Synapsin E-domain is essential for α-synuclein function
Figures

Screening for synapsin isoforms that allow α-syn functionality.
(A) Schematic showing pH-sensitive sensor sypHy and principle of pHluorin experiments to quantitatively evaluate the SV cycle (see main text and methods for more details). (B) Elimination of all synapsins block α-syn functionality at synapses. Left: Schematic showing design of pHluorin experiments. WT or synapsin TKO cultured hippocampal neurons were co-transduced at 5 days in vitro (DIV) with h-α-syn:mCherry (or mCherry as control) and sypHy, and imaged at 14–15 DIV. Right: Stimulation-induced sypHy fluorescence traces (300 action potentials at 20 Hz, delivered at t=0 s – for clarity, symbols only mark every other mean ± SEM ΔF/F0 value in all sypHy traces). Note that while h-α-syn over-expression (orange) attenuated sypHy fluorescence in WT neurons, there was no effect in neurons from mice lacking all synapsins (TKO). All sypHy data quantified in (C). (C) Quantification of peak ΔF/F0 sypHy values (bars: mean ± SEM). A total of 12–19 coverslips were analyzed for each condition, from at least three separate cultures (***p=9e-7, ns p=0.45, student’s t-test). (D) Domain structure of the five main synapsin isoforms. (E) Experimental design to identify the synapsin isoform that reinstated α-syn functionality, Synapsin TKO neurons were co-transduced at 5 DIV with each synapsin isoform, h-α-syn, and sypHy; and imaged at 14–15 DIV. (F) SypHy fluorescence traces (mean ± SEM). Note that h-α-syn(orange) attenuates SV recycling only if the neurons are also co-expressing the ‘a’ isoforms – synapsins Ia, IIa, and IIIa (300 action potentials at 20 Hz, delivered at t=0 sec). Data quantified in G. (G) Quantification of peak ΔF/F0 sypHy values (bars: mean ± SEM). 13–26 coverslips from at least three separate cultures were analyzed for each condition (from left to right: ***p=0.0009, ns p=0.62, ***p=0.00005, ns p=0.99, **p=0.004, student’s t test).
-
Figure 1—source data 1
Tabular data and statistical analyses for graphs presented in panels B, C, F, G.
- https://cdn.elifesciences.org/articles/89687/elife-89687-fig1-data1-v1.xlsx
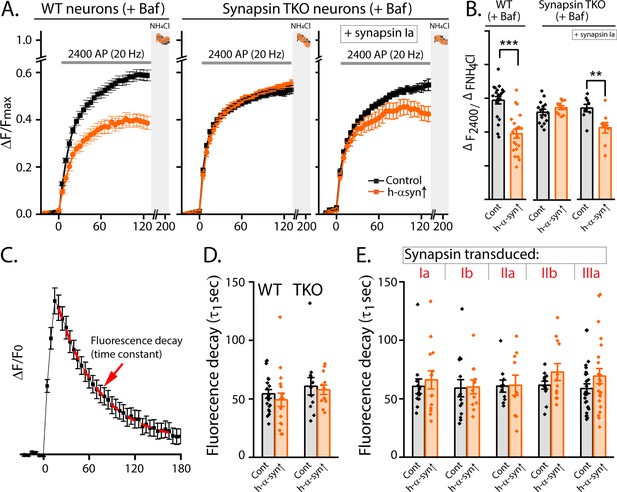
Effects of h-α-syn over-expression are largely due to suppression of exocytosis.
(A) Data from sypHy experiments where reacidification was blocked by bafilomycin (Baf), allowing isolated evaluation of exocytosis only (also see results). Note that h-α-syn over-expression attenuated synaptic exocytosis in WT neurons (left), while there was no effect in synapsin TKO neurons (middle). Reintroduction of tagBFP:synapsin Ia reinstated the h-α-syn-mediated synaptic attenuation (right). All pHluorin data quantified in (B). 9–22 coverslips from at least three independent cultures (***p=1.9e-5 Mann-Whitney test, p=0.22 Student’s t-test, **p=0.008 Student’s t-test). (C) A representative trace showing how the fluorescence decay was quantified to evaluate endocytosis in the sypHy experiments (also see Results). (D–E) Fluorescence decay analyses of h-α-syn over-expression in WT and synapsin TKO neurons (D), as well as in synapsin TKO neurons where each synapsin isoform was reintroduced (E). Note that there were no significant differences in any of these groups. All data in this figure are represented as mean +/- SEM. Ten to 26 coverslips from at least three independent cultures (D: p=0.36; E: p=0.85 both Kruskal Wallis ANOVA).
