Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture
Figures
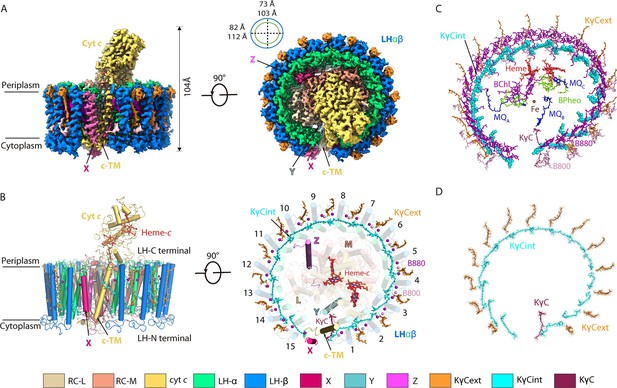
Overall structure of the native reaction center (RC)-light harvesting (LH) complex from R. castenholzii.
(A) A cryo-electron microscopy (cryo-EM) map of the native RC-LH (nRC-LH) complex is shown from the side (left panel) and the bottom (right panel). The dimensions of the RC-LH complex and LH ring are represented. The positions of subunit X, proteins Y and Z, and the cytochrome (cyt) c transmembrane (c-TM) domain are labeled. (B) Side and top views of the nRC-LH complex are presented in cartoon form. LH subunits are numbered clockwise from the gap formed by subunit X and c-TM. Heme-c (red) and keto-γ-carotene (KγC) molecules (orange, cyan, ruby) are shown in stick forms; Mg atoms of the bacteriochlorophylls B800 (pink) and B880 (purple) are shown as spheres. (C) The cofactors bound in the nRC-LH complex. All cofactors are shown in stick forms except for the interior KγC (KγCint) in LH, the iron bound in the RC are shown as spheres. (D) The structural models of the KγCint, exterior KγC (KγCext), and KγC in the nRC-LH complex are fitted in the EM density map. The color scheme: lime green, α-polypetides; marine, β-polypetides; yellow-orange, cyt c; wheat, L subunit; salmon, M subunit; pale cyan, protein Y; hot pink, subunit X; light magenta, protein Z; cyan, KγCint; orange, KγCext; ruby, KγC; purple, B880; pink, B800; tv-red, heme-c; chartreuse, bacteriopheophytins (BPheos); blue, menaquinone-11 (MQ); brown, iron.

Purification and verification of the native reaction center-light harvesting (nRC-LH) and carotenoid (Car)-depleted RC-LH (dRC-LH) complexes from R. castenholzii.
(A) Appearance of the native R. castenholzii (left) and the fifth subculture of diphenylamine (DPA)-treated (right) cells, which were cultured anaerobically under high (180 μmol m–2 s–1), medium (32 μmol m–2 s–1), and low (2 μmol m–2 s–1) illuminations (ILLs), respectively. (B) Growth curves of the native R. castenholzii and the fifth subculture of DPA-treated cells grown under high, medium, and low illuminations. Data are shown as the mean ± standard deviations (n=3). (C) Representative gel filtration chromatography, blue native PAGE, and SDS-PAGE of the nRC-LH complex from R. castenholzii cultured under high illumination. The chromatography diagram of absorption at 280 nm (red) and 800 nm (green) of nRC-LH is shown. The subunits L, M, and cytochrome (cyt) c subunits of RC and the αβ apoproteins of the LH are labeled. (D) Absorption spectrum analyses of RC-LH complexes purified from R. castenholzii grown under high illumination. Black corresponds to untreated cells; brown, gray, orange, blue, and green correspond to Car-depleted cells from five consecutive subcultures of DPA-treated cells, respectively. The characteristic Qy bands of B800 and B880, the Qx and Soret bands of bacteriochlorophylls (BChls), and the absorption peaks of Cars are labeled. (E) Absorption spectrum analyses of RC-LH complexes purified from R. castenholzii cells with or without DPA treatment under high, medium, and low illuminations. (F) Representative gel filtration chromatography, blue native PAGE, and SDS-PAGE of the dRC-LH complex purified from R. castenholzii cultured under high illumination.
-
Figure 1—figure supplement 1—source data 1
Raw figures of the full uncropped blue native PAGE of nRC-LH with and without the relevant bands labelled.
- https://cdn.elifesciences.org/articles/88951/elife-88951-fig1-figsupp1-data1-v1.zip
-
Figure 1—figure supplement 1—source data 2
Raw figures of the full uncropped SDS PAGE of nRC-LH with and without the relevant bands labelled.
- https://cdn.elifesciences.org/articles/88951/elife-88951-fig1-figsupp1-data2-v1.zip
-
Figure 1—figure supplement 1—source data 3
Raw figures of the full uncropped blue native PAGE of dRC-LH with and without the relevant bands labelled.
- https://cdn.elifesciences.org/articles/88951/elife-88951-fig1-figsupp1-data3-v1.zip
-
Figure 1—figure supplement 1—source data 4
Raw figures of the full uncropped SDS PAGE of dRC-LH with and without the relevant bands labelled.
- https://cdn.elifesciences.org/articles/88951/elife-88951-fig1-figsupp1-data4-v1.zip
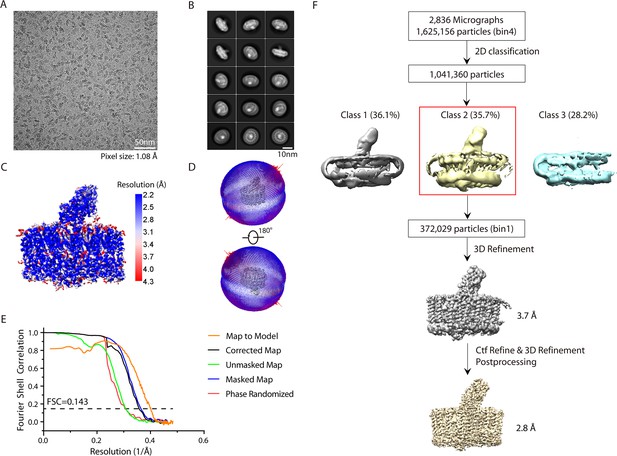
Cryo-electron microscopy (cryo-EM) analysis of the native reaction center-light harvesting (nRC-LH) complex from R. castenholzii.
(A) A representative raw cryo-EM micrograph of the nRC-LH complex purified from the cells grown under high illumination (180 μmol m–2 s–1). (B) Representative reference-free 2D class averages of the nRC-LH complex. (C) Local resolution of the cryo-EM map estimated by ResMap. (D) The different views of angular distribution of the nRC-LH complex particles in final reconstruction. The direction and length of each cylinder represent the direction and amount of particles. (E) The gold-standard Fourier shell correlation (FSC) curve of the cryo-EM map (corrected map, black; unmasked, green; masked, blue; phase randomized, red) and the FSC curve between the map and the final refined model (orange). (F) Cryo-EM data processing workflow for the nRC-LH complex. All structural figures here are generated with ChimeraX.

Cryo-electron microscopy (cryo-EM) analysis of the native reaction center-light harvesting (nRC-LH) complexes purified from R. castenholzii grown under medium (32 μmol m–2 s–1) or low (2 μmol m–2 s–1) illuminations.
(A, B) Cryo-EM data processing workflow for the native RC-LH purified from R. castenholzii grown under medium (A) and low (B) illuminations. The local resolution of the cryo-EM map and the angular distribution of particles in final reconstruction are shown in upper right panel. The gold-standard Fourier shell correlation (FSC) curve of the cryo-EM map and the FSC curve between the map and the final refined model are shown in lower right panel.
