Forkhead transcription factor FKH-8 cooperates with RFX in the direct regulation of sensory cilia in Caenorhabditis elegans
Figures
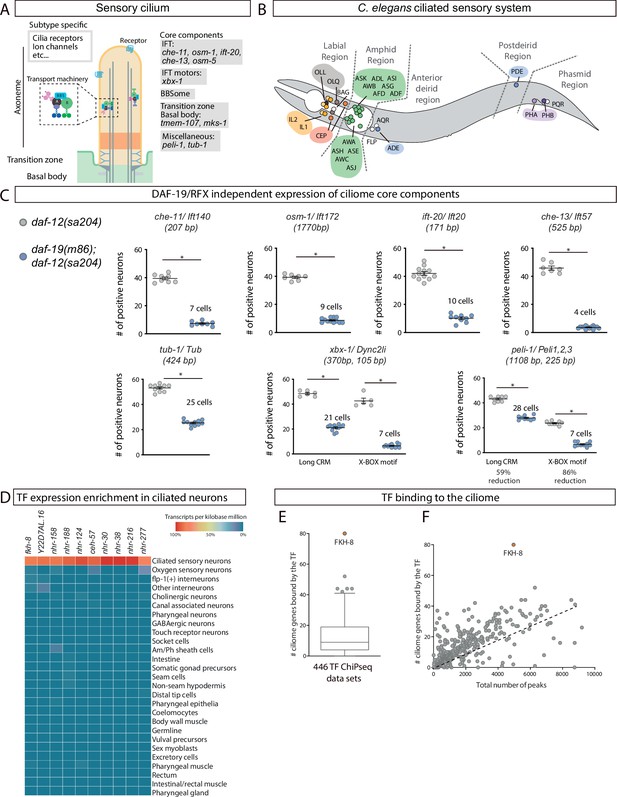
FKH-8 is a candidate for direct regulation of ciliome gene expression in C. elegans.
(A) Schema for a sensory cilium. Cilia components (ciliome) can be divided into core and subtype-specific categories. Core genes whose reporters are analyzed in panel C and Figure 1—figure supplement 2 are indicated by their function. (B) Lateral view of C. elegans hermaphrodite ciliated system. Sixty ciliated neurons from 25 different classes are distributed in five distinct anatomical regions. (C) Ciliome core components show persistent expression in double daf-12(sa204); daf-19(m86) mutants. The same extrachromosomal line was analyzed in the different genetic backgrounds. Each dot represents the total number of reporter-positive neurons in a single animal. Mean and standard error are represented. The mean number of remaining reporter-positive neurons in double daf-12; daf-19 mutants is indicated. Sample sizes for each genetic background: che-11: n=8; osm-1: n≥7; ift-20: n≥9; che-13: n≥7; tub-1: n≥10; xbx-1: n≥5; peli-1: n≥8. See Figure 1—source data 1 for raw data and Figure 1—figure supplements 1 and 2 for details on construct lengths and additional reporter scorings. (D) sc-RNA-seq data analysis identifies 10 TFs specifically enriched in ciliated sensory neurons. These TFs belong to FKH, ZF, NHR and HD families. See Figure 1—figure supplement 3 for detailed description of TF expression in each ciliated neuron type. (E) ChIP-seq data analysis of 259 available TFs shows that FKH-8 ranks first in direct binding to regulatory regions assigned to the ciliome gene list. See Figure 1—source data 2 for gene lists and Figure 1—figure supplement 3 for core ciliome or subtype specific binding analysis. (F) Correlation of total number of peaks versus ciliome-gene peaks shows FKH-8 behaves as an outlier, demonstrating high binding to ciliome genes is not merely due to the high number of FKH-8 binding-events.
-
Figure 1—source data 1
Raw quantification data of ciliome gene reporters in daf-19(m86); daf-12(sa204) included in Figure 1 and Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/89702/elife-89702-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Gene lists and raw data for bioinformatics analysis in Figure 1 and Figure 1—figure supplement 3.
- https://cdn.elifesciences.org/articles/89702/elife-89702-fig1-data2-v2.xlsx
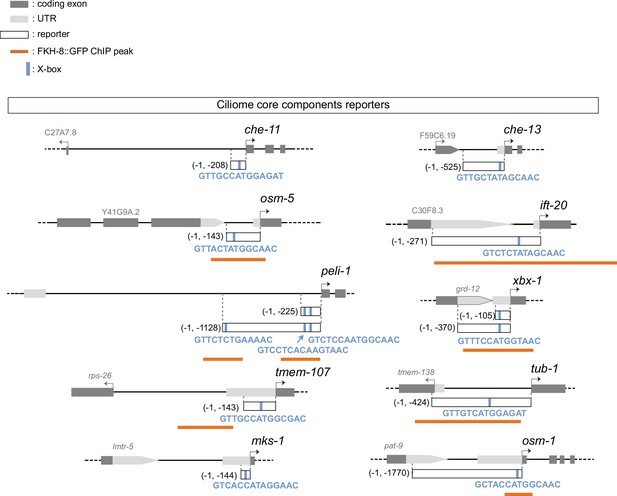
Ciliome reporters used in this work.
Schematic representation of reporter constructs used in the manuscript. Selected core cilia components contain at least one experimentally validated X-box motif in their sequences (marked as a blue bar). For che-11, che-13, osm5, ift-20, tub-1, mks-1 and osm-1 see Efimenko et al., 2005; for peli-1 see Chu et al., 2012, for xbx-1 see Schafer et al., 2003; for tmem-107 see Lambacher et al., 2016. Overlap between x-boxes and FKH-8 binding sites is found for osm-5, ift-20, peli-1, xbx-1, tub1 and osm-1.
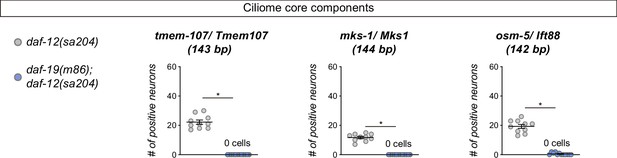
Reporter expression for some core ciliome genes is abolished in daf-19(m86) mutant.
Expression of short reporters for the core cilia components tmem-107, mks-1 and osm-5 is completely abolished in daf-12(sa204); daf-19(m86) double mutants. The same extrachromosomal line was analyzed in the different genetic backgrounds. n≥10. Each dot represents the total number of reporter-positive neurons scored in a single animal. Mean and standard error are represented. See Figure 1—source data 1 for raw scoring data.
