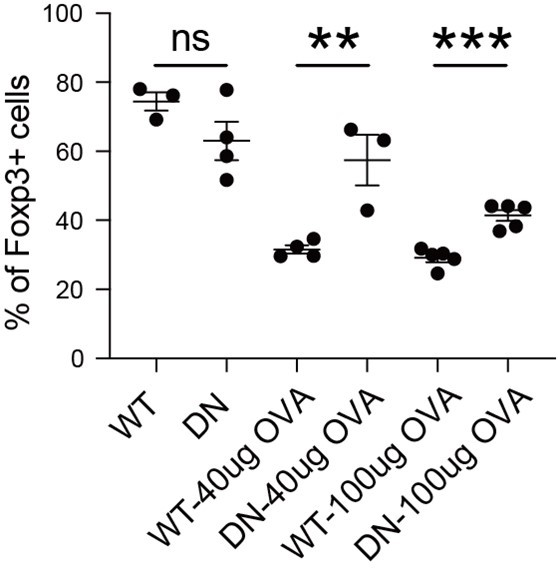
Stability of DN-ORAI iTreg in vivo with or without OVA immunization.
WT-ORAI/DN-ORAI-GFP+-transfected CD45.2+ Foxp3-RFP+ OT-II iTregs were transferred i.v. into CD45.1 mice. Recipients were left or immunized with OVA323-339 in Alum adjuvant. On day 5, mLN were harvested and analyzed for Foxp3 expression by intracellular staining.
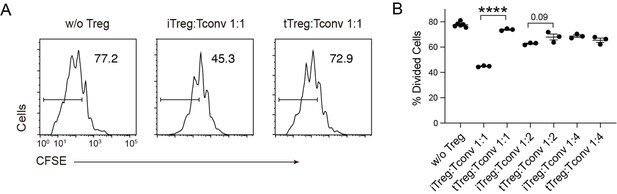
Compare multiple dose of Tconv:Treg ratio in suppression functionCFSE-labelled OT-II T cells were stimulated with OVA-pulsed DC, then different number of Foxp3-GFP+ iTregs and tTregs were added to the culture to suppress the OT-II proliferation.
After 4 days, CFSE dilution were analyzed. Left, Representative histograms of CFSE in divided Tconvs. Right, graph for the percentage of divided Tconvs.
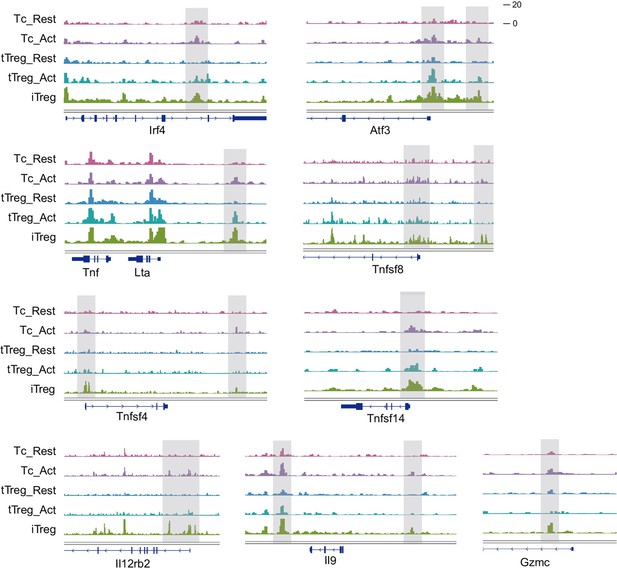
Chromatin accessibility of some “Activation Region”.
Genomic track showing chromatin accessibility of Irf4, Atf3, Lta, Tnfsf8, Tnfsf4, Tnsfsf14, Il12rb2, Il9, Gzmc in activated Tconv and iTreg.
Relationship of cell death and Foxp3 stability in restimulated iTregs.
Sorted Foxp3-GFP+ iTregs were rested for 1d, then restimulated by anti-CD3 and CD28 in the presence of CsA or CM-4620. After 2d restimulation, live cell percentage were analyzed by staining of Live/Dead fixable Aqua, and percentages of Foxp3+ cells were analyzed by intracellular staining of Foxp3. Upper, live cell percentage of iTregs. Lower, percentages of Foxp3 in iTregs.