-
Figure 1—figure supplement 1—source data 1
Tabular data and statistical analyses for graphs presented in panels A, B, D, and E.
- https://cdn.elifesciences.org/articles/89687/elife-89687-fig1-figsupp1-data1-v1.xlsx
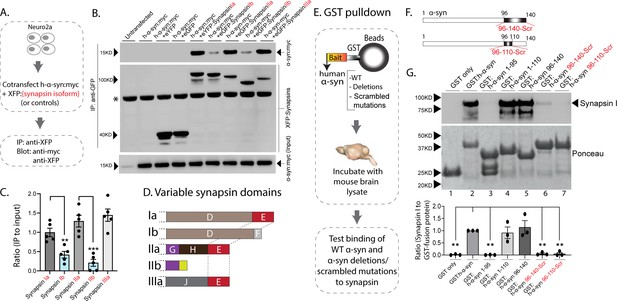
Interaction of synapsin isoforms with h-α-syn.
(A) Workflow for co-immunoprecipitation experiments in neuro2a cells. (B) Western blots from co-immunoprecipitation experiments show that the synapsin isoforms Ia, IIa, and IIIa associate more robustly with h-α-syn (top panel), when compared to synapsins Ib and IIb (a non-specific band is marked with an asterisk). (C) Quantification of blots in (B) n=5, all data presented as mean ± SEM (a vs. b isoform, **p=0.003, ***p=0.0003, Student’s t-test). (D) Schematic showing synapsin isoforms and their variable domains. Note that the E-domain is common between synapsins Ia, IIa, and Iia. (E) Workflow for pulldown of GST-tagged h-α-syn WT/deletions/scrambled mutations after incubation with mouse brain lysates. Equivalent amounts of immobilized GST α-syn variants were used. (F) Schematic showing α-syn regions that were scrambled (amino acids between 96–140 and 96–110). (G) Top: Samples from GST-pulldown were analyzed by NuPAGE and immunoblotted with an antibody against synapsin I (top panel). Bottom: Ponceau staining shows equivalent loading of fusion proteins. Note that full-length h-α-syn bound synapsin I from mouse brains (lane 2), while deletion of the h-α-syn C-terminus (amino acids 96–140, lane 3) eliminated this interaction. Lanes 4–7 show that the sequence within amino acids 96–110 of h-α-syn is critical for binding to synapsin I. All western blots are quantified below (n=3). Data presented as mean ± SEM (**p=0.003, **p=0.002, ns p=0.99, ns p=0.98, **p=0.004, **p=0.004, comparing to full-length h-α-syn, one-way ANOVA with Tukey’s posthoc test).
-
Figure 2—source data 1
Tabular data and statistical analyses for graphs shown in panels C and G.
- https://cdn.elifesciences.org/articles/89687/elife-89687-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Full western blots for segments shown in panel B.
- https://cdn.elifesciences.org/articles/89687/elife-89687-fig2-data2-v1.pdf
-
Figure 2—source data 3
Full western blots for segments shown in panel G.
- https://cdn.elifesciences.org/articles/89687/elife-89687-fig2-data3-v1.pdf

The synapsin E-domain is necessary and sufficient for enabling α-syn functionality.