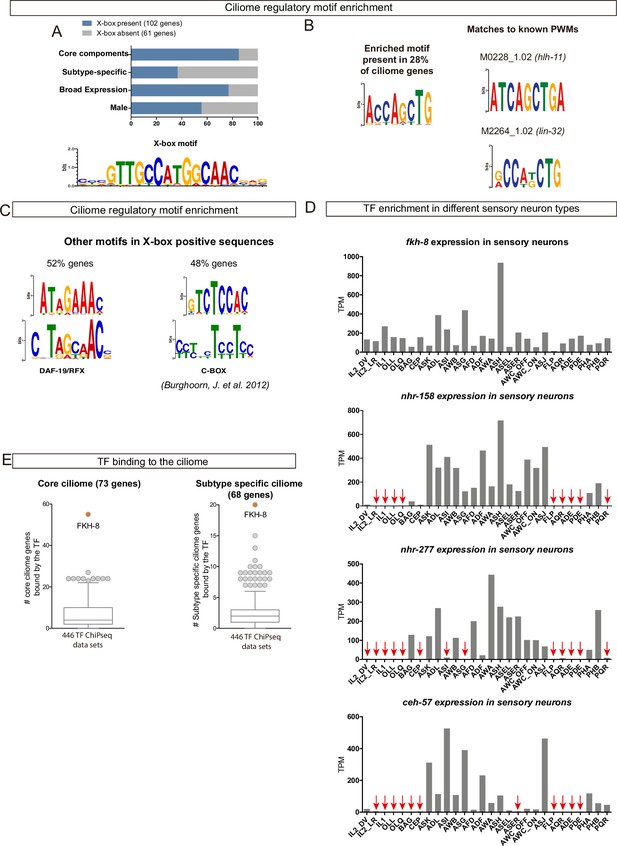
Available -omics data identifies FKH-8 as a candidate transcriptional regulator of ciliome genes in C.elegans.
(A) DAF-19/RFX motifs (X-box) are enriched in regulatory sequences of core and broadly expressed ciliome genes more prominently than in subtype-specific features. Motif logo for the palindromic X-box motif. (B) De novo motif enrichment analysis of putative regulatory sequences of ciliome genes identifies a motif matching known binding site for the pro-neural bHLH TFs lin-32 and hlh-11. (C) Two additional motifs enriched in regulatory sequences of ciliome genes containing X-box sites, show partial similarity to the DAF-19/RFX motif and to the ciliome-related motif C-BOX motif (Burghoorn et al., 2012). (D) sc-RNA-seq data of FACS-isolated neurons from L4 hermaphrodites (Taylor et al., 2021) show broad expression for ceh-57, fkh-8, nhr-158 and nhr-277 TFs across the whole ciliated system of C. elegans. Only fkh-8 expression is detected in all ciliated neuron types. Red arrows indicate values lower than 10 TPM (transcripts per million). (E) ChIP-seq data analysis shows FKH-8 ranks first among 259 TFs directly binding to either core ciliome genes (left) or subtype-specific ciliary features (right).
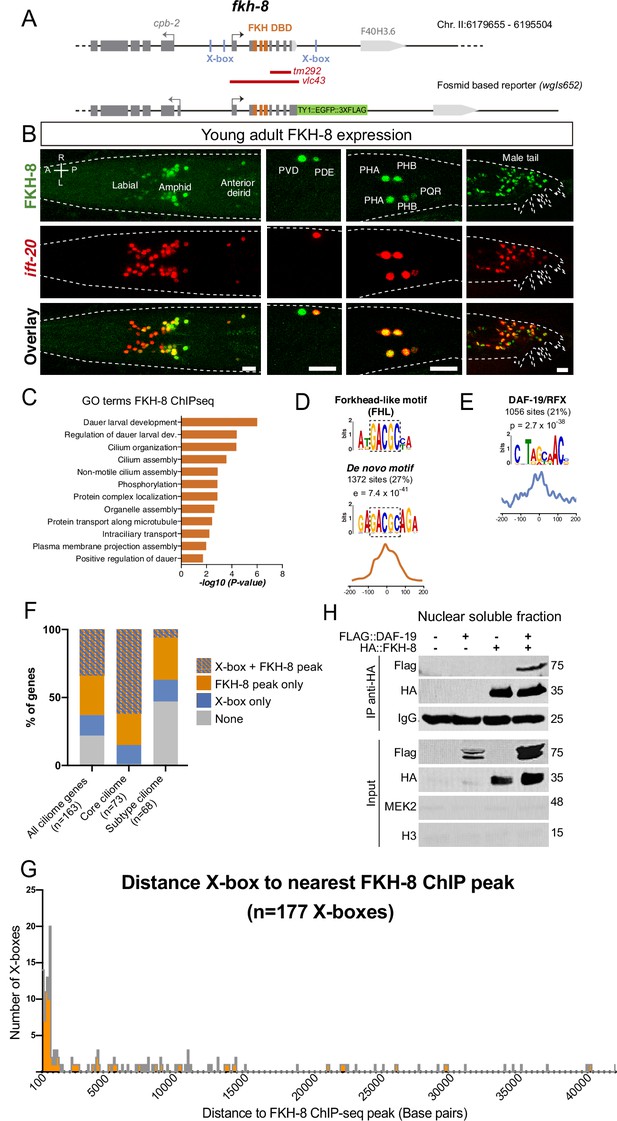
FKH-8 is expressed in sensory ciliated neurons, binds ciliome genes near DAF-19 X-boxes and physically interacts with DAF-19.
(A) fkh-8 locus (top) and fosmid based fkh-8 reporter (bottom). Grey boxes represent exons and orange boxes correspond to exons coding for the FKH DNA binding domain (DBD). Putative daf-19/RFX binding sites (X-boxes) are depicted with blue lines. Red bars indicate extension for the corresponding deletion alleles. (B) Dorso-ventral views of young adult animals expressing both the fosmid-based FKH-8::GFP reporter (in green) and an integrated reporter for the panciliary marker ift-20 (in red). A: anterior, P: posterior, R: right, L: left. Scale bar = 10 µm. See Figure 2—source data 1 for quantification and Figure 2—figure supplement 1 for embryonic expression patterns and expression correlation with DAF-19 and ciliome genes. (C) Genes associated to nearby FKH-8 binding events enrich Gene Ontology terms related to cilia regulated processes and/or functions. Data correspond to adjusted p-value. See Figure 2—source data 2 for gene lists associated to GO terms (D) De novo motif analysis of FKH-8 ChIP-seq data identifies a motif present in 27% of peaks, enriched at central positions, that matches a Forkhead like (FHL) motif. (E) DAF-19/RFX binding motifs (PWM M1534_1.02) are present in 21% of the FKH-8 bound regions and are enriched at central positions. See Figure 2—figure supplement 2 for similar analysis on additional FKH ChIP-seq data sets. (F) Distribution of ciliome genes in four different categories: (1) genes with both X-box motifs and FKH-8 binding events; (2) genes with only FKH-8 binding; (3) Genes with X-box motifs only and (4) Genes with neither FKH-8 binding or X-boxes. Most ciliome genes contain both X-boxes and FKH-8 peaks, this dual signature is highly prevalent in core ciliome genes while is minoritary in subtype ciliome genes. See Figure 2—source data 2 for gene lists associated to each signature. (G) Distance between X-boxes found in ciliome genes and the center of the nearest FKH-8 ChIP-seq peak. 42% of X-boxes are located less than 600 bp from a FKH-8 ChIP-seq peak. See Figure 2—figure supplement 2 for differential analysis of core and subtype ciliome genes. (H) Co-immuno precipitation of HA tag FKH-8 and FLAG tag DAF-19 expressed in HEK293 cells shows physical interaction between both transcription factors in the soluble fraction of nuclear extracts. MEK2 is used to assess for the presence of cytoplasmic components and Histone H3 to assess the presence of chromatin. See Figure 2—source data 3 for original blots and Figure 2—figure supplement 2 for additional analysis of interaction in chromatin associated fractions.