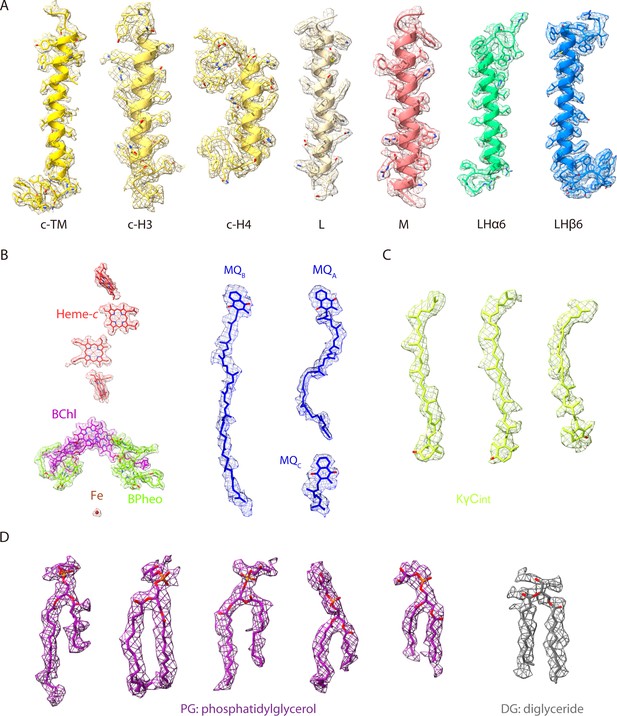
Cryo-electron microscopy (cryo-EM) densities and structural models of the reaction center-light harvesting (RC-LH) complex from R. castenholzii.
(A) EM density maps of representative α-helices in the native RC-LH (nRC-LH) complex are shown in mesh forms. The corresponding atomic models are shown in stick forms. (B) The cofactors in the native RC were docked into the EM density map. (C) A density map of the keto-γ-carotenes (KγC) (interior KγC [KγCint]) bound in LH ring interior of the carotenoid-depleted RC-LH (dRC-LH) complex. The corresponding atomic models are colored in limon. (D) EM map densities fitted with resolved lipids for the nRC-LH.
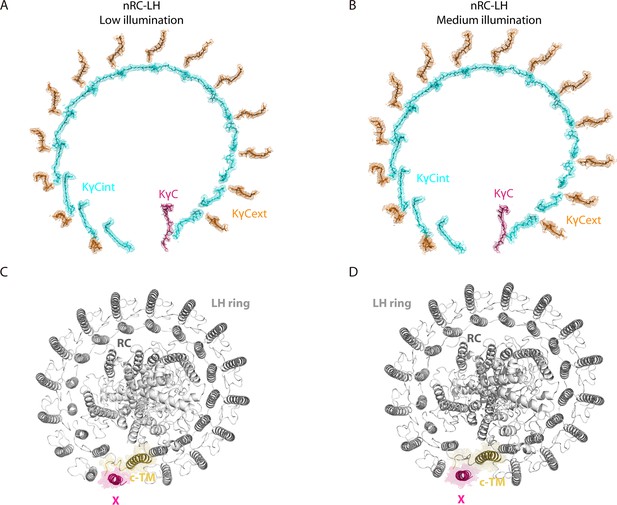
The structures of native reaction center-light harvesting (RC-LH) (nRC-LH) complexes obtained from R. castenholzii grown under low (2 μmol m–2 s–1) and medium (32 μmol m–2 s–1) illuminations.
(A, B) The structural models of the interior keto-γ-carotenes (KγCint), exterior keto-γ-carotenes (KγCext), and a KγC at the LH ring opening in the nRC-LH complexes obtained at low (A) and medium (B) illuminations were fitted in the electron microscopy (EM) density map. (C, D) The structural models of proteins in the nRC-LH complexes obtained at low (C) and medium (D) illuminations. The corresponding EM densities of subunit X and cyt c transmembrane (c-TM) are shown in surface forms.
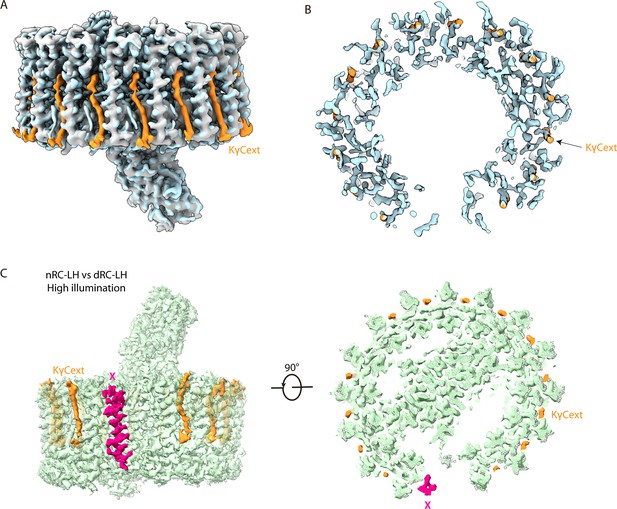
Comparison of the cryo-electron microscopy (cryo-EM) maps of the native reaction center-light harvesting (nRC-LH) and carotenoid-depleted RC-LH (dRC-LH) complexes obtained at medium (32 μmol m–2 s–1) and high (180 μmol m–2 s–1) illuminations.
(A) Superposition of cryo-EM maps of the reported 4.1 Å RC-LH model and the low-pass filtered nRC-LH map that was obtained at the same medium illumination. The cryo-EM map of nRC-LH complex (blue) was low-pass filtered to the same 4.1 Å resolution as the reported RC-LH (gary). The cryo-EM densities for the exterior keto-γ-carotenes (KγCext) were highlighted in orange. (B) The global density differences between the reported 4.1 Å RC-LH model and the nRC-LH complex obtained at medium illumination. The difference map of KγCext is shown in orange. (C) Superposition of cryo-EM maps of the nRC-LH (surface) and dRC-LH (mesh) complexes obtained at high illumination. The cryo-EM densities of subunit X and KγCext were highlighted in hot pink and orange, respectively. All structural figures here are generated with ChimeraX.
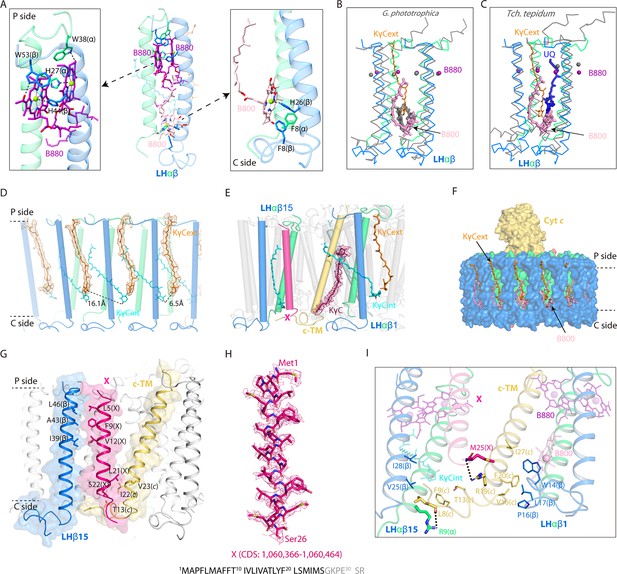
Interactions of the keto-γ-carotenes (KγC), bacteriochlorophylls (BChls), and subunit X with the light harvesting (LH) ring.
