Rapid cloning-free mutagenesis of new SARS-CoV-2 variants using a novel reverse genetics platform
Figures
Rescue and characterization of recombinant SARS-CoV-2.
(A) Schematic representation of the SARS-CoV-2 genome and the infectious subgenomic amplicons (ISA)-based method for virus recovery. Eight respectively four overlapping fragments covering the whole SARS-CoV-2 genome were PCR amplified. The heterologous CMV promoter was cloned upstream of the 5′ untranslated region (UTR) and a poly(A) tail, HDV ribozyme, and SV40 termination signal downstream of the 3′ UTR. (B) Infectious virus reconstituted from four fragments (rCoV2-4fr) assessed by cytopathic effect (CPE, top) on susceptible Vero E6 cells by supernatant transfer. Plaque size was compared by standard plaque assay 2 d after inoculation on Vero E6 cells (bottom). (C) Workflow for the rescue of recombinant SARS-CoV-2. Four fragments were PCR amplified, mixed in equimolar ratios, transfected into HEK293T cells, and infectious virus was recovered 7 d post-transfection. Commercially available SARS-CoV-2 rapid antigen tests can be used for a quick non-quantitative analysis. (D) Detection of intracellular SARS-CoV-2 nucleocapsid protein (N, green) and nuclei (Hoechst, blue) in Vero E6 cells infected with parental wild-type or recombinant virus by immunocytochemistry. (E) Growth kinetics of recombinant virus and its parental wild-type virus. Vero E6 cells were infected in triplicates at a multiplicity of infection (MOI) of 0.01, supernatant was collected 12, 24, 48, and 72 hr post-infection and analyzed by plaque assay. Cell layers were washed 2 hr post-infection. Data represents mean ± SEM, analyzed with multiple t-tests and Benjamini, Krieger, and Yekutieli correction (N = 3 individual biological replicates, n = 3 technical replicates). (F) Cryo-transmission electron microscope pictures of parental wild-type virus and recombinant virus in glutaraldehyde-fixed samples. Scale bar is 100 µm (top) and 2 mm (bottom) in (B), 20 µm in (D), and 100 nm in (F).
-
Figure 1—source data 1
Uncropped agarose gel image of Figure 1C.
- https://cdn.elifesciences.org/articles/89035/elife-89035-fig1-data1-v1.zip
Clonal virus populations and reconstitution efficiency.
(A) Schematic representation of the workflow to generate clonal virus populations. Right after transfection, cells were diluted to less than 0.5 virus-producing cells/well in 96-well plates. Then, 7 d post-transfection, the supernatant was transferred onto Vero E6-TMPRSS2 cells. Plates were observed until a cytopathic effect (CPE) became apparent. (B) Clonal virus populations arising from a single virus-producing cell were identified after supernatant transfer onto Vero E6-TMPRSS2 cells. CPE of infectious virus was assessed by microscopy and virus was collected for further analysis. Thereafter, plates were fixed and stained for the fast enumeration of positive wells (in light blue).
Sequence integrity using CLEVER.
(A) Schematic representation of the sequence alignment of recombinant viruses sequenced by next-generation sequencing (NGS), mutations with a relative abundance of >10% are indicated with a star. A total of eight bulk (gray) and five clonal (green) populations were analyzed. (B) Details on substitution and position in the genome. For a more detailed analysis see Supplementary file 2.
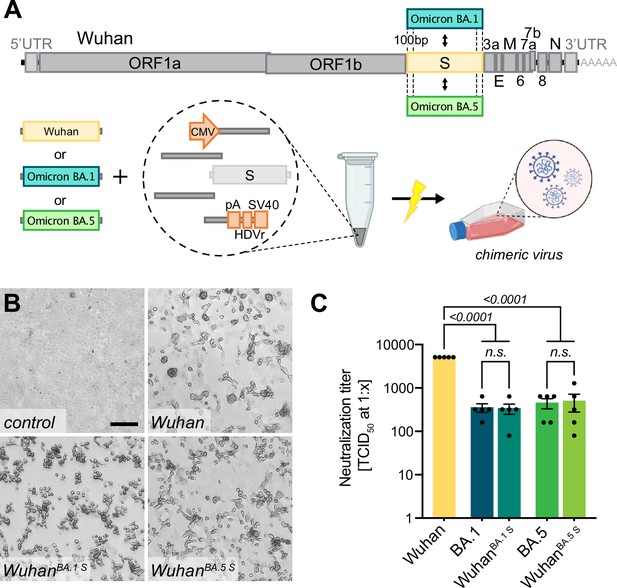
Creating chimeric virus by fragment exchange.
