Genetic basis of Arabidopsis thaliana responses to infection by naïve and adapted isolates of turnip mosaic virus
Figures
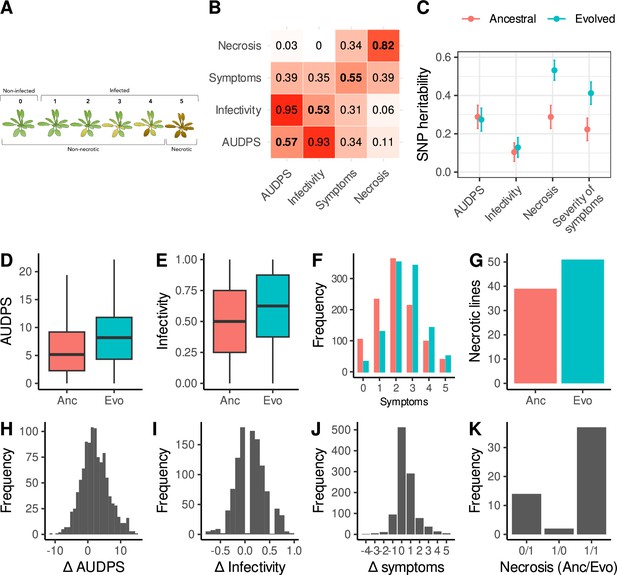
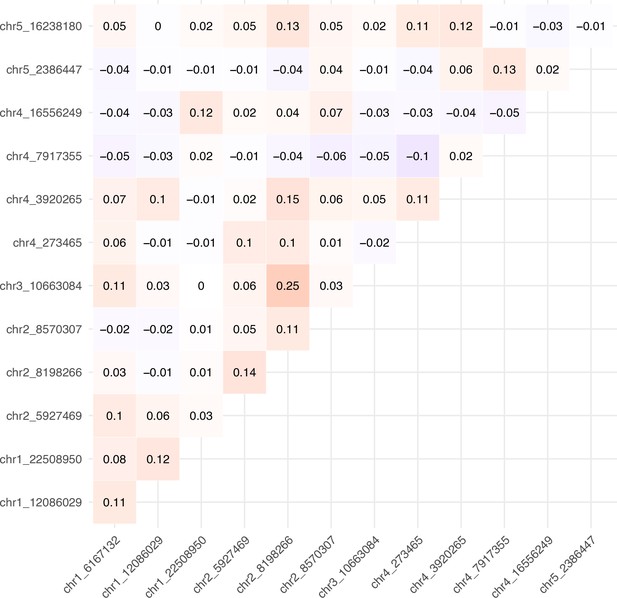
Disease phenotypes.
(A) Illustration of the scale used to evaluate the severity of symptoms: (0) no symptoms or healthy plant (1) mild symptoms without chlorosis, (2) visible chlorosis, (3) advanced chlorosis, (4) strong chlorosis and incipient necrosis, (5) necrosis and death of the plant. (B) Correlation matrix between disease phenotypes in response to the ancestral (upper left) and evolved (lower right) isolates. The diagonal shows correlations between the same phenotype in response to each viral isolate. (C) SNP heritability for each trait in response to each virus. (D–G) Disease phenotypes across lines in response to the ancestral (red) and evolved (blue) turnip mosaic virus (TuMV) isolates. (H–J) Differences in disease phenotypes in response to each viral isolate (evolved minus ancestral). (K) Number of lines showing necrosis in response to the ancestral or/and evolved isolates; 1 indicates the presence of necrosis. For clarity, the 997 lines showing no necrosis for either isolate (0/0) are not shown.