(A) Interactions between the LHαβ heterodimer and the bound BChls. Close-up views of amino acid residues that coordinate the LH-bound B880s (left) and B800 (right) are shown on the periplasmic (P) and the cytoplasmic (C) side. The BChls and interacting amino acid residues are shown in stick forms. (B, C) Superposition of LHαβ heterodimer from nRC-LH (colored) with Gemmatimonas (G.) phototrophica LHh (B, gray) and Tch. tepidum LH1 (C, gray). The LH-bound B800 and exterior KγC (KγCext) in nRC-LH are shown as pink and orange sticks, respectively. Mg atoms of LH-bound B880 are shown in spheres. The LHh-bound B800 in G. phototrophica is shown in gray sticks, and Tch. tepidum LH1-bound ubiquinone (UQ) is shown in blue sticks. (D, E) KγC organization. Interior KγC (KγCint) are shown in cyan, KγCext are shown in orange, and the KγC inserted between cytochrome c transmembrane (c-TM) and LHαβ is shown in ruby. (F) Incorporation of the KγCext and B800s at the cytoplasmic side blocked the LHαβ interface. (G, I) Interactions between the assigned subunit X (hot pink), c-TM (yellow-orange), and neighboring LHαβ1 and LHαβ15 in the nRC-LH. The N-terminus (N-ter) and C-terminus (C-ter) of subunit X, c-TM and LHβ15 are indicated. The hydrogen bonding and hydrophobic interactions between the amino acid residues are labeled and indicated with dashed lines. The BChls B880 and B800 are shown as purple and pink sticks, respectively. (H) The assigned subunit X (hot pink) are fitted in the EM density map. Location of the coding sequence (CDS) in R. castenholzii genomic DNA, and the amino acid sequence of subunit X are indicated, with the modeled amino acid residues colored in black.

Interactions between LHαβ heterodimers in the native reaction center-light harvesting (RC-LH) (nRC-LH) complex from R. castenholzii.
(A) The left panel shows the interactions between LHα and LHβ polypeptides. Close-up views of interactions between the C-terminus (top) and N-terminus (bottom) of LHα and LHβ are shown on the right. Interacting amino acid residues are shown in stick forms. (B) Structure-based sequence alignment of the LHα and LHβ subunits from R. castenholzii and the representative purple bacteria. Tch. tepidum (PDB ID: 5Y5S), G. phototrophica (PDB ID: 7O0U), Rhodopila (Rpi.) globiformis (PDB ID: 7XXF), and Rba. sphaeroides (PDB ID: 7F0L). (C) Structural comparison of LHαβ heterodimers from R. castenholzii nRC-LH with Rba. sphaeroides LH2 (left, PDB ID: 7PBW) and Rhodopseudomonas (R.) acidophila LH3 (right, PDB ID: 1IJD) that both bind B800s. The LH-bound B800 and exterior keto-γ-carotenes (KγCext) in nRC-LH are shown as pink and orange sticks, respectively. The B800 bound in Rba. sphaeroides LH2 (violet-purple) and R. acidophila LH3 (salmon) are also shown in stick forms. Mg atoms of LH-bound B880 are shown in spheres. The periplasmic (P) and cytoplasmic (C) sides are labeled.
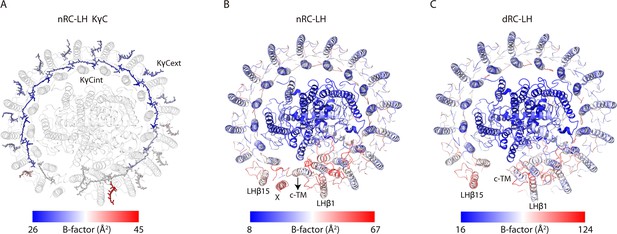
B-factors distribution of the native reaction center-light harvesting (nRC-LH) and carotenoid-depleted RC-LH (dRC-LH) complexes from R. castenholzii.
(A) B-factors distribution of the keto-γ-carotenes in nRC-LH. B-factor values are indicated by color. Protein subunits of the nRC-LH are shown in gray. (B) B-factors distribution of protein subunits in the nRC-LH complex. (C) B-factor distribution of protein subunits in the dRC-LH complex. B-factor values are indicated by color. The subunit X, cytochrome (cyt) c transmembrane (c-TM) of cyt c subunit, LHβ1 and LHβ15 with higher B-factor are indicated.
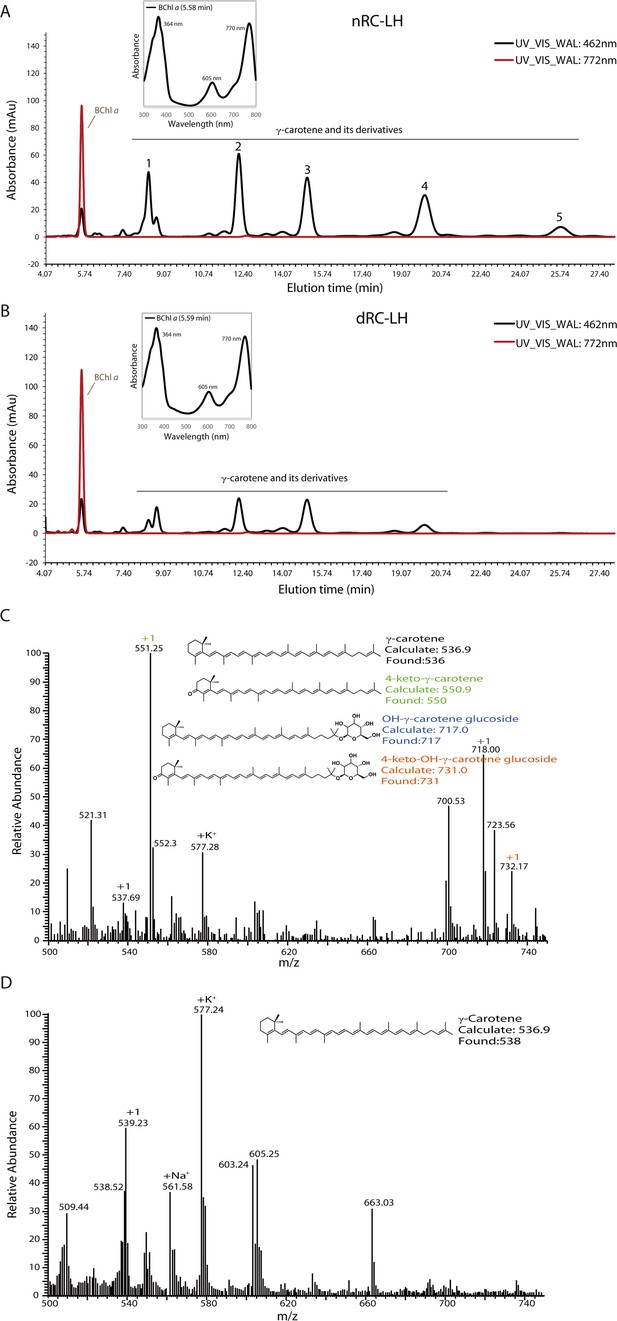
High-performance liquid chromatography (HPLC)-mass spectrometry (MS) analyses of the pigments in reaction center-light harvesting (RC-LH) complex from R. castenholzii.
(A, B) HPLC analysis of the extracted pigments of native RC-LH (nRC-LH) (A) and carotenoid-depleted RC-LH (dRC-LH) (B) that were isolated from the fifth sub-culture of diphenylamine (DPA)-treated R. castenholzii cells. The pigments were detected by their absorbance at 462 nm (black line) and 772 nm (red line), based on the characteristic absorption spectrum and LC-MS. The first peak at retention time of 5.58 min was identified to be the bacteriochlorophyll (BChl) a , which was characterized with absorption peaks at 364 nm, 605 nm, and 770 nm. The following peaks 1–5 were all characterized to be γ-carotene and its derivatives. (C, D) MS analysis of the HPLC peak 1 (C) and peaks 2–5 (D) using atmospheric pressure chemical ionization (APCI) method. The calculated and found molecular weights of γ-carotene, 4-keto-γ-carotene, OH-γ-carotene glucoside, and 4-keto-OH-γ-carotene glucoside were ~536 Da, ~550 Da, ~717 Da, and ~731 Da, respectively. Both the measured m/z values and the chemical formula of γ-carotene and its derivatives were indicated. All the experiments were repeated three times.

Structural comparison of the native reaction center-light harvesting (nRC-LH) from R. castenholzii with the RC-LH1s from Rba. sphaeroides and Tch. tepidum.