(A) Schematic representation of the exchange of individual fragments. Shown is the replacement of the Wuhan S sequence by the sequence encoding for the Omicron BA.1 or BA.5 S gene. The genetic background (outside of S) is kept in the original Wuhan sequence. All fragments needed to reconstitute the virus were transfected and chimeric virus was rescued. (B) Successful rescue of infectious chimeric virus was assessed by cytopathic effect (CPE) formation on Vero E6 cells. Scale bar represents 100 μm. (C) Titers of neutralizing antibodies against different SARS-CoV-2 S gene variants were validated in sera from vaccinated individuals. Sera were incubated with parental wild-type virus (Wuhan), Omicron BA.1 or BA.5 clinical isolates (BA.1, BA.5), as well as chimeric viruses having the Wuhan background combined with either the Omicron BA.1 S gene (WuhanBA.1 S) or Omicron BA.5 S gene (WuhanBA.5 S). Neutralizing titers were determined with a neutralization assay and TCID50 read-out. Data represents mean ± SEM, analyzed with one-way ANOVA followed by Bonferroni’s test (N = 5).
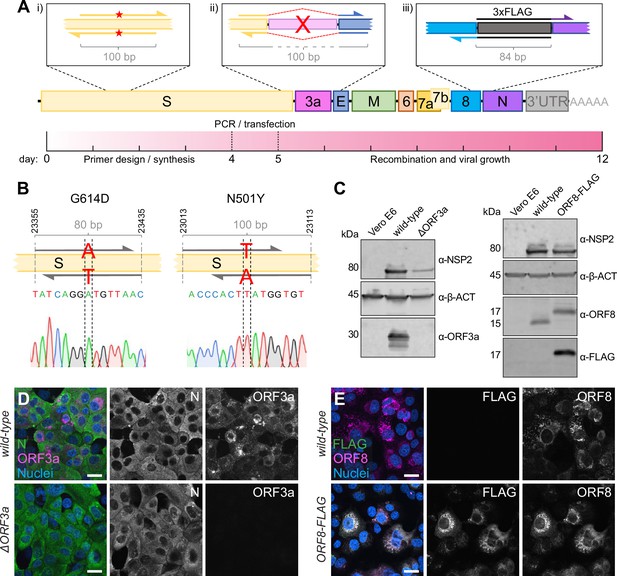
Direct mutagenesis using the CLEVER primer design.
(A) Schematic representation of the CLEVER primer design for direct mutagenesis. Shown is the (i) introduction of small nucleotide changes, (ii) the deletion of larger sequences, here shown for ORF3a, and (iii) the insertion of nucleotide stretches such as 3xFLAG as well as a timeline showing the expected work-flow needed from in silico design to virus rescue. (B) Details on the G614D and N501Y substitution within the S gene. Shown is position, primer design, and the integration into the viral genome confirmed by Sanger sequencing. (C) Validation of mutations by immunoblot. Shown is the validation of the ∆ORF3a (left) and ORF8-3xFLAG virus (right). Vero E6 cells were assessed with α-β-actin (α-β-ACT) and viral infection was detected using α-NSP2. ORF3a expression or ORF8/FLAG expression, respectively, was compared to wild-type infected cells and uninfected controls. (D) Validation of ∆ORF3a by immunocytochemistry. ∆ORF3a virus created by direct mutagenesis was compared to its parental wild-type virus. Shown is the expression of ORF3a (magenta) in both viruses. Nucleocapsid (N, green) expression was used to assess viral infection, nuclei were stained with Hoechst (blue). (E) Validation of ORF8-3xFLAG by immunocytochemistry. C-terminal tagging of ORF8 with 3xFLAG was achieved with direct mutagenesis. Shown is the expression of ORF8 (magenta) and FLAG (green) in both viruses. Nuclei were stained with Hoechst (blue). Scale bar is 20 µm in (D, E).