-
Figure 2—source data 1
Raw quantification data of FKH-8 and ift-20 reporter co-expression represented in Figure 2B.
- https://cdn.elifesciences.org/articles/89702/elife-89702-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Raw data for bioinformatics analysis in Figure 2 and Figure 2—figure supplements 1 and 2.
- https://cdn.elifesciences.org/articles/89702/elife-89702-fig2-data2-v2.xlsx
-
Figure 2—source data 3
Co-IP original files of unedited gels in Figure 2.
- https://cdn.elifesciences.org/articles/89702/elife-89702-fig2-data3-v2.zip
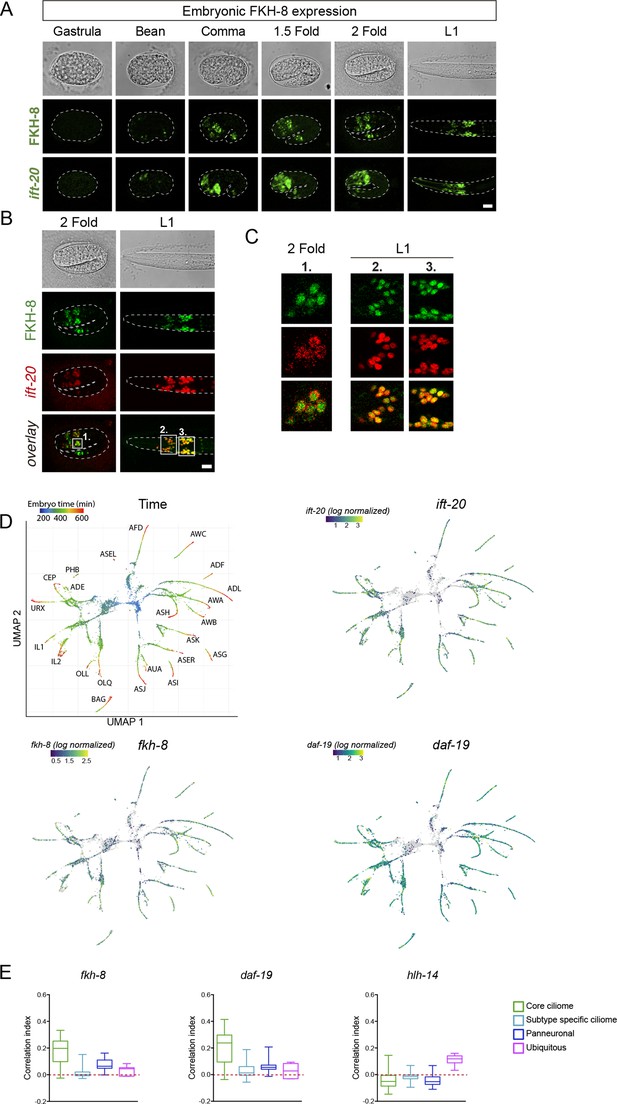
fkh-8 expression along ciliated system development.
(A) Representative Z projections of developmental embryonic milestones until hatching (L1) shows FKH-8::GFP fosmid reporter expression matches in time and space panciliary reporter ift-20::gfp expression. Scale bar = 10 µm. (B) Representative Z projections of two fold embryo and fist larval stage (L1) animals expressing both FKH-8::GFP fosmid reporter (in green) and an integrated reporter for the panciliary marker ift-20:: tagRFP (in red). Note that due to long maturation time of the tag-RFP reporter, ift-20::tagRFP expression is only detected from the two fold stage, while ift-20::gfp reporter in (A) is first detected at bean stage, similar to fkh-8 expression. Scale bar = 10 µm. (C) Single Z-plane from regions indicated in (B) show colocalization of FKH-8::GFP and RFP in the ciliated sensory neurons. (D) Embryonic sc-RNA-data (Packer et al., 2019) from C. elegans ciliated neurons and their progenitor cells. Pseudo-time (left pannel) shows the maturation trajectory of ciliated neurons that coincides with increasing ift-20, fkh-8, and daf-19 expression. (E) Correlation index of fkh-8, daf-19 and hlh-14 TF scRNAseq expression and four different gene categories (core ciliome, subtype ciliome, panneuronal or ubiquitous) in all ciliated lineages (Packer et al., 2019). fkh-8 and daf-19 expression shows high correlation index with core ciliome genes but not with other gene categories, while hlh-14, bHLH TF not involved in ciliogenesis shows low correlation values in all categories. See Figure 2—source data 2 f for raw data.
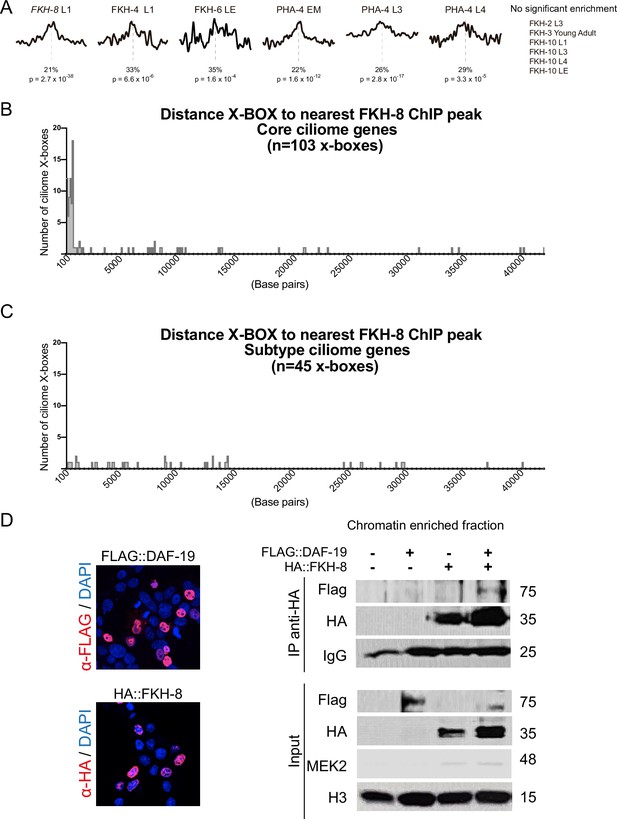
FKH-8 binds near X-BOX motifs.