(A, B) Overall structural comparison of R. castenholzii nRC-LH (colored) with Rba. sphaeroides RC-LH1 (white, PDB ID: 7F0L) and Tch. tepidum RC-LH1 (white, PDB ID: 5Y5S). (C, D) Superposition of LHαβ heterodimers from R. castenholzii nRC-LH with Rba. sphaeroides RC-LH1 (C) and Tch. tepidum RC-LH1 (D). R. castenholzii nRC-LH is shown in cartoon form, except for the Mg atoms of LH-bound BChls (B800/B880) are shown in spheres. Interior keto-γ-carotenes (KγCint) and exterior keto-γ-carotenes (KγCext) in the nRC-LH are shown as cyan and orange sticks, respectively. The carotenoids in Rba. sphaeroides and Tch. tepidum RC-LH1 are shown as white sticks. The ubiquinone (UQ) bound in Tch. tepidum LHαβ is colored blue. (E) Superposition of R. castenholzii nRC-LH with Rba. sphaeroides RC-LH1 at the LHαβ15 heterodimer to show the spatial organizations of subunit X, cytochrome c transmembrane (c-TM), and Y with that of PufX and PufY (or protein-U). The right panel shows top view of the superposed region near PufY. Arrow shows the distance (~17 Å) between the N-terminus of the transmembrane helices of c-TM and PufY. The model of R. castenholzii nRC-LH is colored same as Figure 1. The model of Rba. sphaeroides RC-LH1 is colored in white, with PufX colored in arsenic and PufY colored in light pink.

Assignment of the amino acid sequences and coding sequences of subunit X and protein Y in Roseiflexus sp. and R. castenholzii DSM 13941/HLO8.
(A) Amino acid sequences of the KatS3mg058_1126 and KatS3mg058_1154 proteins, and the coding sequence of KatS3mg058_1154 in Roseiflexus sp. genome. (B) Amino acid sequences of subunit X and protein Y, and the coding sequence of protein Y in the genome of R. castenholzii DSM 13941/HLO8 strain. The non-conserved amino acid residues in protein Y and KatS3mg058_1154 are colored in blue and underlined. (C) The structural model of protein Y in the native reaction center-light harvesting (RC-LH) were docked into the electron microscopy (EM) density map. Arrows indicate the non-conserved amino acid residues of protein Y compared with Roseiflexus sp. KatS3mg058_1154.
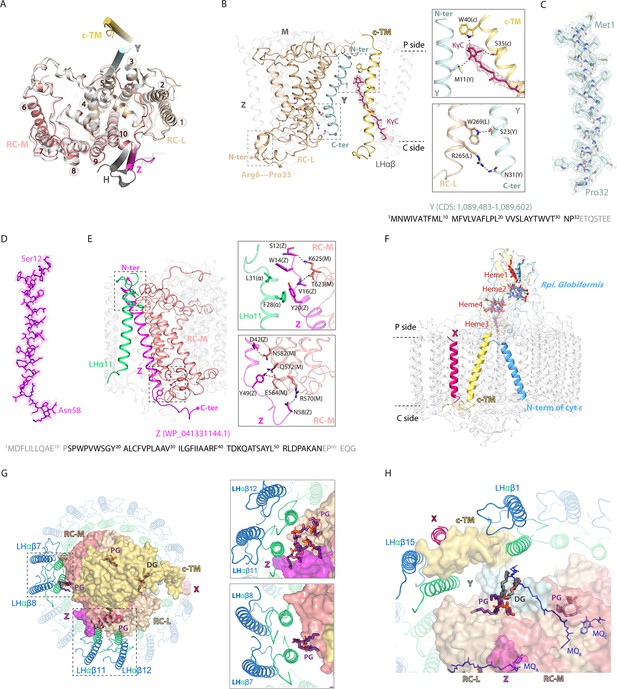
Stabilizing the reaction center (RC)-light harvesting (LH) interactions.
(A) Superposition of R. castenholzii RC structure (colored) with that of Rba. sphaeroides (white, PDB ID: 7F0L) showed excellent match at the L (wheat) and M (salmon) subunits, each of which contains five transmembrane helices (TM1–5 for L and TM6–10 for M). The newly assigned TM helices from protein Y (pale cyan) and Z (light magenta) are located on the two sides of the RC. The only TM helix of Rba. sphaeroides H subunit (gray) does not match with that of protein Z. (B) Interactions between the assigned protein Y (pale cyan), cytochrome c transmembrane (c-TM) (yellow-orange), and the RC-L (wheat). The N-terminus (N-ter) and C-terminus (C-ter) of Y, and RC-L N-terminal extension (Arg6-Pro35) that located at the periplasmic (P) and the cytoplasmic (C) side are indicated. The hydrogen bonding interactions between the amino acid residues and KγC (ruby sticks) are labeled and indicated with dashed lines. (C, D) The assigned TM helix of protein Y (C, pale cyan) and protein Z (D, light magenta) are fitted in the electron microscopy (EM) density map. Location of the coding sequence (CDS) of Y in R. castenholzii genomic DNA and the protein accession number of protein Z are indicated. The amino acid sequences of protein Y and Z are indicated below, with the modeled amino acid residues colored in black. (E) Interactions between the assigned protein Z (light magenta), LHα11 (lime green), and the RC-M (salmon). The N-terminus (N-ter) and C-terminus (C-ter) of Z that located at the periplasmic (P) and the cytoplasmic (C) side are indicated. The hydrogen bonding interactions are shown in the dashed lines. (F) Superposition of R. castenholzii RC-bound cytochrome (cyt) c (yellow-orange) with that of Rpi. globiformis (cornflower blue, PDB ID: 7XXF) showed excellent match at the tetra-heme binding domain. The c-TM and N-ter of Rpi. globiformis cyt c directed into opposite directions. (G, H) Interactions of the lipids (phosphatidylglycerol, PG; and diglyceride, DG) with the LH and RC. The L, M, and cyt c subunits of RC are shown in surface, and LHαβs are shown in cartoon forms, the lipids and RC-bound menaquinone-11s (MQs) are shown in deep purple and blue sticks, respectively.
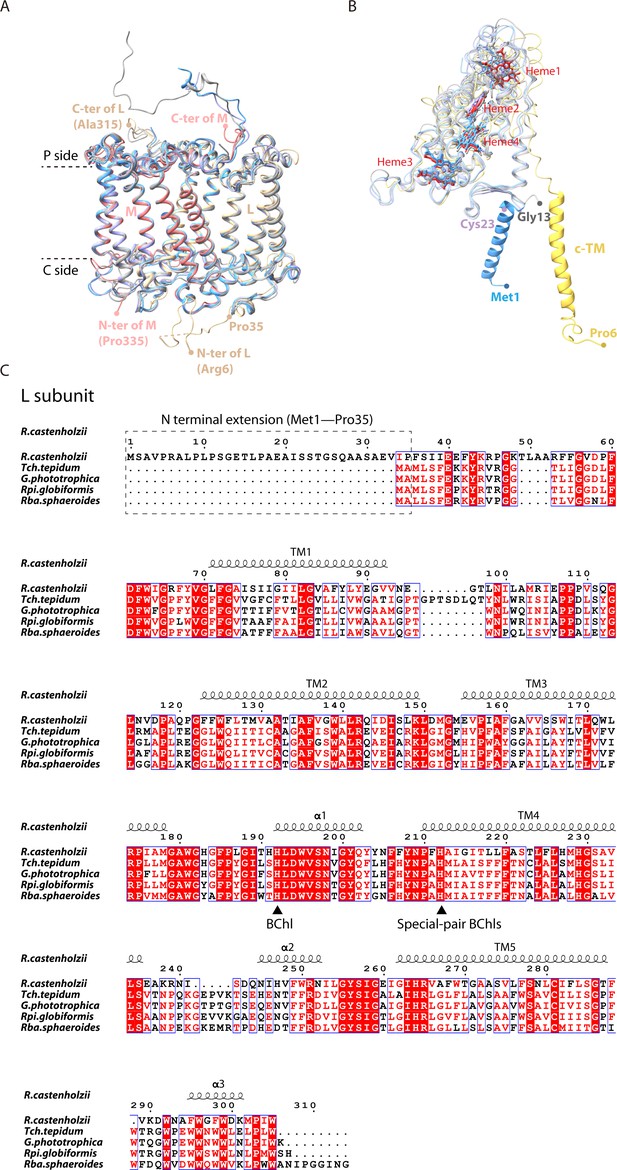
Comparison of R. castenholzii reaction center (RC) with the reported RCs from Tch. tepidum (PDB ID: 5Y5S), G. phototrophica (PDB ID: 7O0U), Rpi. globiformis (PDB ID: 7XXF).