(A) Schematic showing synapsin Ia scrambled E-domain sequence (synapsin Iascr-E). Numbers depict amino acid positions, letters in the inset depict amino-acids. Note that the WT amino acids are randomized in the scrambled mutant. (B) Design of sypHy experiments co-expressing synapsin Iascr-E and h-α-syn in cultured neurons from synapsin TKO mice. (C) Stimulation-induced sypHy fluorescence traces (300 action potentials at 20 Hz, delivered at t=0 sec). Note that while h-α-syn attenuated sypHy fluorescence in synapsin TKO neurons expressing synapsin Ia, h-α-syn had no effect in neurons expressing synapsin Iascr-E. Insets: Quantification of peak ΔF/F0 sypHy values (bars: mean ± SEM). Ten to 16 coverslips from at least three separate cultures were analyzed for each condition (***p=0.0007, ns p=0.67, one-way ANOVA with Tukey’s posthoc analysis). (D) Top: Schematic for co-immunoprecipitation experiments, to test the interaction of h-α-syn with WT synapsin Ia or synapsin Iascr-E. Neuro2a cells were co-transfected with myc-tagged α-syn and respective YFP-tagged synapsin Ia, and the YFP was immunoprecipitated. Bottom: Note that h-α-syn co-immunoprecipitated with synapsin Ia, but not synapsin Iascr-E; quantification of the gels below (n=4, all data are means ± SEM ***p<0.001, Student’s t test – a non-specific band is marked with an asterisk). (E) Schematic of experiments to test if the synapsin E-domain is sufficient to enable α-syn functionality in synapsin TKO neurons. Synapsin-E (a 46 amino acid sequence) was fused to the C-terminus of sypHy, so that upon expression in neurons, the E-domain would be present on the cytosolic surface of Svs. (F) SypHy fluorescence traces (mean ± SEM). Note that while h-α-syn (orange) was unable to attenuate SV recycling in synapsin TKO neurons (as expected), diminished synaptic responses were seen when the E-domain was present. Insets: Quantification of peak ΔF/F0 sypHy values (bars: mean ± SEM). Twelve 19 coverslips from at least three separate cultures were analyzed for each condition (ns p=0.89, ***p=2.8e-7, one-way ANOVA with Tukey’s posthoc analysis).
-
Figure 3—source data 1
Tabular data and statistical analyses for graphs shown in panels C, D and F.
- https://cdn.elifesciences.org/articles/89687/elife-89687-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Full western blots for segments shown in panel D.
- https://cdn.elifesciences.org/articles/89687/elife-89687-fig3-data2-v1.pdf
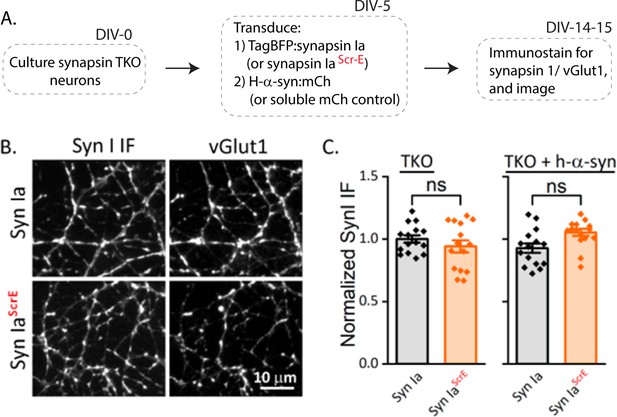
Similar synaptic of localization of synapsin Ia and synapsin IaScr-E in synapsin TKO neurons.
(A) Schematic of experiments to evaluate quantitative localization of tagBFP:synapsin Ia and tagBFP:synapsin IaScr-E in synapsin TKO neurons (with and without h-α-syn over-expression). Neurons were immunostained for synapsin I (for reliable visualization of the transduced synapsin constructs), as well as for the SV-marker vGlut1 (to confidently identify synapses). (B) Representative images showing equivalent immunofluorescence of synapsin Ia and synapsin IaScr-E at synapses. Over-expression of h-α-syn did not affect their synaptic fluorescence. (C) Quantified synaptic fluorescence data, represented as mean +/- SEM, 15–16 coverslips from at least three independent cultures were analyzed for each condition. ns p=0.52, ns p=0.14, one-way ANOVA.
-
Figure 3—figure supplement 1—source data 1
Tabular data and statistical analyses for graph shown in panel C.
- https://cdn.elifesciences.org/articles/89687/elife-89687-fig3-figsupp1-data1-v1.xlsx
-
Figure 3—figure supplement 1—source data 2
Raw images.
Full images of anti-synapsin I and anti-vGlut1 immunofluorescence channels containing the details shown in panel B (marked with white dashed square).
- https://cdn.elifesciences.org/articles/89687/elife-89687-fig3-figsupp1-data2-v1.zip
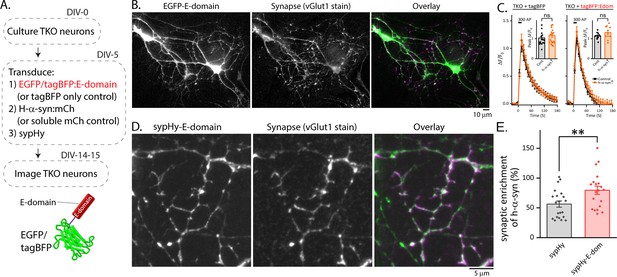
Synaptic targeting of synapsin E-domain constructs in synapsin null neurons.