-
Figure 4—source data 1
Uncropped western blot images of Figure 4C.
- https://cdn.elifesciences.org/articles/89035/elife-89035-fig4-data1-v1.zip
CLEVER primer design for direct mutagenesis.
Shown are the different approaches of primer design to guarantee a 100 bp overlap between the neighboring fragments. Primers can either be separated in distance to ensure homology regions or additional nucleotides must be added to the 5′ end of the primer. Small nucleotide changes, deletions, or insertions can directly be introduced by adjusting the primer annealing sites and the homology regions, as depicted.
Direct rescue and mutagenesis of clinical isolates.
(A) Schematic representation of the circular assembly within the eukaryotic cell with the linker fragment. The heterologous elements needed downstream of the 3′ untranslated region (UTR) (pA, HDVr, SV40) and upstream of the 5′ UTR (CMV) are assembled in one fragment, separated by a spacer sequence and flanked by homologous regions needed for intracellular recombination. (B) Representative agarose gel pictures from PCR fragments amplified by one-step RT-PCR from viral RNA and the linker fragment (L). Recombinant virus was rescued from five (top) or eight fragments (bottom), plus the linker fragment. Asterisks mark fragments harboring the introduced changes within their homology region. (C, D) Validation of (C) Omicron BA.5 ∆ORF3a and (D) XBB.1.5 ∆ORF3a by immunocytochemistry. The expression of ORF3a (magenta) in Omicron BA.5 and XBB.1.5 clinical isolates and recombinant ∆ORF3a viruses is shown. Nucleocapsid (N, green) expression was used to assess viral infection, nuclei were stained with Hoechst (blue). Scale bar represents 20 μm.
-
Figure 5—source data 1
Uncropped agarose gel images of Figure 5B.
- https://cdn.elifesciences.org/articles/89035/elife-89035-fig5-data1-v1.zip

Cloning-free rescue of chimeric virus.
(A) All fragments were directly amplified by one-step RT-PCR from viral RNA of Wuhan, Omicron BA.1, and Omicron BA.5. Eight fragments were amplified to prove high flexibility in exchanging fragments. (B) In addition to the chimeric viruses described in Figure 3, the genetic background (region outside of S) was replaced by the sequence of Omicron BA.1 or Omicron BA.5 and combined with a heterologous S sequence of Wuhan, Omicron BA.1 or BA.5, respectively. (C) Infectious chimeric virus was rescued and assessed via cytopathic effect (CPE) formation on Vero E6 cells. Scale bar represents 100 μm. (D) Scheme for the rapid distinguishment between Wuhan, Omicron BA.1, BA.5, and XBB.1.5 variants. Indicated regions (S or M) were Sanger sequenced to discriminate variants or confirm chimeric viruses. Amino acids are highlighted in yellow for the clear identification of the S gene variant and/or the background (within M).
-
Figure 5—figure supplement 1—source data 1
Uncropped agarose gel images of Figure 5—figure supplement 1A.
- https://cdn.elifesciences.org/articles/89035/elife-89035-fig5-figsupp1-data1-v1.zip
Cloning-free rescue of Chikungunya virus (CHIKV) and Dengue virus (DENV).
(A) Schematic representation of the CHIKV genome and the design for the cloning-free rescue. The genome was divided into three overlapping fragments, and a silent SNP was introduced by PCR (red asterisk). (B, C) Successful recombination of the four PCR products (B) within the eukaryotic cell leads to a circular product and virus production (C), cytopathic effect (CPE) on BHK-21 cells for rCHIKV and negative control. (D) Schematic representation of the DENV genome and the design for the cloning-free rescue. The genome has been divided into two overlapping fragments and a silent SNP has been introduced by PCR (red asterisk). The protocol has been tested on two different clinical isolates (DENV1 and DENV3), whereas two different recombination sites were tested for DENV1 (rDENV1-A and rDENV1-B, only one schematically represented). (E, F) Overlapping PCR products (E) were transfected into BHK-21 cells, and CPE (F) was assessed on VeroE6-TMPRSS2 cells (rDENV1-A, rDENV1-B, rDENV3, and negative control). Scale bar is 100 µm in (C) and (F).