(A) Comparison of R. castenholzii native RC structure (colored) with that of G. phototrophica RC-dLH (gary), Tch. tepidum RC-LH1 (purple), and Rpi. globiformis RC-LH1 (cornflower blue). The N- and C-terminus of the L and M subunits are labeled and indicated. The periplasmic (P) and cytoplasmic (C) sides are labeled. (B) Comparison of R. castenholzii cytochrome (cyt) c structure (yellow-orange, heme-c colored in red) with the tetraheme-bound cyt c subunit form of G. phototrophica RC-dLH (gary), Tch. tepidum RC-LH1 (purple), and Rpi. globiformis RC-LH1 (cornflower blue). The N-terminus of each cyt c subunit is labeled. (C) Structure-based sequence alignment of the L subunit from R. castenholzii and the representative purple bacteria. Tch. tepidum (PDB ID: 5Y5S), G. phototrophica (PDB ID: 7O0U), Rpi. globiformis (PDB ID: 7XXF), and Rba. sphaeroides (PDB ID: 7F0L). The amino acid residues essential for coordinating the BChl-Mg are indicated with black triangles.

Structure-based sequence alignment of the M subunit from R. castenholzii and the representative purple bacteria.
Tch. tepidum (PDB ID: 5Y5S), G. phototrophica (PDB ID: 7O0U), Rpi. globiformis (PDB ID: 7XXF), and Rba. sphaeroides (PDB ID: 7F0L). The amino acid residues essential for coordinating the BChl-Mg are indicated with black triangles.

Structure-based sequence alignment of the cyt c subunit from R. castenholzii and the representative purple bacteria.
Tch. tepidum (PDB ID: 5Y5S), G. phototrophica (PDB ID: 7O0U), Rpi. globiformis (PDB ID: 7XXF), and Rba. sphaeroides (PDB ID: 7F0L). The amino acid residues essential for coordinating the heme-c are indicated with black triangles.
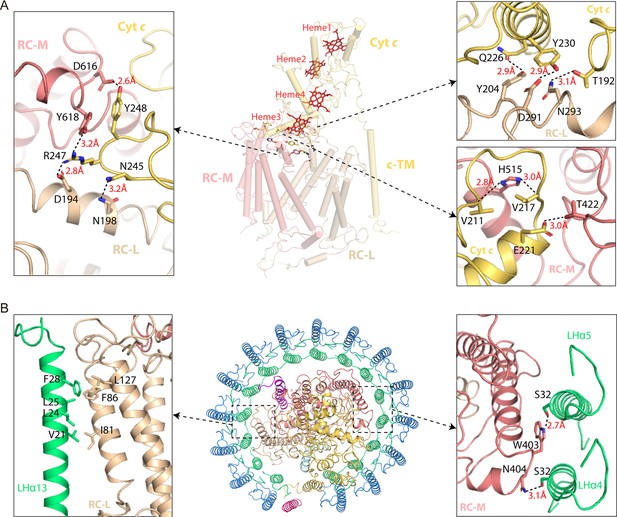
Interactions between the light harvesting (LH) ring and reaction center (RC) in native RC-LH (nRC-LH) complex from R. castenholzii.
(A) The interactions between cytochrome (cyt) c (yellow orange) and L (wheat) and M (salmon) subunits of the RC. The side view of RC is shown in middle. L, M, and cyt c subunits in the RC are shown in cartoon forms, and the c-type hemes (red) are shown in stick forms. The zoom-in views of the interactions are shown on the two sides. The amino acid residues involved in the interactions are shown in stick forms. The distances of hydrogen bonding interactions are labeled and indicated with dashed lines. (B) The amino acid residues involved in the LH and RC (which is composed of L, M, cyt c subunits) interactions are shown in stick forms. The general location of the LH and RC contacting area is shown in middle, zoom-in views of the interactions are shown on the two sides.
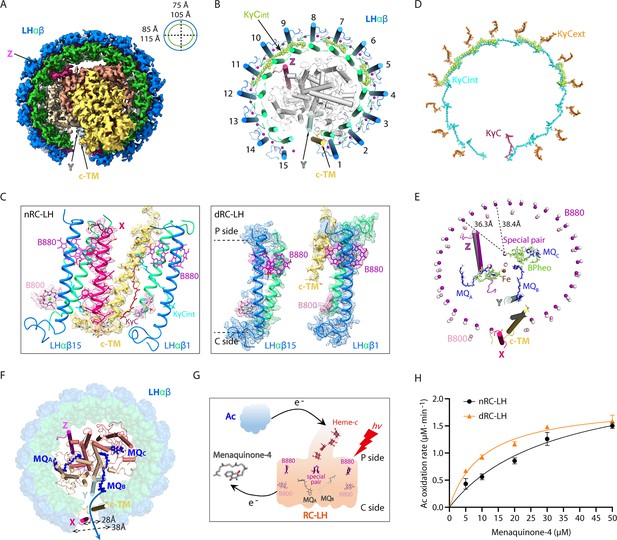
Cryo-electron microscopy (cryo-EM) structure of the carotenoid-depleted reaction center-light harvesting (dRC-LH) complex of R. castenholzii and its conformational changes that accelerated quinone/quinol exchange.
(A) Cryo-EM map of dRC-LH seen from the bottom with LH ring dimensions indicated. (B) Cartoon representation of the dRC-LH complex from the bottom. The interior keto-γ-carotenes (KγCint) are shown in limon and bacteriochlorophyll (BChl) Mg atoms are shown as spheres. (C) Comparison of the LH ring openings in the nRC-LH (left) and dRC-LH (right). The cryo-EM maps of the subunit X (hot pink), cytochrome c transmembrane (c-TM) (yellow-orange), and neighboring LHαβ1 and LHαβ15 are shown to indicate the conformational changes. The LH-bound B880s (purple), B800s (pink), and KγC (ruby) are shown in sticks and fitted in the EM map. The periplasmic (P) and cytoplasmic (C) sides are labeled. (D) Comparison of the KγC arrangement between the nRC-LH and dRC-LH complexes. The KγCint, KγCext, and KγC in nRC-LH are shown as cyan, orange, and ruby sticks, respectively. The five KγCint molecules bound in the dRC-LH complex are shown as limon spheres. (E) Comparison of the central BChl-Mg atoms in nRC-LH and dRC-LH. The B880 and B800 Mg atoms are shown as purple and pink spheres, respectively, in nRC-LH, and as white spheres in dRC-LH. The two structures are superposed at the TM helices of the L and M subunits. The distances between the central Mg atoms of B880 and the nearest special pair BChls are labeled and indicated with dashed lines. The cofactors bound in the RC are shown in stick form; the iron is shown as spheres. TM helices of subunit X (hot pink), protein Y (pale cyan), Z (light magenta), c-TM (yellow-orange in nRC-LH and white in dRC-LH), and LHαβ1 and LHαβ15 (colored in nRC-LH and white in dRC-LH) are shown in ribbon form to demonstrate the spatial organization. (F) Comparison of the LH ring opening and quinone channels in nRC-LH and dRC-LH. The LH ring of dRC-LH is shown in surface form; the RC (including Y and Z), c-TM, and subunit X in nRC-LH are shown in cartoon forms; and menaquinones (MQs) are shown in blue sticks. Dashed lines indicate the dimensions of the LH ring openings in the two structures. The blue arrow represents the putative quinone shuttling path. (G) Model diagram of the auracyanin (Ac) oxidation assay. Upon illumination, light energy absorbed by the LH-bound BChls (B800 and B880) is transferred to RC. The primary charge separation occurs and initiates sequential electron transfer that reduces the MQs. The generated hydroquinone diffuses out of the RC-LH and exchanges with the menaquinone-4 in the solution. Once the reduced Ac is oxidized, the released electrons can be transferred back to reduce the photo-oxidized special pair through the c-type hemes. (H) The rate of Ac oxidation at various starting concentrations of menaquinone-4, in presence of the nRC-LH (black) or dRC-LH (orange). Data are shown as the mean ± standard deviations (n=3).