(A) Schematic of experiments to evaluate synaptic targeting of synapsin E-domain constructs in synapsin TKO neurons. Note that EGFP is tagged to the E-domain in these experiments. (B) The EGFP:E-domain construct was diffusely distributed in neurons and not enriched to synapses (marked by immunostaining of VGlut1). A representative image showing that the E-domain construct is not targeted to synapses (green: EGFP:E-domain, magenta: vGlut1). (C) Over-expression of synapsin E-domain in the context of excessive α-syn did not have any effect on SV recycling (as determined by sypHy experiments), presumably because the E-domain fails to enrich at synapses. Data shown as mean mean ± SEM, 11–20 coverslips from at least 3 independent cultures were analyzed for each condition (p=0.16, Kruskal-Wallis ANOVA). (D) Representative images illustrating synaptic localization of the E-domain tagged to sypHy (green: sypHy:E-domain, magenta: vGlut1). (E) Expression of sypHy:E-domain in synapsin TKO neurons enhances the synaptic enrichment of h-α-syn. Synaptic enrichment (see Methods section) of h-α-syn was measured in synapsin TKO neurons expressing either sypHy or sypHy-E-domain. We observed significantly higher enrichment of h-α-syn in the latter. Data shown as mean ± SEM, 23 to 25 coverslips from three independent cultures were analyzed for each condition (**p=0.009, Mann-Whitney U-test).
-
Figure 3—figure supplement 2—source data 1
Tabular data and statistical analyses for graphs shown in panels C and E.
- https://cdn.elifesciences.org/articles/89687/elife-89687-fig3-figsupp2-data1-v1.xlsx
-
Figure 3—figure supplement 2—source data 2
Raw images.
Full images of EGFP:E-domain and anti-vGlut1 immunofluorescence channels containing the details shown in panel B, and the full images of sypHy:E-domain and anti-vGlut1 immunofluorescence channels containing the details shown in panel D (marked with white dashed square).
- https://cdn.elifesciences.org/articles/89687/elife-89687-fig3-figsupp2-data2-v1.zip

Synapsin-dependent redistribution of synaptic vesicles by α-syn overexpression.
(A) Representative images from WT or synapsin TKO neurons immunostained with an SV marker (vGlut1); zoomed insets marked by yellow boundaries. Note that the compact clustering of SVs is lost in synapsin-null neurons. (B) FWHM as a quantitative means to determine spreading of fluorophores at synapses (also see Results). Note that an increase in FWHM corresponds to a decrease in intensity and increased spreading of fluorescence within a bouton. (C) Quantification of synaptic fluorescence in WT and synapsin TKO neurons. Overall intensities are decreased in TKO synapses (left), and FWHM is increased (right), compared to WT synapses; consistent with a spreading of SVs in the synapsin null setting. (D) Experimental plan to determine effects of h-α-syn over-expression on the overall distribution of SVs in WT and synapsin TKO neurons. (E) FWHM of vGlut1 staining at synapses is augmented by h-α-syn over-expression in WT neurons, but not in neurons from synapsin TKO mice. Reintroduction of synapsins Ia/IIa (but not Ib/IIb) in the setting of h-α-syn over-expression rescues the changes in vGlut1-FWHM (F). All data in this figure are represented as mean +/-SEM. Nine to 28 coverslips from at least three independent cultures were analyzed for C, E, and F (C, left: ***p=0.0006, Mann-Whitney U-test; right: see E; E: ***p=4e-8, ns p=0.92, one-way ANOVA with Tukey’s posthoc analysis; F, left: ***p=2.7e-4, ns p=1.0, Kruskal-Wallis ANOVA with Dunn’s posthoc test; F, right: **p=0.001, ns p=0.52, one-way ANOVA with Tukey’s posthoc analysis).
-
Figure 4—source data 1
Raw images.
Full images of anti-vGlut1 immunofluorescence channels in WT and synapsin TKO neurons, containing the details shown in panel A (marked with white dashed square).
- https://cdn.elifesciences.org/articles/89687/elife-89687-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Tabular data and statistical analyses for graphs shown in panels C, E, and F.