(A) Analysis of DAF-19/RFX binding motifs (PWM M1534_1.02) in peaks for ChIP-seq datasets of other C. elegans FKH TFs present in ENCODE database. There is no significant enrichment for DAF-19 motif in FKH-2, FKH-3, and FKH-10 datasets. Motif enrichment in FKH-4, FKH-6, and PHA-4 datasets is less significant and shows less defined enrichment in the centre of the peaks. p represents p Value associated to motif enrichment and % refers to the number of peaks with the motif present. EM: Embryo mixed stage; L1: Larval stage 1; L3: Larval stage 3, L4: Larval stage 4, LE: Late embryo. See Figure 2—source data 2 for detailed data. (B) Distance between X-boxes found in the promoter regions of core ciliome genes and the center of the nearest FKH-8 ChIP peak. 63% of X-boxes are located less than 600 bp from a FKH-8 ChIP peak. (C) Distance between X-boxes found in the promoter regions of subtype ciliome genes and the center of the nearest FKH-8 ChIP peak. 9% of X-boxes are located less than 600 bp from a FKH-8 ChIP peak. (D) Co-immuno precipitation of FKH-8 and DAF-19 expressed in HEK293 cells. Micrographs shows nuclear localization of transfected HA::FKH-8 and FLAG::DAF-19. In addition to the interaction detected in the soluble nuclear fraction (Figure 2), both factors also interact bound to DNA (chromatin fraction). See Figure 2—figure supplement 2—source data 1 for original blots.
-
Figure 2—figure supplement 2—source data 1
Co-IP original files of unedited gels for the chromatin fraction.
- https://cdn.elifesciences.org/articles/89702/elife-89702-fig2-figsupp2-data1-v2.zip
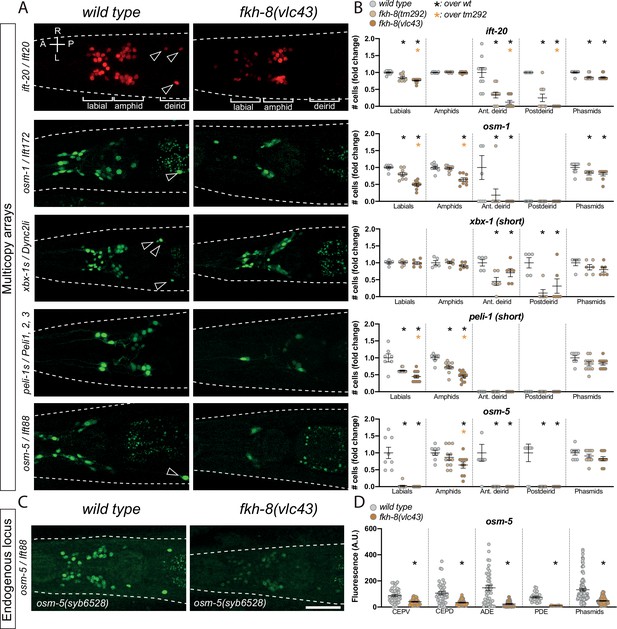
FKH-8 TF and FKH-binding sites are required for correct core ciliome gene reporter expression.
(A) Dorso-ventral images from young adult heads expressing different core ciliome multicopy array gene reporters in wild type and fkh-8(vlc43) null mutant animals. All reporters are extrachromosomal arrays except for ift-20 reporter which is integrated. Arrow heads point deirid expression lost in the mutant. A: anterior, P: posterior, R: right, L: left. (B) Quantification of the number of gfp-positive cells in five distinct anatomical regions for each reporter in wild type, fkh-8(tm292) hypomorphic allele and fkh-8(vlc43) null mutant. To facilitate comparisons, values in each region are normalized to controls. The same extrachromosomal line was analyzed in the different genetic backgrounds. Each dot represents the number of reporter-expressing neurons scored in a single animal. Mean and standard error are represented. Black asterisk denotes significantly different from wild type and orange asterisk indicates vlc43 is significantly different from tm292 allele. Sample sizes for each genetic background: ift-20: n≥10; osm-9: n=9; osm-5: n≥8; peli-1: n≥7; xbx-1: n≥5. See Figure 3—source data 1 for raw scoring data, Figure 3—figure supplement 1 for analysis of the hypomorphic recessive nature of the tm292 allele and quantification of additional reporters not affected in fkh-8 mutants, see Figure 3—figure supplement 2 and Figure 3—source data 2 for functional characterization of predicted FKH binding sites in ift-20 and xbx-1 regulatory regions. (C) Dorso-ventral images from young adult heads expressing GFP from the endogenously tagged osm-5 locus [osm-5(syb6528), osm-5::SL2::GFP::H2B] in wild type and fkh-8(vlc43) null mutant. A global decrease in fluorescence intensity is detected in fkh-8(vlc43) animals compared to wild type. Scale bar = 25 µm. (D) Fluorescence intensity level quantification in specific ciliated neuron populations shows significant reduction of expression in fkh-8(vlc43) animals. A. U.: arbitrary units. See Figure 3—source data 1 for raw scoring data. n≥20 for each cell type and genetic background.
-
Figure 3—source data 1
Raw quantification data of ciliome gene reporter expression defects in fkh-8 mutants and cis-regulatory point mutation analysis corresponding to Figure 3 and Figure 3—figure supplements 1 and 2.
- https://cdn.elifesciences.org/articles/89702/elife-89702-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Motif enrichment analysis of xbx-1 and ift-20 reporters.
- https://cdn.elifesciences.org/articles/89702/elife-89702-fig3-data2-v2.xlsx
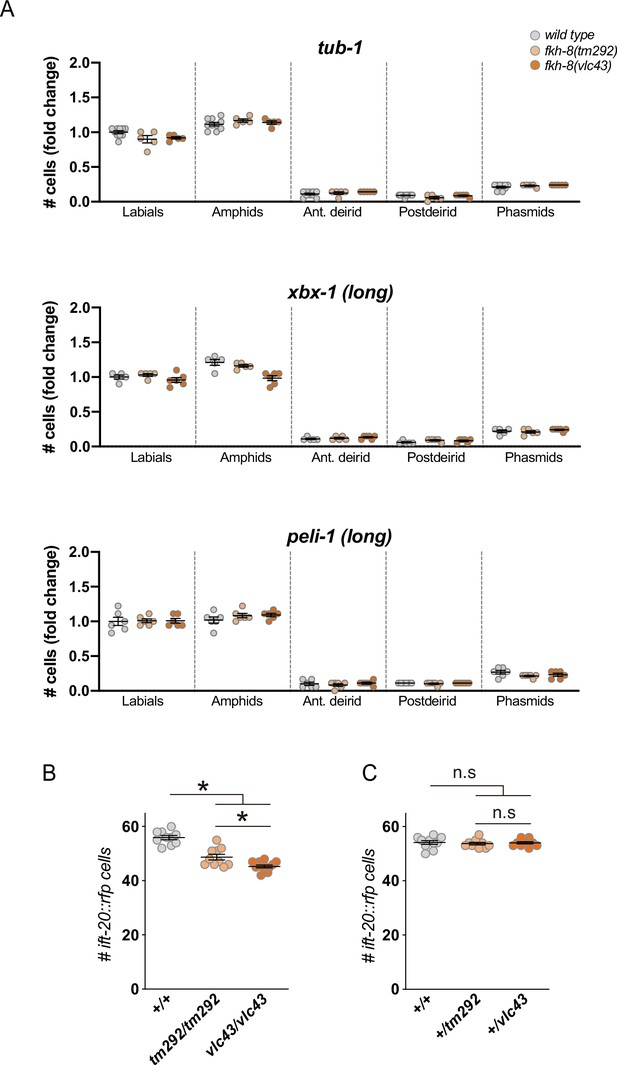
f. fkh-8(tm292) is a hypomorphic recessive allele.