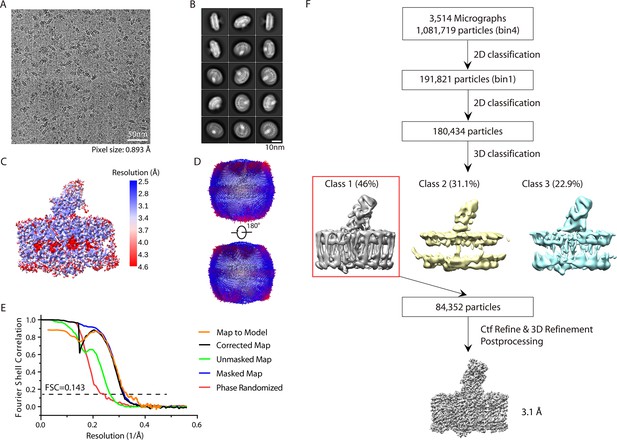
Cryo-electron microscopy (cryo-EM) analyses of the carotenoid-depleted reaction center-light harvesting (dRC-LH) complex from R. castenholzii.
(A) A representative raw cryo-EM micrograph of the dRC-LH complex extracted from the fifth sub-culture of diphenylamine (DPA)-treated R. castenholzii cells grown under high illumination (180 μmol m–2 s–1). (B) Representative reference-free 2D class averages of the dRC-LH complex. (C) Local resolution of the cryo-EM map estimated by ResMap. (D) The different views of angular distribution of the dRC-LH particles in final reconstruction. (E) The gold-standard Fourier shell correlation (FSC) curve of the cryo-EM map (corrected map, black; unmasked, green; masked, blue; phase randomized, red) and the FSC curve between the map and the final refined model (orange). (F) Workflow of the cryo-EM data processing for the dRC-LH complex. All structural figures here are generated with ChimeraX.

Structural comparisons of LHαβ heterodimers with bound and unbound keto-γ-carotenes (KγC) in R. castenholzii reaction center-light harvesting (RC-LH) complexes.
(A) The density map of LH-bound KγC in the carotenoid-depleted RC-LH (dRC-LH) complex. The corresponding atomic models of KγC (interior KγC [KγCint]) bound in LHαβ5, -7, -9, -10, and -11 are colored in limon. (B) Superposition of LHαβ5, -6, -7, and -8 from dRC-LH. LHαβs are shown in ribbon form, the Mg atoms of LH-bound BChls (B800/B880) are shown in spheres. KγCint and Phe28 sidechains are shown as stick forms. (C, D) The LHα-Phe28 sidechain orientations in dRC-LH (C) and native RC-LH (nRC-LH) (D). LHαβs are shown in cartoon form. The Phe28 sidechain and KγC are shown as sticks.
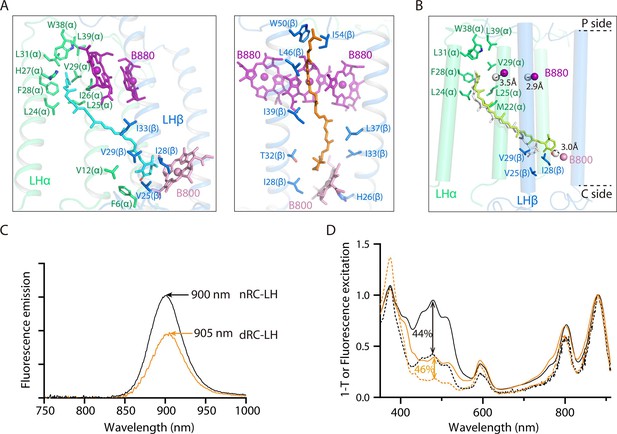
Binding conformation of the interior and exterior keto-γ-carotenes (KγCint and KγCext, respectively) and measurement of the Car-to-BChl energy transfer efficiency in native reaction center-light harvesting (nRC-LH) and carotenoid-depleted RC-LH (dRC-LH) complexes.
(A) Coordination of representative KγCint (cyan) and KγCext (orange) molecules in the nRC-LH complex. Shown as stick forms are the amino acid residues from LHα (lime green) and LHβ (marine) surrounding the 4-oxo-β-ionone ring; the ψ-end group of the KγC; and the BChls B880 (purple) and B800 (pink) in the nearby LHαβ. (B) Coordination of the KγCint molecules, which are shown in limon and white in dRC-LH and nRC-LH, respectively. Amino acid residues from the nearby LHα (lime green) and LHβ (marine) and the B800 molecule that covers the KγCint molecule are shown as stick forms. The distance deviations of the central Mg atoms in B880 (purple) and B800 (pink) in the two structures are labeled and indicated with dashed lines. The periplasmic (P) and cytoplasmic (C) sides are labeled. (C) Spectral analysis of the RC-LH complex. Fluorescence emissions are shown for nRC-LH (black) and dRC-LH (orange) complexes isolated from R. castenholzii after excitation at 470 nm. (D) Fluorescence excitation and absorption (1−T) spectra are shown as dotted and solid lines, respectively, for nRC-LH (black) and dRC-LH (orange). The Car-to-BChl energy transfer efficiency (vertical dashed line) was calculated by normalizing the fluorescence excitation and absorption spectra at 880 nm to 1.0.
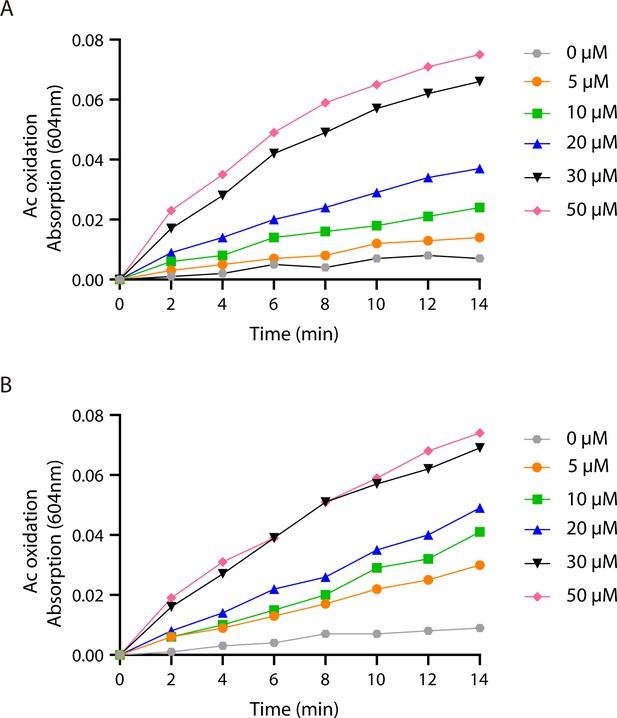
The auracyanin (Ac) oxidation activities of the native reaction center-light harvesting (nRC-LH) and carotenoid-depleted RC-LH (dRC-LH) complexes from R. castenholzii.