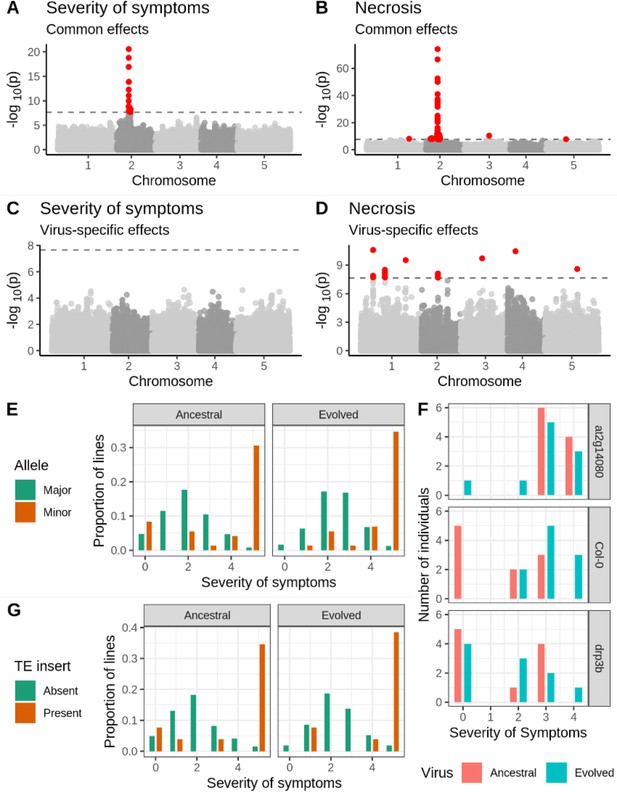
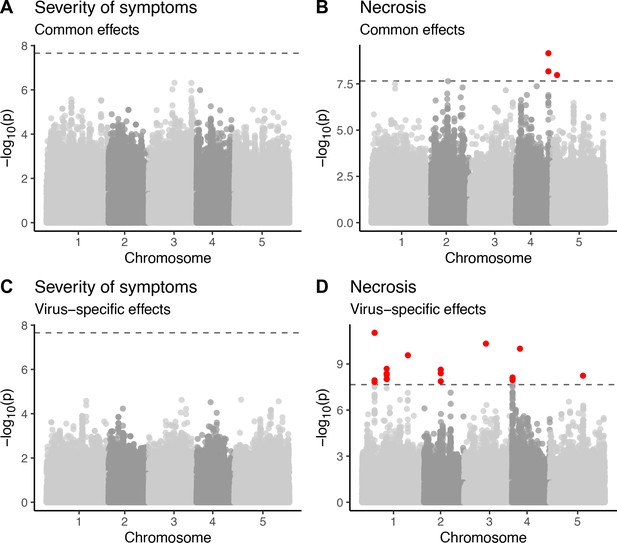
Genetic associations with severity of symptoms and systemic necrosis.
(A, B) associations with a common response to both viral isolates. (C, D) associations with isolate-specific responses. SNPs in red indicate markers with −log10p-values above the Bonferroni-corrected significance threshold. (E) Severity of symptoms in response to each viral isolate for accessions with the major and minor alleles at the most strongly associated SNP. (F) Severity of symptoms in response to each virus for plants of T-DNA knockout mutants for two major candidate genes and Col-0 wild-type controls. (G) Severity of symptoms in response to each virus for lines with and without a TE insertion in AT2G14080.
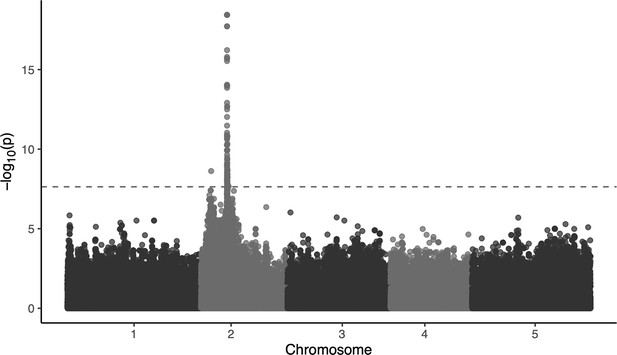
Manhattan plots for the association between SNPs and necrosis in the replicate dataset of 118 lines.
Manhattan plots for associations with severity and symptoms and necrosis in a model conditioned on the most strongly associated SNP (5,927,469 in chromosome 2).