(A) Quantification of the number of gfp-positive cells in five distinct anatomical regions for each reporter in wild type, fkh-8(tm292) hypomorphic allele and fkh-8(vlc43) null mutant. To facilitate comparisons, values in each region are normalized to controls. The same extrachromosomal line was analyzed in the different genetic backgrounds. Each dot represents the number of reporter-expressing neurons scored in a single animal. Mean and standard error are represented. fkh-8(tm292) and fkh-8(vlc43) show similar expression values than wild type for these markers. See Figure 3—source data 1 for raw scoring data. (B) Quantification of the number of ift-20::rfp-positive cells in wild type, fkh-8(tm292) and fkh-8(vlc43) mutants shows stronger defects in vlc43 null allele.: n≥5. (C) Heterozygote cross progeny from wild type, fkh-8(tm292) and fkh-8(vlc43) show similar number of ift-20::rfp-positive cells denoting the recessive nature of both alleles. n≥10.
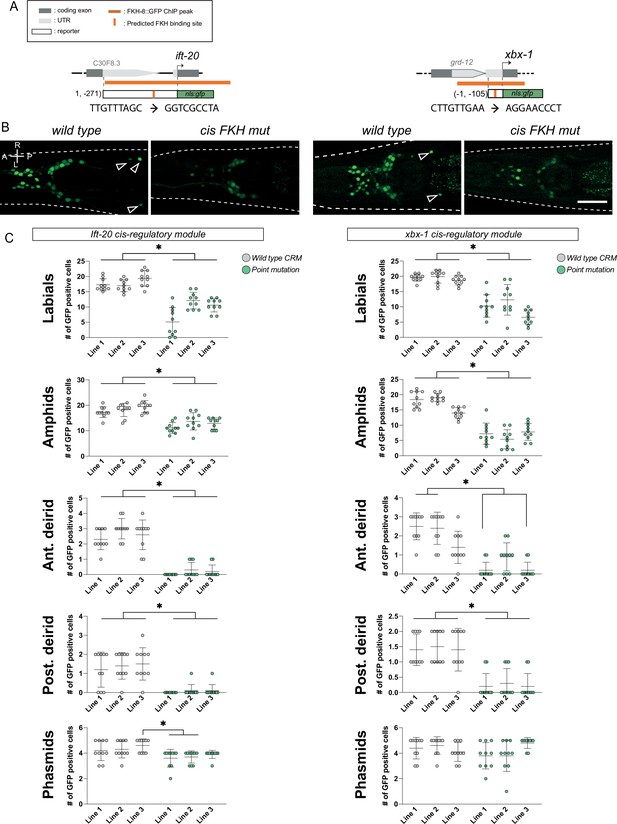
f. Functional characterization of putative FKH sites in cis-regulatory modules of two core ciliome components.
(A) Schematics for the ift-20 and xbx-1 loci and reporters. Dark grey boxes represent exons whereas light grey boxes correspond to UTRs. FKH-8 Chip-seq peaks are depicted with an orange horizontal line while predicted FKH DNA binding motifs are indicated with a vertical orange bar. Sequences corresponding to wild type and mutated putative FKH sites are indicated. See Figure 3—source data 2 for FKH putative binding site assignment. (B) Cis-regulatory mutation of putative FKH sites greatly reduces ciliome gene reporter expression. Representative dorso-ventral images from young adult heads expressing wild type or FKH-site-mutated reporters for core ciliome genes ift-20 (left) and xbx-1 (right). A: anterior, P: posterior, R: right, L: left. Scale bar = 25 µm. (C) Quantification of total number of gfp-positive cells for wild type and point mutated gfp reporters. Each graph represents one of the five anatomical regions scored. Three different extrachromosomal lines were analyzed for each construct. Each dot represents the number of reporter-expressing neurons scored in a single animal. Statistically significant differences are indicated with asterisks. See Figure 3—source data 1 for raw scoring data. n=10.
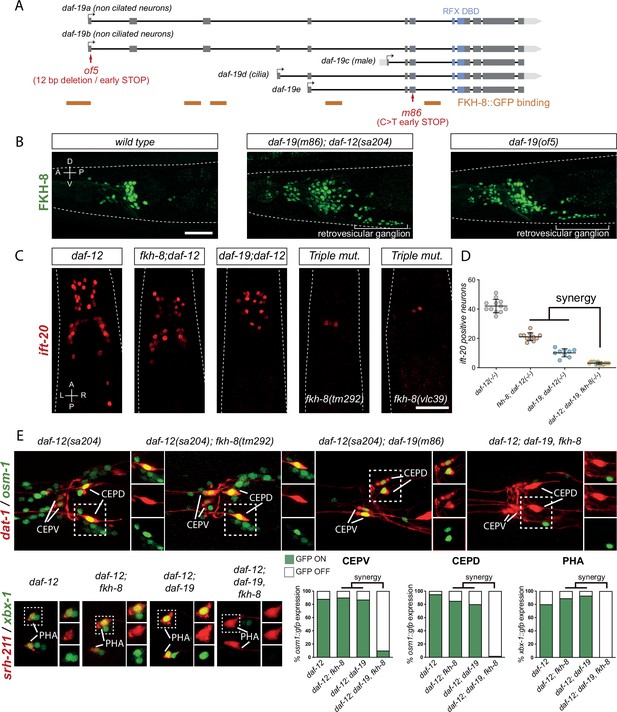
FKH-8 and DAF-19 exhibit crosstalk and synergistic effects in the transcriptional regulation of ciliome genes.