(A, B) Ac oxidation assays of the nRC-LH and dRC-LH complexes. The absorbance of Ac at 604 nm in presence of the nRC-LH (A) or dRC-LH (B) complex was recorded every 2 min, at 323 K and under illumination of 180 μmol m–2 s–1. Reduced Ac was used as electron donor, and varied concentrations of menaquinone-4 was used as electron acceptor.
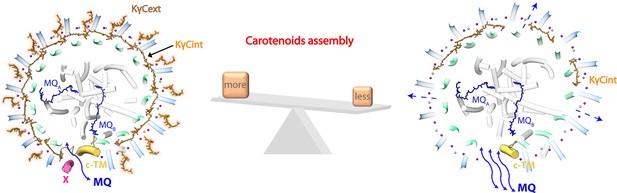
Schematic diagram of the carotenoid (Car) assembly-related structural dynamics of R. castenholzii reaction center-light harvesting (RC-LH) complex.
In native RC-LH, incorporation of the external keto-γ-carotenes (KγCext) and LH-bound B800s blocked the LHαβ interface. Alternatively, the subunit X disrupts the ring and forms a potential quinone channel with the cytochrome c transmembrane (c-TM), facilitating controlled quinone/quinol binding and shuttling. In Car-depleted RC-LH (dRC-LH), less Car assembly exposed the LHαβ interface, absence of the subunit X and cytoplasmic region of c-TM concomitantly broadened the LH opening, which together accelerated the quinone/quinol exchange.
Videos
Top view of the conformational changes between native reaction center-light harvesting (RC-LH) (nRC-LH) and carotenoid-depleted RC-LH (dRC-LH) complexes from R. castenholzii.
Conformational changes of the light harvesting (LH) ring opening between native reaction center-LH (RC-LH) (nRC-LH) and carotenoid-depleted RC-LH (dRC-LH) complexes from R. castenholzii.
The color scheme is same as Figure 4C.
Tables
Peptide mass fingerprinting (PMF) analysis of the R. castenholzii in reaction center-light harvesting (RC-LH) that are separated by blue-native PAGE.
| Subunit | Accession | Description | Score | Coverage |
|---|---|---|---|---|
| cyt c | BAC76415.1 | Cytochrome subunit of photosynthetic reaction center (R. castenholzii) | 131.79 | 40.94% |
| L and M | BAC76414.1 | Precursor for L and M subunits of photosynthetic reaction center (R. castenholzii) | 195.79 | 28.39% |
| LHα subunit | BAC76413.1 | Alpha subunit of light harvesting 1 (R. castenholzii) | 97.78 | 100% |
| LHβ subunit | BAC76412.1 | Beta subunit of light harvesting 1 (R. castenholzii) | 49.11 | 100% |
| Z subunit | WP_041331144.1 | Hypothetical protein (R. castenholzii) | 19.05% |
Cryo-electron microscopy (cryo-EM) data collection, refinement, and validation statistics.
| Native RC-LH at 180 μmol m–2 s–1(nRC-LH)(EMD-34838)(PDB 8HJU) | Carotenoid-depleted RC-LH at 180 μmol m–2 s–1(dRC-LH)(EMD-34839)(PDB 8HJV) | |
|---|---|---|
| Data collection and processing | ||
| Magnification | 64,000 | 81,000 |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e–/Å2) | 50 | 49.65 |
| Defocus range (μm) | –1.0 to –2.3 | –1.1 to –1.7 |
| Pixel size (Å) | 1.08 | 0.893 |
| Symmetry imposed | C1 | C1 |
| Initial particle images (no.) | 1,625,156 | 1,081,719 |
| Final particle images (no.) | 372,029 | 84,352 |
| Map resolution (Å) FSC threshold | 2.8 0.143 | 3.1 0.143 |
| Refinement | ||
| Initial model used (PDB code) | 5YQ7 | Native RC-LH |
| Model resolution (Å) FSC threshold | 2.8 0.5 | 3.1 0.5 |
| Map sharpening B factor (Å2) | 98 | 120 |
| Model composition Non-hydrogen atoms Protein residues Ligands | 23,917 2330 97 | 22,193 2265 67 |
| B factors (Å2) Protein Ligand | 35.81 32.21 | 52.46 55.37 |
| R.m.s. deviations Bond lengths (Å) Bond angles (°) | 0.011 1.509 | 0.010 1.205 |
| Validation MolProbity score Clashscore Poor rotamers (%) | 2.00 16.11 0.71 | 2.17 18.87 0.84 |
| Ramachandran plot Favored (%) Allowed (%) Disallowed (%) | 95.83 4.17 0.00 | 94.16 5.75 0.09 |
| Native RC-LH at 2 μmol m–2 s–1(EMD-35988)(PDB 8J5O) | Native RC-LH at 32 μmol m–2 s–1(EMD-35989)(PDB 8J5P) | |
| Data collection and processing | ||
| Magnification | 81,000 | 81,000 |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e–/Å2) | 50 | 50 |
| Defocus range (μm) | –1.2 to –2.0 | –1.2 to –2.0 |
| Pixel size (Å) | 0.893 | 0.893 |
| Symmetry imposed | C1 | C1 |
| Initial particle images (no.) | 779,594 | 686,082 |
| Final particle images (no.) | 322,595 | 272,617 |
| Map resolution (Å) FSC threshold | 2.9 0.143 | 3.1 0.143 |
| Refinement | ||
| Initial model used (PDB code) | 8HJU | 8HJU |
| Model resolution (Å) FSC threshold | 2.8 0.5 | 3.1 0.5 |
| Map sharpening B factor (Å2) | 98 | 120 |
| Model composition Non-hydrogen atoms Protein residues Ligands | 23,691 2318 93 | 23,737 2331 92 |
| B factors (Å2) Protein Ligand | 27.54 24.73 | 39.01 37.37 |
| R.m.s. deviations Bond lengths (Å) Bond angles (°) | 0.018 1.915 | 0.017 1.850 |
| Validation MolProbity score Clashscore Poor rotamers (%) | 1.97 18.79 0.00 | 1.98 17.69 0.30 |
| Ramachandran plot Favored (%) Allowed (%) Disallowed (%) | 96.79 3.21 0.00 | 96.50 3.46 0.04 |
Edge-to-edge distance (Å) of interior keto-γ-carotenes (KγCint) to light harvesting (LH)-bound B800/B880s in the native reaction center-LH (nRC-LH) and carotenoid-depleted RC-LH (dRC-LH) complexes from R. castenholzii.
| nRC-LH | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B800 | – | 3.6 | 4.2 | 3.7 | 3.6 | 4.1 | 3.5 | 3.4 | 3.7 | 3.9 | 4.0 | 3.8 | 4.2 | 3.7 | – |
| B880 | – | 4.3 | 3.8 | 3.8 | 3.9 | 3.9 | 4.2 | 4.0 | 3.9 | 4.3 | 4.0 | 4.1 | 3.6 | 3.6 | 3.7 |
| dRC-LH | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
| B800 | – | – | – | – | 4.2 | – | 3.4 | – | 4.1 | 3.3 | 3.9 | – | – | – | – |
| B880 | – | – | – | – | 4.0 | – | 4.2 | – | 4.4 | 3.8 | 4.0 | – | – | – | – |
Edge-to-edge distance (Å) of exterior keto-γ-carotenes (KγCext) to light harvesting (LH)-bound B800/B880s in the native reaction center-LH (nRC-LH) complex from R. castenholzii.
| nRC-LH | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B800 | – | 3.7 | 3.4 | 3.9 | 3.8 | 3.8 | 3.7 | 3.3 | 3.5 | 3.2 | 3.3 | 3.4 | 3.5 | 3.5 | 4.2 |
| B880 | – | 4.8 | 5.0 | 5.0 | 5.0 | 4.1 | 4.2 | 4.3 | 4.8 | 4.8 | 5.1 | 4.7 | 4.2 | 4.8 | 4.6 |
Distances (Å) between the LHαβ transmembrane (TM) helices of the native reaction center-light harvesting (nRC-LH) and carotenoid-depleted RC-LH (dRC-LH) complexes from R. castenholzii.