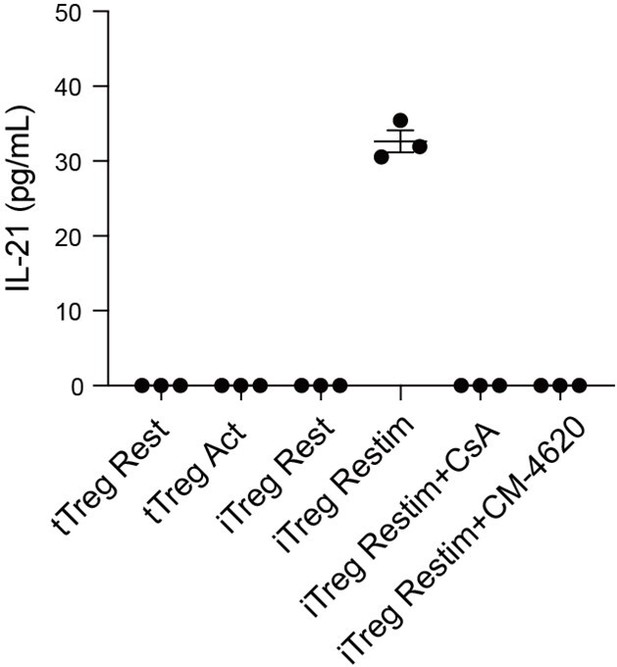
IL-21 secretion in tTreg and iTreg upon activation.
iTregs and tTregs were sorted and restimulated with anti-CD3 and anti-CD28 antibodies, in the presence of CsA and CM-4620. Cell culture supernatant were harvested after 2 d restimulation and IL-21 secretion was analyzed by ELISA.
Videos
Imaging of store-operated calcium entry (SOCE) signal in Tconvs.
Imaging of store-operated calcium entry (SOCE) signal in tTregs.
Imaging of store-operated calcium entry (SOCE) signal in iTregs.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57BL6/J | Jackson Laboratory | Strain #:000664 from Jackson Laboratory | |
| Strain, strain background (Mus musculus) | Foxp3-GFP (B6.Cg-Foxp3tm2Tch/J) | Hai Qi of School of Medicine, Tsinghua University | Strain #:006772 from Jackson Laboratory | |
| Strain, strain background (Mus musculus) | Foxp3-RFP | Zhou Xuyu of Institute of Microbiology, Chinese Academy of Sciences | Foxp3-RFP | |
| Strain, strain background (Mus musculus) | CD45.1 | Jackson Laboratory | Strain #:002014 from Jackson Laboratory | |
| Strain, strain background (Mus musculus) | OT-II | Hai Qi of School of Medicine, Tsinghua University | Strain #:004194 from Jackson Laboratory | |
| Antibody | Ultra-LEAF Purified anti-mouse CD3ε Antibody, Armenian Hamster monoclonal | Biolegend | 100340 | 0.5 μg/ml |
| Antibody | Ultra-LEAF Purified anti-mouse CD28 Antibody, Syrian Hamster monoclonal | Biolegend | 102116 | 1 μg/ml |
| Antibody | NFATc1 Antibody (7A6), mouse monoclonal | Santa Cruz | sc-7294 | 1:2000 |
| Antibody | NFATc2 Antibody (4G6-G5), mouse monoclonal | Santa Cruz | sc-7296 | 1:2000 |
| Antibody | Lamin A/C (4C11) Mouse mAb | CST | 4777 | 1:2000 |
| Antibody | β-Actin (13E5) Rabbit mAb | CST | 4970 | 1:1000 |
| Antibody | GAPDH (D4C6R) Mouse mAb | CST | 97166 | 1:1000 |
| Antibody | Monoclonal ANTI-FLAG M2 antibody produced in mouse | Sigma | F1804 | 1:50 |
| Antibody | Goat Anti-Mouse IgG Secondary Antibody, monoclonal | Sino Biological | SSA021 | 1:50 |
| Antibody | Histone H3 (D2B12) XP Rabbit mAb (ChIP Formulated) | CST | 4620 | 1:50 |
| Antibody | Anti-Histone H3 (tri methyl K4) antibody - ChIP Grade, Rabbit polyclonal | Abcam | ab8580 | 1:50 |
| Antibody | Goat Anti-Rabbit IgG Secondary Antibody, monoclonal | Sino Biological | SSA018 | 1:50 |
| Antibody | Biotin anti-mouse CD3ε Antibody, Armenian Hamster monoclonal | Biolegend | 100303 | 0.5 μg/ml |
| Antibody | FOXP3 Monoclonal Antibody (150D/E4), PE, mouse monoclonal | eBioscience | 12-4774-42 | 1:500 |
| Antibody | FOXP3 Monoclonal Antibody (150D/E4), Alexa Fluor 488, mouse monoclonal | eBioscience | 53-4774-41 | 1:500 |
| Antibody | CD4 Monoclonal Antibody (GK1.5), APC, rat monoclonal | eBioscience | 17-0041-82 | 1:500 |
| Antibody | CD25 Monoclonal Antibody (PC61.5), PE, rat monoclonal | eBioscience | 12-0251-82 | 1:500 |
| Antibody | CD44 Monoclonal Antibody (IM7), APC, rat monoclonal | eBioscience | 17-0441-83 | 1:500 |
| Antibody | PE anti-mouse CD62L, rat monoclonal | Biolegend | 104408 | 1:500 |