(A) daf-19 locus codes for five different daf-19 isoforms. Grey boxes represent exons whereas blue boxes correspond to exons coding for the RFX DNA binding domain (DBD). FKH-8 binding events are depicted as orange lines. Red arrows locate mutations of the corresponding alleles. (B) Lateral views from young adult hermaphrodite heads expressing fkh-8 fosmid-based reporter (wgIs652). Lack of all daf-19 isoforms (m86 allele) derepresses fkh-8 in non-ciliated neurons. This phenotype is mimicked by the specific absence of long daf-19a/b isoforms (of5 allele). Scale bar = 25 µm. See Figure 4—figure supplement 1 for unaffected DAF-19 expression in ciliated neurons in fkh-8(vlc43) mutants. (C) Dorso-ventral images from young adult hermaphrodites showing core ciliome ift-20 reporter expression in different genetic backgrounds. Scale bar = 25 µm. (D) Mean number of ift-20 reporter-expressing neurons in daf-12(sa204); daf-19(m86), fkh-8(tm292) triple mutants is significantly different from each of the double mutants and significantly lower than the expected from the multiplicative effect of both daf-12(sa204); fkh-8(tm292) and daf-12(sa204); daf-19(m86) animals. The same extrachromosomal line was analyzed in the different genetic backgrounds. Each dot represents the number of reporter-expressing neurons scored in a single animal. Mean and standard error are represented. See Figure 4—figure supplement 2 for quantification of FKH-8 and DAF-19 synergistic effects in xbx-1 and peli-1 reporter expression and Figure 4—source data 1 for raw data and statistics for all analyzed genetic backgrounds. n≥10 for each genetic background. (E) Analysis of osm-1 and xbx-1 ciliome reporters in specific subpopulations of ciliated neurons. CEPV and CEPD are labeled with dat-1::mcherry (otIs181) and srh-211:: tagRFP (vlcEx1365) is expressed in PHA neuron, both reporters are unaffected in all genetic backgrounds. Quantification of ciliome reporters is depicted in the corresponding graphs. Figure 4—source data 1 for raw data and statistics for all analyzed genetic backgrounds, n=30 worms per genotype and reporter construct.
-
Figure 4—source data 1
Raw quantification data of synergistic actions of fkh-8 and daf-19 depicted in Figure 4, Figure 4—figure supplements 1 and 2.
- https://cdn.elifesciences.org/articles/89702/elife-89702-fig4-data1-v2.xlsx
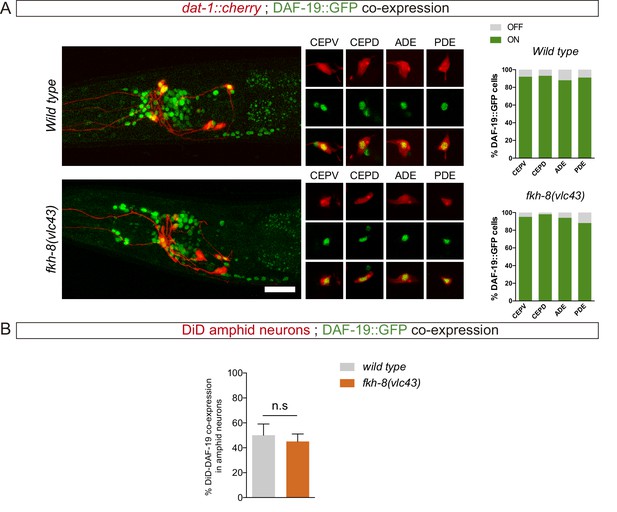
Lack of FKH-8 has no major effect on DAF-19 expression.
(A) Representative lateral views from heads of young adult hermaphrodites co-expressing a fosmid-based DAF-19::GFP reporter and dat-1::mcherry reporter labeling the dopaminergic neurons. Lack of FKH-8 does not affect DAF-19::GFP expression pattern. Co-localization analysis shows normal expression in the dopaminergic ciliated neurons (CEPV, CEPD, ADE, PDE), quantified in the graphs. Scale bar = 20 µm. See Figure 4—source data 1 for raw data. N=50. (B) daf-19 expression is largely unaffected in the subpopulation of DiD-positive ciliated amphid neurons in null fkh-8 mutant animals. DAF-19::GFP is consistently detected in the ASI, ADL and AWB neurons in both wild type and null fkh-8 mutant backgrounds. Mean and standard deviation are represented. n=10 animals.
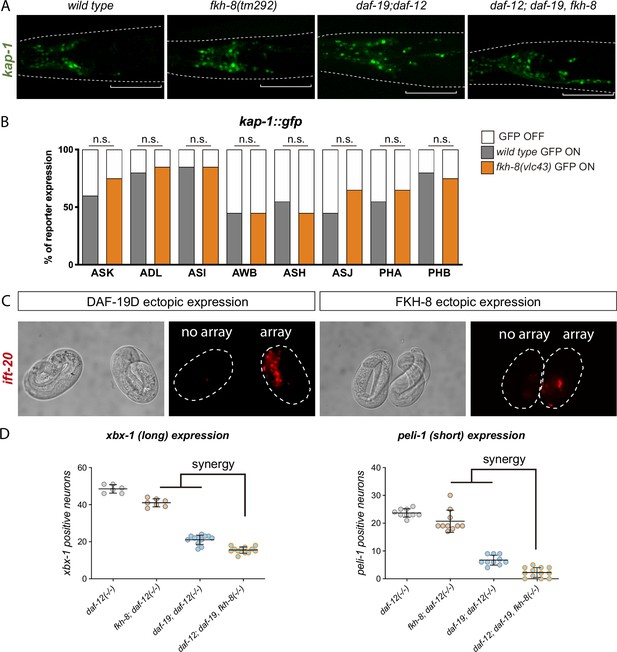
FKH-8 and DAF-19 show synergistic effects in the transcriptional regulation of the ciliome.
(A) Representative micrographs of kap-1 reporter expression in different genetic backgrounds. kap-1::gfp is unaffected in fkh-8(tm292) and ectopically expressed both in daf-19(m86); daf-12(sa204) and daf-12(sa204); daf-19(m86), fkh-8(tm292). Ectopic expression is more evident in the posterior part of the head, labeled with a white line. (B) Quantification of kap-1 reporter expression in different subpopulations of sensory ciliated neurons shows similar expression in fkh-8(vlc43) null mutants and wild type animals. N≥19. (C) Representative micrographs of embryonic ift-20::rfp expression after ectopic expression of DAF-19D and FKH-8 through a heatshock inducible promoter. Two-cell stage embryos were grown at 20 °C for 4 hours and then heat shocked at 37 °C for 20 min, embryos were scored for ectopic ift-20::rfp 20 hours after heatshock. Two different lines were analyzed for each construct. 100% of embryos with DAF19D ectopic expression show ectopic ift-20::rfp expression, while heatshocked embryos without array had normal ift-20::rfp expression (Line 1 n=24 embryos with array, n=11 embryos without array; Line 2 n=21 embryos with array, n=12 embryos without array). Ectopic FKH-8 expression had no obvious effect on ift-20::rfp. (Line 1 n=10 embryos with array, n=5 embryos without array; Line 2 n=15 embryos with array, n=3 embryos without array). (D) Quantification of ciliome reporters in fkh-8 and daf-19 mutants. The same extrachromosomal line was analyzed in the different genetic backgrounds. Each dot represents the number of reporter-expressing neurons scored in a single animal. Mean and standard error are represented. For both reporters, triple mutant is significantly different from each of the double mutants and significantly lower than the expected from the multiplicative effect of both daf-12(sa204); fkh-8(tm292) and daf-12(sa204); daf-19(m86) animals. n≥6.