-
Figure 5—figure supplement 2—source data 1
Uncropped agarose gel images of Figure 5—figure supplement 2B and E.
- https://cdn.elifesciences.org/articles/89035/elife-89035-fig5-figsupp2-data1-v1.zip
Overview of reverse genetics methods for SARS-CoV-2.
The most commonly used reverse genetics systems for the rescue of recombinant SARS-CoV-2 are listed and the prominent intermediate steps are depicted. Note that this is a schematic summary and additional steps (such as purification, linearization before transcription, etc.) or small aberrations of the protocol (e.g., different starting material) can apply. The first groups reporting the successful adaptation to SARS-CoV-2 are mentioned. For the CLEVER method, additionally the direct mutagenesis within the initial RT-PCR step is depicted (mutated sites marked with red asterisks). Repeated icons are only labeled once. DNA fragments are represented as blue lines. T7, T7 RNA polymerase; BAC, bacterial artificial chromosome; YAC, yeast artificial chromosome; CPER, circular polymerase extension reaction; ISA, infectious subgenomic amplicons; CLEVER, CLoning-free and Exchangeable system for Virus Engineering and Rescue.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | DH5α E. coli | NEB | Cat# C2987 | Competent cells |
| Strain, strain background (E. coli) | One Shot Stbl3 Chemically Competent E. coli | Invitrogen | Cat# C737303 | Competent cells |
| Strain, strain background (SARS-CoV-2) | SARS-CoV-2 Wuhan (SARS-CoV_FR-3) | Other | GenBank OR018857 | Provided by G. Kochs, Freiburg, Germany |
| Strain, strain background (SARS-CoV-2) | SARS-CoV-2 Wuhan (SARS-CoV_Muc) | Other | GenBank OR018856 | Provided by G. Kochs, Freiburg, Germany |
| Strain, strain background (SARS-CoV-2) | SARS-CoV-2 Omicron variant BA.1 | Human donors | GenBank OR018858 | Approved by Ethikkommission Nordwest- und Zentralschweiz #2022-00303 |
| Strain, strain background (SARS-CoV-2) | SARS-CoV-2 Omicron variant BA.5 | Human donors | GenBank OR018859 | Approved by Ethikkommission Nordwest- und Zentralschweiz #2022-00303 |
| Strain, strain background (SARS-CoV-2) | SARS-CoV-2 Omicron variant XBB.1.5 | Human donors | GenBank OX393614 | Approved by Ethikkommission Nordwest- und Zentralschweiz #2022-00303 |
| Strain, strain background (CHIKV) | Chikungunya virus (CHIKV) | Other | – | Provided by K. Leuzinger, Basel, Switzerland |
| Strain, strain background (DENV1) | Dengue virus serotype 1 (DENV1) | Other | – | Provided by K. Leuzinger, Basel, Switzerland |
| Strain, strain background (DENV3) | Dengue virus serotype 3 (DENV3) | Other | – | Provided by K. Leuzinger, Basel, Switzerland |
| Cell line (Chlorocebus sabaeus) | African green monkey kidney cells (Vero E6) | Other | – | Provided by V. Thiel, Bern, Switzerland |
| Cell line (Homo sapiens) | Adenocarcinomic human alveolar basal epithelial cells (A549) | NIBSC | Cat# 101006 | – |
| Cell line (Mesocricetus auratus) | Baby hamster kidney cells (BHK-21) | Thermo Scientific | ATCC CCL-10 | – |
| Cell line (H. sapiens) | Human embryonic kidney cells (HEK293T) | Other | – | Provided by D. Pinschewer, Basel, Switzerland |
| Cell line (H. sapiens) | Human embryonic kidney cells (HEK293) | Batavia | Cat# 110-025; Lot# 19D006 | – |