| Peptide, recombinant protein | Recombinant Human TGF-beta 1 Protein | R&D | 240-B-010 | |
| Peptide, recombinant protein | Recombinant Human IL-2 | R&D | 202-IL | |
| Peptide, recombinant protein | Purified Streptavidin | Biolegend | 405150 | |
| Commercial assay, kit | LIVE/DEAD Fixable Aqua Dead Cell Stain Kit, for 405 nm excitation | Invitrogen | L34965 | |
| Commercial assay, kit | Propidium Iodide | Beyotime | C1062M-3 | |
| Commercial assay, kit | Mouse CD4+ T Cell Isolation Kit | Stem cell | 19852 | |
| Commercial assay, kit | Mouse CD25 Treg Cell positive selection Kit | Stem cell | 18782 | |
| Commercial assay, kit | Foxp3/Transcription Factor Staining Buffer Set | eBioscience | 00-5523-00 | |
| Commercial assay, kit | NE-PER Nuclear and Cytoplasmic Extraction Reagents | Thermo | 78835 | |
| Commercial assay, kit | TruePrep DNA Library Prep Kit V2 for Illumina | Vazyme | TD501 | |
| Commercial assay, kit | Hyperactive Universal CUT&Tag Assay Kit for Illumina | Vazyme | TD903 | |
| Commercial assay, kit | TruePrep Index Kit V2 for Illumina | Vazyme | TD202 | |
| Commercial assay, kit | VAHTS DNA Clean Beads | Vazyme | N411 | |
| Commercial assay, kit | Mouse IL-21 Uncoated ELISA | Invitrogen | 88–8210 | |
| Chemical compound, drug | Cyclosporin A | MCE | HY-B0579 | |
| Chemical compound, drug | CM-4620 | MCE | HY-101942 | |
| Chemical compound, drug | Thapsigargin | Invitrogen | T7459 | |
| Chemical compound, drug | Ionomycin | beyotime | S1672 | |
| Chemical compound, drug | Caffeine | Aladdin | C106953 | |
| Chemical compound, drug | 4-CMC | Sigma | C55402 | |
| Chemical compound, drug | Fluo-4, AM, cell permeant | Thermo | F14201 | |
| Chemical compound, drug | Cal RedTM R525/650 AM | AAT Bioquest | 20591 | |
| Chemical compound, drug | Indo-1 AM | BD | 565879 | |
| Chemical compound, drug | Pluronic F-127 | Sigma | P2443 | |
| Chemical compound, drug | CellTak 1 MG WI, 1/CS | BD | 354240 | |
| Chemical compound, drug | Hanks’ Solution | Coolaber | SL6080 | |
| Chemical compound, drug | HBSS, 10× (without Calcium) | Macgene | CC016 | |
| Chemical compound, drug | Imject Alum | Thermo | 77161 | |
| Chemical compound, drug | OVA Peptide (323-339) | GenScript | RP10610-1 | |
| Chemical compound, drug | OVA | Sigma | A5503 | |
| Chemical compound, drug | Cell Trace CFSE | Thermo | C34554 | |
| Chemical compound, drug | Retinoic Acid | Sigma | R2625 | |
| Chemical compound, drug | InSolution Rapamycin | Sigma | 553211 | |
| Chemical compound, drug | L-Ascorbic acid (Vitamin C) | Sigma | A4403 | |
| Chemical compound, drug | AS2863619 | MCE | HY-126675A | |
| Chemical compound, drug | BAY 11-7082 | MCE | HY-13453 | |
| Chemical compound, drug | SP600125 | MCE | HY-12041 | |
| Chemical compound, drug | T-5224 | MCE | HY-12270 | |
| Software, algorithm | FlowJo | FlowJo LLC | ||
| Software, algorithm | Prism | GraphPad | ||
| Software, algorithm | IGV | Broad Institute | ||
| Software, algorithm | Imaris | Andor | ||
| Software, algorithm | R | R studio | ||
| Software, algorithm | JPK Data Processing | JPK |