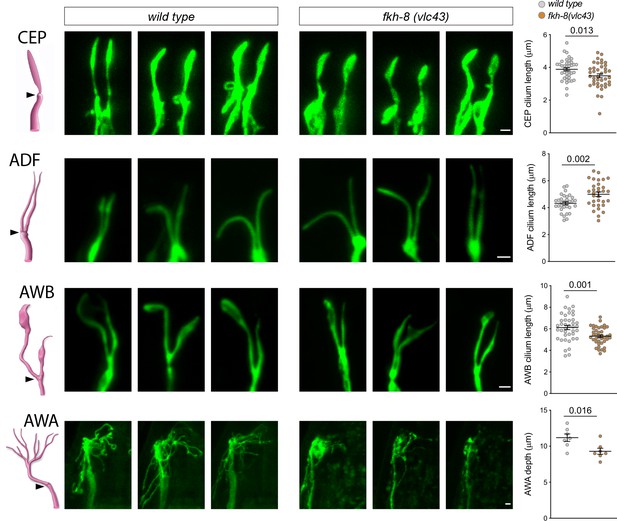
fkh-8(vlc43) null mutants display morphological defects in cilia.
Integrated reporters unaffected in fkh-8 mutant are used to label the cilia of several distinct subpopulations of ciliated neurons. CEP: otIs259(dat-1::gfp); ADF: zdIs13(tph-1::gfp); AWB: kyIs104(str-1::gfp); AWA: pkIs583(gpa-6::gfp). Panels show representative images from three animals in wild type and fkh-8(vlc43) mutant backgrounds. Cilium length of CEP and AWB neurons is significantly reduced in the absence of FKH-8 whereas ADF cilia length is increased. Depth of AWA cilium arborization is significantly reduced in fkh-8(vlc43) null mutants. Each dot in the graphs represents measures for a single cilium. Mean and standard error are represented. See Figure 5—figure supplement 1 for cilia morphology analysis in worms immobilized in polystyrene beads and Figure 5—source data 1 for raw data and statistics. CEP and AWB n=40; ADF n=32; AWA n=7.
-
Figure 5—source data 1
Raw quantification data of cilia morphology analysis depicted in Figure 5 and Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/89702/elife-89702-fig5-data1-v2.xlsx
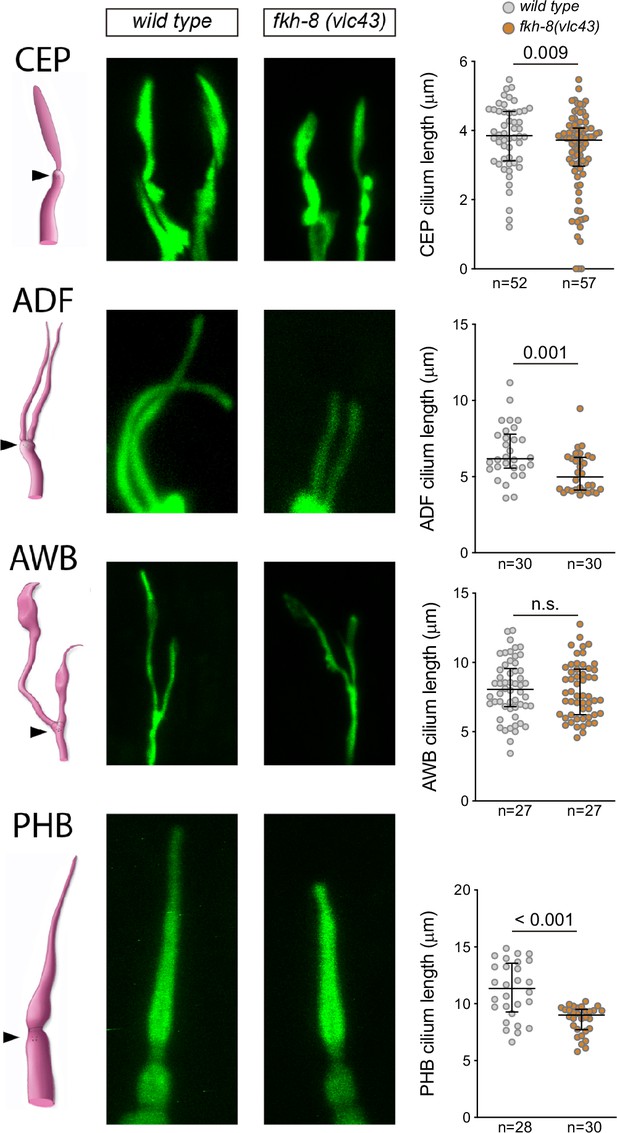
fkh-8(vlc43) null mutants display morphological defects in cilia using physical immobilization with polystyrene beads.
Integrated reporters unaffected in fkh-8 mutant are used to label the cilia of several distinct subpopulations of ciliated neurons. CEP: otIs259(dat-1::gfp); ADF: zdIs13(tph-1::gfp); AWB: kyIs104(str-1::gfp); AWA: pkIs583(gpa-6::gfp). Panels show representative images from wild type and fkh-8(vlc43) mutant backgrounds. Cilium length of CEP ADF and PHB is significantly reduced in the absence of FKH-8, whereas AWB cilia length is unaffected. Mean and standard error are represented. Please note that different fixation methods are used compared to Figure 5. CEP, AWB: n≥50, ADF: n≥30, PHB: n≥28.
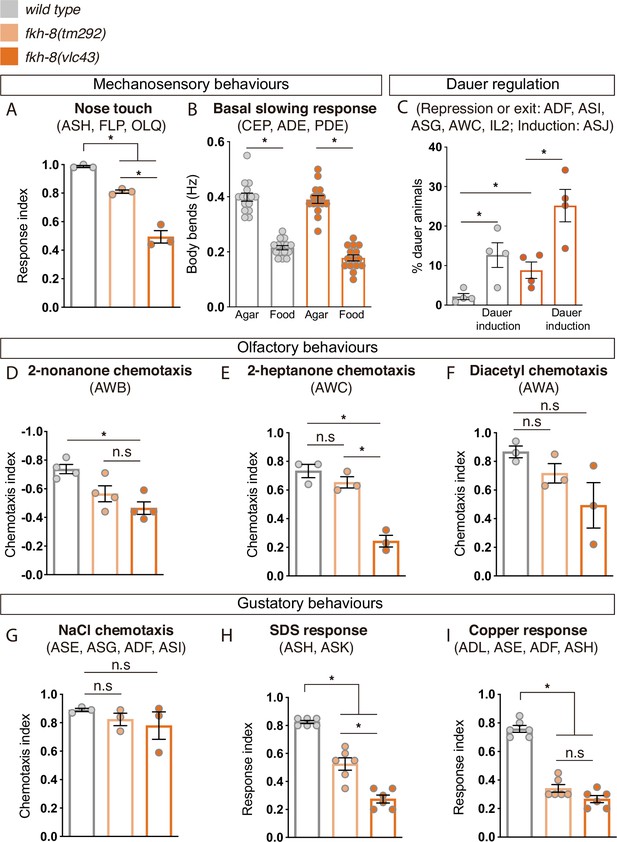
FKH-8 is required for the correct display of several sensory mediated behaviors.