The distance is measured between LHαβn and LHαβ (n+1).
| nRC-LH | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15* | 16† |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LHα | 14.8 | 14.7 | 14.6 | 14.3 | 14.4 | 14.7 | 15.4 | 14.9 | 15.4 | 14.6 | 14.7 | 15.0 | 14.5 | 14.4 | 19.4 | 9.7 |
| LHβ | 20.0 | 19.8 | 20.0 | 20.4 | 20.2 | 20.0 | 19.8 | 19.9 | 19.7 | 20.1 | 20.0 | 19.9 | 20.1 | 20.1 | 11.2 | 28.1 |
| dRC-LH | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15* | 16† |
| LHα | 15.4 | 15.4 | 15.3 | 15.0 | 14.9 | 15.3 | 16.2 | 15.5 | 16.1 | 15.2 | 15.4 | 15.6 | 15.3 | 14.9 | 19.6 | 10.2 |
| LHβ | 21.0 | 20.7 | 20.9 | 21.2 | 20.9 | 21.0 | 20.5 | 20.9 | 20.6 | 20.8 | 21.0 | 20.7 | 20.9 | 20.9 | – | – |
-
*
The distances from LHα15 Met-22 to c-TM Val-33, and LHβ15 Ile-39 to subunit X.
-
†
The distances from LHα1 Val-29 to c-TM Tyr-44, and LHβ1 Leu-37 to subunit X.
The distances (Å) between adjacent light harvesting (LH)-bound B880s of the native reaction center-LH (nRC-LH) and carotenoid-depleted RC-LH (dRC-LH) complexes from R. castenholzii.
| nRC-LH | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Edge-to-edge* | 3.3 | 3.1 | 3.4 | 3.4 | 3.6 | 3.8 | 3.7 | 3.8 | 3.7 | 3.6 | 3.5 | 3.6 | 3.5 | 3.6 | 3.8 |
| Edge-to-edge† | 3.5 | 3.4 | 3.6 | 3.6 | 3.4 | 4.0 | 3.6 | 3.8 | 3.5 | 3.5 | 3.3 | 3.7 | 3.6 | 3.6 | 19.1 |
| Mg-to-Mg* | 10.2 | 9.9 | 9.8 | 9.9 | 9.7 | 9.8 | 9.9 | 10.0 | 9.9 | 9.9 | 9.8 | 9.6 | 9.7 | 9.3 | 9.8 |
| Mg-to-Mg† | 7.6 | 7.5 | 7.7 | 7.7 | 7.6 | 7.6 | 7.6 | 7.5 | 7.6 | 7.8 | 7.7 | 7.9 | 7.9 | 7.9 | 26.2 |
| dRC-LH | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
| Edge-to-edge* | 3.5 | 3.5 | 3.5 | 3.7 | 3.5 | 3.7 | 3.7 | 3.8 | 3.9 | 3.8 | 3.6 | 3.5 | 3.6 | 3.3 | 3.7 |
| Edge-to-edge† | 4.1 | 3.8 | 4.0 | 3.8 | 3.9 | 3.8 | 4.1 | 4.0 | 4.1 | 3.9 | 3.9 | 3.9 | 3.9 | 3.8 | 20.2 |
| Mg-to-Mg* | 9.7 | 9.7 | 9.5 | 9.6 | 9.6 | 9.6 | 9.4 | 9.7 | 9.7 | 9.6 | 9.5 | 9.4 | 9.5 | 9.3 | 9.2 |
| Mg-to-Mg† | 9.0 | 8.7 | 8.8 | 8.8 | 8.4 | 8.9 | 8.8 | 8.6 | 8.8 | 8.8 | 8.9 | 9.0 | 9.0 | 8.7 | 27.2 |
-
*
Distance between the B880 bound by the same transmembrane pairs of LH.
-
†
Distance between the B880 bound by adjacent transmembrane pairs of LH.
The distances (Å) between light harvesting (LH)-bound B800s of the native reaction center-LH (nRC-LH) and carotenoid-depleted RC-LH (dRC-LH) complexes from R. castenholzii.
| nRC-LH | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Edge-to-edge | 15.4 | 15.5 | 15.5 | 15.4 | 15.4 | 15.6 | 15.6 | 15.4 | 15.6 | 15.5 | 15.5 | 15.6 | 15.6 | 15.2 | 33.4 |
| Mg-to-Mg | 18.4 | 18.3 | 18.4 | 18.5 | 18.5 | 18.5 | 18.5 | 18.3 | 18.4 | 18.4 | 18.5 | 18.5 | 18.4 | 18.3 | 38.5 |
| dRC-LH | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
| Edge-to-edge | 15.8 | 15.9 | 16.0 | 15.7 | 15.8 | 16.2 | 15.9 | 15.8 | 16.1 | 15.8 | 16.0 | 16.1 | 15.8 | 15.8 | 33.2 |
| Mg-to-Mg | 19.1 | 19.1 | 19.3 | 19.1 | 19.2 | 19.4 | 19.1 | 19.2 | 19.2 | 19.1 | 19.3 | 19.3 | 19.1 | 19.2 | 38.3 |
The Mg-to-Mg distances between light harvesting (LH)-bound B880s and the nearest special pair of bacteriochlorophyll (BChls) in the reaction center (RC) of R. castenholzii native RC-LH (nRC-LH) and carotenoid-depleted RC-LH (dRC-LH) complexes.
| nRC-LH | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mg-to-Mg* | 40.3 | 42.4 | 44.7 | 47.2 | 46.8 | 43.9 | 40.2 | 37.3 | 36.5 | 37.7 | 40.6 | 43.7 | 44.5 | 44.8 | 44.2 |
| Mg-to-Mg† | 41.5 | 43.8 | 46.3 | 48.1 | 45.6 | 42.0 | 38.5 | 36.8 | 36.8 | 39.6 | 42.6 | 44.5 | 44.9 | 44.8 | 43.6 |
| dRC-LH | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
| Mg-to-Mg* | 42.0 | 44.8 | 47.2 | 49.9 | 49.0 | 45.8 | 42.0 | 39.1 | 38.3 | 39.7 | 43.0 | 46.0 | 46.2 | 46.4 | 45.4 |
| Mg-to-Mg† | 43.1 | 45.8 | 48.5 | 50.0 | 47.2 | 43.8 | 40.2 | 38.4 | 38.6 | 41.3 | 44.5 | 46.0 | 46.3 | 46.1 | 44.6 |
-
*
The distances from Mg2+ of the first LH-bound B880 to the nearest special pair of BChls.
-
†
The distances from Mg2+ of the second LH-bound B880 to the nearest special pair of BChls.
Apparent Michaelis constants of menaquinone-4 as electron acceptor in the auracyanin (Ac) oxidation assay, in presence of the R. castenholzii native reaction center-light harvesting (nRC-LH) or carotenoid-depleted RC-LH (dRC-LH) complex.
| nRC-LH | dRC-LH | |
|---|---|---|
| Km (μM) | 32.24±6.84 | 10.43±1.23 |
| kcat (min–1) | 82.61±9.09 | 63.83±2.49 |
| kcat/Km | 2.56±0.49 | 6.12±0.62 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Roseiflexus castenholzii) | DSM 13941/HLO8 | Hanada et al., 2002 | / | / |
| Chemical compound, drug | N-Dodecyl-β-D-maltoside (β-DDM) | Anatrace | D310 | / |
| Chemical compound, drug | Bacteriochlorophyll a | Sigma-Aldrich | B5906 | / |
| Chemical compound, drug | γ-Carotene | Sigma-Aldrich | 54765 | / |
| Chemical compound, drug | Menaquinone-4 | Sigma-Aldrich | PHR2271 | / |
| Software, algorithm | RELION 3.1 | Zivanov et al., 2018 | / | / |