| Cell line (Cricetulus griseus) | Chinese hamster ovary cells (CHO-K1) | Thermo Scientific | ATCC CCL-61 | – |
| Biological sample (H. sapiens) | Serum samples | Human donors | – | Approved by Ethikkommission Nordwest- und Zentralschweiz #2022-00303 |
| Antibody | Anti-β-actin (mouse monoclonal) | Cell Signaling Technology | Cat# 3700; RRID:AB_2242334; Lot# 20 | WB (1:1000) |
| Antibody | Anti-FLAG (rabbit polyclonal) | Cell Signaling Technology | Cat# 14793; RRID:AB_2572291; Lot# 5 | WB (1:1000) |
| Antibody | Anti-FLAG (rat monoclonal) | BioLegend | Cat# 637301; RRID:AB_1134266; Lot# B318853 | ICC (1:1000) |
| Antibody | Anti-SARS-CoV-2 NSP2 (rabbit polyclonal) | GeneTex | Cat# GTX135717; RRID:AB_2909866; Lot# B318853 | WB (1:5000) |
| Antibody | Anti-SARS-CoV-2 Nucleocapsid protein (mouse monoclonal, 4F3C4) | Sven Reiche (doi: 10.1016/j.virusres.2006.07.005) | – | ICC (1:500) |
| Antibody | Anti-SARS-CoV-2 ORF3a (sheep polyclonal) | MRC PPU reagents (doi:10.1371/journal.pbio.3001091) | – | WB (1:1000) ICC (1:500) |
| Antibody | Anti-SARS-CoV-2 ORF8 (rabbit polyclonal) | Novus Biologicals | Cat# NBP3-07972; Lot# 25966-2102 | WB (1:1000) ICC (1:1000) |
| Recombinant DNA reagent | pLEX307-TMPRSS2-blast (plasmid) | Addgene | Cat# 158458 | – |
| Recombinant DNA reagent | pUC19_CoV-2_Linker | This study | Addgene plasmid #211731 | Plasmid expressing 5’ and 3’ regions for direct rescue |
| Recombinant DNA reagent | pUC19_CoV-2_frA (plasmid) | This study | – | Plasmid encoding SARS-CoV-2 ‘fragment A,’ see Supplementary file 7 |
| Recombinant DNA reagent | pUC19_CoV-2_frB (plasmid) | This study | – | Plasmid encoding SARS-CoV-2 ‘fragment B,’ see Supplementary file 7 |
| Recombinant DNA reagent | pUC19_CoV-2_frC (plasmid) | This study | – | Plasmid encoding SARS-CoV-2 ‘fragment C,’ see Supplementary file 7 |
| Recombinant DNA reagent | pUC19_CoV-2_frD (plasmid) | This study | – | Plasmid encoding SARS-CoV-2 ‘fragment D’ Wuhan isolate, see Supplementary file 7 |
| Recombinant DNA reagent | pUC19_CoV-2_frD_S Omicron BA.1 (plasmid) | This study | – | Plasmid encoding SARS-CoV-2 ‘fragment D’ Omicron BA.1 isolate, see Supplementary file 7 |
| Recombinant DNA reagent | pUC19_CoV-2_frD_S Omicron BA.5 (plasmid) | This study | – | Plasmid encoding SARS-CoV-2 ‘fragment D’ Omicron BA.5 isolate, see Supplementary file 7 |
| Recombinant DNA reagent | pUC57_CoV-2_frA (plasmid) | GenScript | – | High-quality DNA ordered from GenScript |
| Recombinant DNA reagent | pUC57_CoV-2_frB (plasmid) | GenScript | – | High-quality DNA ordered from GenScript |
| Recombinant DNA reagent | pUC57_CoV-2_frC (plasmid) | GenScript | – | High-quality DNA ordered from GenScript |
| Recombinant DNA reagent | SARS-CoV-2 Omicron Strain S gene original_pcDNA3.1(+) | GenScript | Cat# MC_0101273 | – |
| Sequence-based reagent | Oligonucleotides (primers) | This study | – | For all primers, see Supplementary file 5 |
| Commercial assay or kit | Maxwell RSC Viral Total Nucleic Acid Purification Kit | Promega | Cat# AS1330 | – |
| Commercial assay or kit | Maxwell RSC miRNA from Plasma or Serum | Promega | Cat# AS1680 | – |
| Commercial assay or kit | SuperScript IV One-Step RT-PCR System | Invitrogen | Cat# 12594100 | – |