- https://cdn.elifesciences.org/articles/89687/elife-89687-fig4-data2-v1.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus, both sexes) | WT mice, C57BL/6JRccHsd | Inotiv (Envigo) | RRID:IMSR_ENV:HSD-043 | |
| Genetic reagent (Mus musculus, both sexes) | Synapsin TKO mice | Gitler et al., 2004; Boido et al., 2010 | RRID:MMRRC_041434-JAX | Rederived on C57Bl/6 background |
| Cell line (Human) | HEK293-T | ATCC | RRID:CVCL_0063 | |
| Cell line (Mouse) | Neuro2A | ATCC | RRID:CVCL_0470 | TKG Cat#TKG 0509 |
| Antibody | anti-vGlut1 (goat polyclonal) | Synaptic Systems | 135 307 | (IF: 1:1000) |
| Antibody | anti-synapsin I (mouse monoclonal) | Synaptic Systems | 106 011 | (IF: 1:1000) |
| Antibody | anti-synapsin-1 (Recombinant) | Abcam | ab254349 | (WB: 1:1000) |
| Antibody | anti-c-myc (mouse monoclonal clone 9E10) | Sigma-Aldrich | M4439 | (WB 1:500) |
| Antibody | anti-GFP (Rabbit polyclonal) | Abcam | ab290 | (WB 1:5000) |
| Antibody | anti-VAMP2 (mouse monoclonal) | Synaptic Systems | 104 211 | (IF 1:1000) |
| Antibody | anti-goat IgG, NL-637 label (Donkey polyclonal) | R&D systems | NL002 | (IF 1:1000) |
| Antibody | anti-mouse IgG, NL-493 label (Donkey polyclonal) | R&D systems | NL009 | (IF 1:1000) |
| Antibody | anti-rabbit IgG H&L, HRP (Goat polyclonal) | Abcam | ab205718 | (WB 1:1000) |
| Antibody | anti-mouse IgG H&L, HRP (Goat polyclonal) | Abcam | ab205719 | (WB 1:1000) |
| Recombinant DNA reagent | pD1 | Tevet and Gitler, 2016 | AAV1 | Cap1, Rep2, E2A, E4, VA |
| Recombinant DNA reagent | pD2 | Tevet and Gitler, 2016 | AAV2 | Cap2, Rep2, E2A, E4, VA |
| Recombinant DNA reagent | pAAV2-hSyn-sypHy | Orenbuch et al., 2012 | DG87 | hSyn (promoter) Synaptophysin I –2XpHluorin |
| Recombinant DNA reagent | pAAV2-hSyn-sypHy:E-domain | This paper | DG189 | hSyn (promoter) Synaptophysin I –2XpHluorin – Synapsin Ia E domain |
| Recombinant DNA reagent | pAAV2-hSyn-tagBFP | This paper | DG138 | hSyn (promoter) tagBFP |
| Recombinant DNA reagent | pAAV2-hSyn-tagBFP:E-domain | This paper | DG201 | hSyn (promoter) tagBFP-Synapsin Ia E domain |
| Recombinant DNA reagent | pAAV2-hSyn-tagBFP:ScrE-domain | This paper | DG200 | hSyn (promoter) tagBFP- Scrambled Synapsin Ia E domain |
| Recombinant DNA reagent | pAAV2-hSyn-EGFP:E-domain | This paper | DG187 | hSyn (promoter) EGFP-Synapsin Ia E domain |
| Recombinant DNA reagent | pAAV2-hSyn-h-α-syn-mCherry | Atias et al., 2019 | DG79 | hSyn (promoter) human-alpha-synuclein-mCherry |
| Recombinant DNA reagent | pAAV2-hSyn- mCherry | Atias et al., 2019 | DG97 | hSyn (promoter) mCherry |
| Recombinant DNA reagent | pEYFPC1-Synapsin Ia | Gitler et al., 2004 | Syn03 | CMV (enhancer +promoter) EYFP-Synapsin Ia |
| Recombinant DNA reagent | pEGFPC1-Synapsin Ib | Gitler et al., 2004 | Syn65 | CMV (enhancer +promoter) EGFP-Synapsin Ib |
| Recombinant DNA reagent | pEGFPC1-Synapsin IIa | Gitler et al., 2004 | Syn50 | CMV (enhancer +promoter) EGFP-Synapsin IIa |
| Recombinant DNA reagent | pEGFPC2-Synapsin IIb | Gitler et al., 2004 | Syn73 | CMV (enhancer +promoter) EGFP-Synapsin IIb |
| Recombinant DNA reagent | pEGFPC1-Synapsin IIIa | Gitler et al., 2004 | Syn59 | CMV (enhancer +promoter) EGFP-Synapsin IIIa |