(A) Mutations in fkh-8 significantly impair appropriate backward response to nose touch, revealing functionality defects for the ASH, FLP and/or OLQ ciliated neurons. This phenotype is stronger in fkh-8(vlc43) null mutants than in the hypomorphic tm292 allele. n=20 animals per replicate, three biological replicates per genotype. (B) Decrease in locomotory rate upon re-entering a bacterial lawn is unaffected in fkh-8 mutants. n=15 worms per genotype and condition. (C) fkh-8 null mutants significantly fail to prevent dauer entry. Pheromones induce dauer in fkh-8 mutants, albeit less efficiently than in controls. Four biological replicates n>295 per replicate and genotype. (D to F) Lack of fkh-8 significantly impairs olfaction-mediated behaviors. Defects are observed for 2-nonanone repulsion mediated by AWB [Wild type n=59, 128, 114, 165; fkh-8(tm292) n=76, 123, 129, 209 and fkh-8(vlc43) n=82, 92, 130, 139] and 2-heptanone attraction mediated by AWC neurons [Wild type n=124, 129, 133; fkh-8(tm292) n=68, 94, 102 and fkh-8(vlc43) n=87, 83, 85]. Diacetyl response, mediated by AWA, is affected but not to a significant level due to high variability in the response [Wild type n=168, 69, 103; fkh-8(tm292) n=57, 85, 110 and fkh-8(vlc43) n=115, 107, 74]. (G to I) Attractive chemotaxis towards NaCl is unaffected in fkh-8 mutant animals. [Wild type n=62, 78, 72; fkh-8(tm292) n=105, 116, 106 and fkh-8(vlc43) n=111, 52, 78]. Avoidance behavior towards toxic SDS and copper anions is significantly impaired. [Six biological replicates, 5 worms per replicate and genotype, 4 tests per worm]. Mean and standard error are represented in all graphs. See Figure 6—figure supplement 1 for quantification of non-cilia mediated behaviors and Figure 6—source data 1 for raw data and statistics.
-
Figure 6—source data 1
Raw quantification data for behavioral analysis in Figure 6 and Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/89702/elife-89702-fig6-data1-v2.xlsx
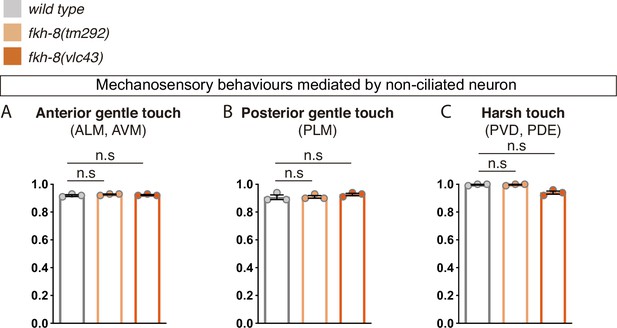
FKH-8 is not required for correct display of mechanosensory behaviors mediated by non-ciliated neurons.
(A to C) fkh-8 mutants show normal avoidance behaviors elicited by mechanical stimuli known as gentle touch and harsh touch paradigms, suggesting FKH-8 is not required for the correct functionality of non-ciliated neurons ALM, AVM, PLM, and PVD. Redundant actions of PVD and PDE controlling scape response to harsh touch prevent to assess defects about the functionality of ciliated PDE neurons. Mean and standard error for three independent replicates are represented. See Figure 6—source data 1 for raw data. n=60 worms per genotype.
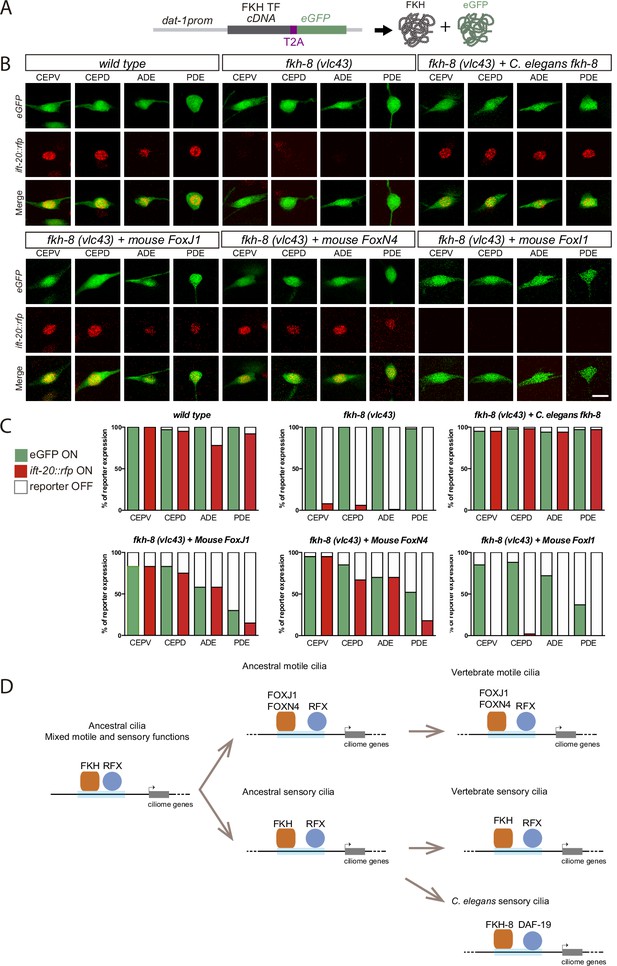
Mammalian FKH TFs with known motile cilia regulatory functions can rescue fkh-8 mutant phenotype.
(A) Rescue strategy: dat-1 promoter, unaffected in fkh-8 mutants, is used to drive FKH TF cDNA and eGFP expression specifically in the dopaminergic ciliated system. (B) Representative images of dopaminergic neurons expressing an integrated reporter for the core ciliome marker ift-20 (in red) in wild type, fkh-8(vlc43) mutants and with the co-expression of different rescuing constructs. Scale bar = 5 µm. (C) Quantification of rescue experiments. ift-20 reporter expression is lost from the dopaminergic neurons in fkh-8(vlc43) null mutants compared to wild type animals. Expression of FKH-8, FOXJ1, and FOXN4 but not FOXI1 is sufficient to recover ift-20 expression in dopaminergic neurons. N=30 animals per transgenic line. See Figure 7—source data 1 for raw data and similar results obtained with two additional transgenic lines per construct. (D) Speculative model on the evolution of ciliome gene regulatory logic. FKH and RFX TFs could have an ancestral role in the direct coregulation of ciliome genes before its functional diversification into motile and primary cilia cell types. Different RFX and FKH TF members could have evolved to regulate ciliome genes in specific cell types in different organisms. Orange squares represent FKH TFs and blue circles RFX TFs, light blue bars represent ciliome enhancers.
-
Figure 7—source data 1
Raw quantification data for rescuing experiments in Figure 7.
- https://cdn.elifesciences.org/articles/89702/elife-89702-fig7-data1-v2.xlsx
Additional files
-
Supplementary file 1
List of reagents: Strains, plasmids and primers.
- https://cdn.elifesciences.org/articles/89702/elife-89702-supp1-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/89702/elife-89702-mdarchecklist1-v2.docx