| Commercial assay or kit | SF Cell Line 4D-Nucleofector-X Kit | Lonza | Cat# V4XC-2012 | – |
| Commercial assay or kit | EasySeq SARS-CoV-2 WGS Library Prep Kit | NimaGen | SKU: RC-COV096 | – |
| Chemical compound, drug | Blasticidin | Sigma-Aldrich | Cat# SBR00022 | – |
| Chemical compound, drug | Hoechst 33342 dye | Sigma-Aldrich | Cat# B2261 | – |
| Chemical compound, drug | RNase A | Thermo Scientific | Cat# EN0531 | – |
| Software, algorithm | ImageJ v2.9.0/1.53t | NHI | https://imagej.nih.gov/ij/download.html; RRID:SCR_003070 | – |
| Software, algorithm | Prism v9 | GraphPad | https://www.graphpad.com/scientific-software/prism/; RRID:SCR_002798 | – |
| Software, algorithm | Omero | Open Microscopy Environment | http://www.openmicroscopy.org/site/products/omero; RRID:SCR_002629 | – |
| Software, algorithm | Adobe Illustrator CC | Adobe | http://www.adobe.com/products/illustrator.html; RRID:SCR_010279 | – |
| Software, algorithm | 7500 Real-Time PCR Software Version 2.0.6 | Applied Biosystems | RRID:SCR_014596 | – |
| Software, algorithm | ImmunoSpot Software Version 7.0.26.0 | ImmunoSpot | RRID:SCR_011082 | – |
| Software, algorithm | Nikon NIS-Elements AR Versions 5.30.07 | Nikon | RRID:SCR_014329 | – |
| Software, algorithm | LI-COR Image Studio Version 2.0 | LI-COR | – | – |
| Other | SARS-CoV-2 Rapid Antigen Test | Roche | Cat# 9901-NCOV-01G | SARS-CoV-2 Antigen Test to check for viral rescue, see Figure 1 |
Additional files
-
Supplementary file 1
Infectivity assessment of recombinant virus on Vero E6 cells.
Vero E6 cells were infected with the indicated recombinant virus and pictures were taken 3 d post-infection. Of note, the virus was not titrated and the development of CPE is not quantitative. Scale bar represents 100 μm.
- https://cdn.elifesciences.org/articles/89035/elife-89035-supp1-v1.pdf
-
Supplementary file 2
Genomic characterization of recombinant SARS-CoV-2 virus based on NGS data.
Mutations with a relative abundance of >10% in the entire virus population are listed. Ambiguities or low coverage are highlighted in italics.
- https://cdn.elifesciences.org/articles/89035/elife-89035-supp2-v1.xlsx
-
Supplementary file 3
Homology regions successfully used for recombinant SARS-CoV-2, CHIKV, and DENV1/DENV3.
Length, GC content, and the hypothetical annealing temperature (according to OligoCalc, salt-adjusted) are listed. Note that homology regions were chosen independently from GC content or annealing temperature and values are only listed for completion.
- https://cdn.elifesciences.org/articles/89035/elife-89035-supp3-v1.xlsx
-
Supplementary file 4
Sanger sequencing data of the region of interest for the generated mutant rSARS-CoV-2, rCHIKV, and rDENV1/rDENV3.
- https://cdn.elifesciences.org/articles/89035/elife-89035-supp4-v1.xlsx
-
Supplementary file 5
Oligonucleotide list.
- https://cdn.elifesciences.org/articles/89035/elife-89035-supp5-v1.xlsx
-
Supplementary file 6
PCR settings for individual fragments.
- https://cdn.elifesciences.org/articles/89035/elife-89035-supp6-v1.xlsx
-
Supplementary file 7
Plasmids.
- https://cdn.elifesciences.org/articles/89035/elife-89035-supp7-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/89035/elife-89035-mdarchecklist1-v1.pdf