| Recombinant DNA reagent | pEYFPC1-Synapsin Ia ScrE | This paper | Syn88 | CMV (enhancer +promoter) EYFP-Synapsin Ia Scrambled E domain |
| Recombinant DNA reagent | pEYFPC1 | Clontech | CMV (enhancer +promoter) EYFP | |
| Recombinant DNA reagent | pEGFPC1 | Clontech | CMV (enhancer +promoter) EGFP | |
| Recombinant DNA reagent | h-α-syn-myc | This paper | pLP351 | EF-1 alpha promoter (pCCL backbone) |
| Recombinant DNA reagent | GST | Parra-Rivas et al., 2023 | Addgene # 209891 | pGEX-KG myc |
| Recombinant DNA reagent | GST-h-α-syn FL | This paper | Addgene # 213498 | pGEX-KG myc backbone |
| Recombinant DNA reagent | GST-h-α-syn 1–95 | This paper | Addgene # 213499 | pGEX-KG myc backbone |
| Recombinant DNA reagent | GST-h-α-syn 1–110 | This paper | Addgene # 213500 | pGEX-KG myc backbone |
| Recombinant DNA reagent | GST-h-α-syn 96–140 | This paper | Addgene # 213501 | pGEX-KG myc backbone |
| Recombinant DNA reagent | GST-h-α-syn FL Scrambled 96–110 | This paper | Addgene # 213503 | pGEX-KG myc backbone |
| Recombinant DNA reagent | GST-h-α-syn FL Scrambled 96–140 | This paper | Addgene # 213502 | pGEX-KG myc backbone |
| Recombinant DNA reagent | pAAV2-hSyn-tagBFP-Synapsin Ia | This paper | DG199 | hSyn (promoter) tagBFP-Synapsin Ia |
| Recombinant DNA reagent | pAAV2-hSyn-tagBFP-Synapsin Ib | This paper | DG150 | hSyn (promoter) tagBFP-Synapsin Ib |
| Recombinant DNA reagent | pAAV2-CMV/CBAP-tagBFP-Synapsin IIa | Orenbuch et al., 2012 | DG18 | CMV-CBAP (promoter) tagBFP-Synapsin IIa |
| Recombinant DNA reagent | pAAV2-hSyn-tagBFP-Synapsin IIb | This paper | DG160 | hSyn (promoter) tagBFP-Synapsin IIb |
| Recombinant DNA reagent | pAAV2-hSyn-tagBFP-Synapsin IIIa | This paper | DG153 | hSyn (promoter) tagBFP-Synapsin IIIa |
| Chemical compound, drug | Bafilomycin A1 | Enzo Life Sciences | BML-CM110-0100 | |
| Chemical compound, drug | 6,7-Dinitroquinoxaline-2,3 (1H,4H-dione), DNQX | Sigma-Aldrich | D0540 | |
| Chemical compound, drug | DL-2-Amino-5-phosphonopentanoic acid, APV | Sigma-Aldrich | A5282 | |
| Software, algorithm | Origin Pro 2023 | Originlab | RRID:SCR_014212 | |
| Software, algorithm | GraphPad Prism 6 | Graphpad | RRID:SCR_002798 | |
| Software, algorithm | NIS-elements AR 5.21.03 | Nikon | RRID:SCR_014329 | |
| Other | N-PER | Thermo Scientific | 87792 | Methods: Biochemical assays and evaluation |
| Other | protease/ phosphatase inhibitors | Cell Signaling | 5872 | Methods: Biochemical assays and evaluation |
| Other | Neurobasal-A medium | Thermo-Fisher Scientific | 10888022 | Methods: Hippocampal Cultures, AAV production, and transduction |
| Other | B27 supplement | Thermo-Fisher Scientific | 17504044 | Methods: Hippocampal Cultures, AAV production, and transduction |
| Other | Glutamax I | Thermo-Fisher Scientific | 35050038 | Methods: Hippocampal Cultures, AAV production, and transduction |
| Other | Fetal Bovine Serum, European Grade | Biological Industries | 04-007-1A | Methods: Hippocampal Cultures, AAV production, and transduction |
| Other | Gentamicin | Biological Industries | 03-035-1C | Methods: Hippocampal Cultures, AAV production, and transduction |
| Other | immumount | Thermo Scientific | 9990402 | Methods: pHluorin assays, analysis, and fluorescence microscopy |