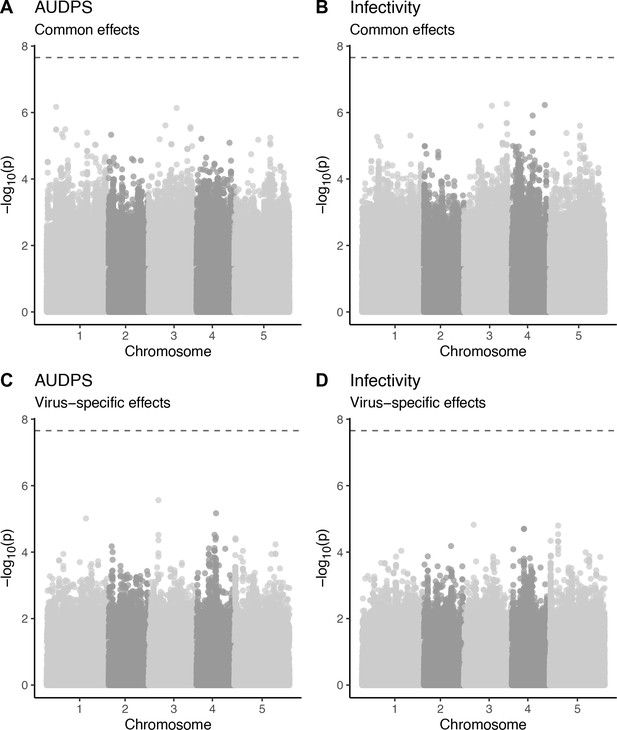
Manhattan plots for common and virus-specific associations with area under the disease progress stairs (AUDPS) and infectivity.
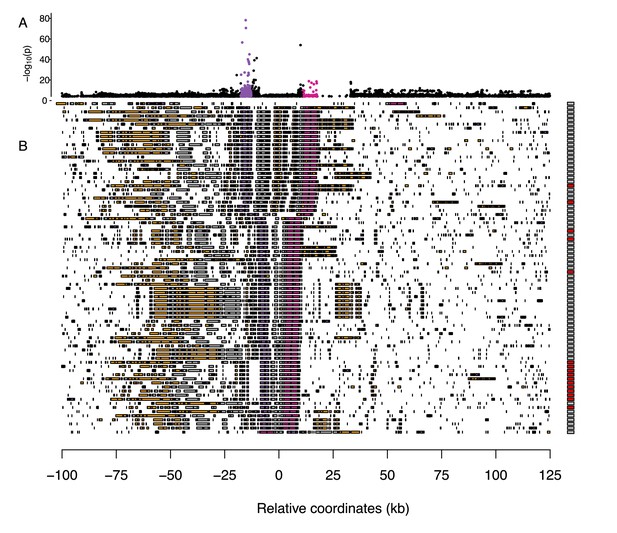
Multiple sequence alignment around the major association with severity of symptoms and necrosis on chromosome 2.
(A) An enlarged view of the peak of association with necrosis in Figure 2B. (B) A summary of structural variation in assembled genomes. Transposable elements are shown in orange and coding genes in gray, with candidate genes AT2G14080 and DRP3B highlighted in purple and pink, respectively. Boxes on the right indicate necrosis (red) in response to either virus or no necrosis (gray). For clarity, only half of the non-necrotic lines are plotted.
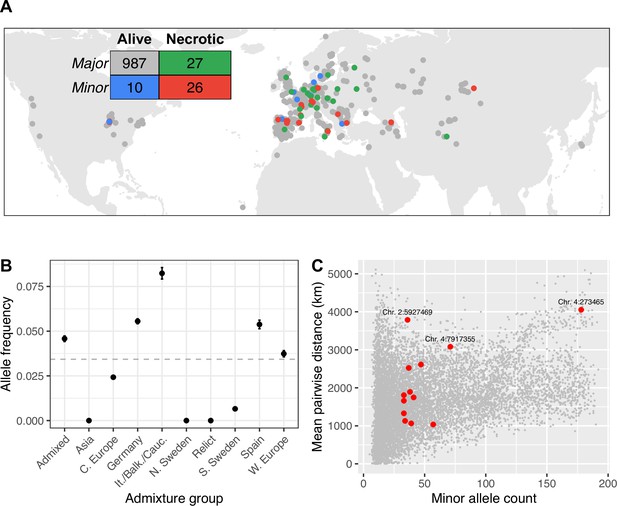
Global distribution of necrotic alleles.
(A) Global distribution of genotypes (minor/major alleles at the SNP most strongly associated with systemic necrosis), and phenotypes (whether lines showed systemic necrosis or not). The inset table shows the numbers of each genotype/phenotype; colors correspond to those on the map. (B) Frequencies of minor alleles at chromosome 2 position 5,927,469 within each admixture group. The horizontal line indicates global frequency. (C) Mean distances between pairs of loci harboring the minor allele at the 13 associated loci (red) and between minor alleles at 10,000 randomly chosen loci.
Additional files
-
Supplementary file 1
Full list of A. thaliana lines for the GWAS analysis. (EXCEL).
- https://cdn.elifesciences.org/articles/89749/elife-89749-supp1-v1.xlsx
-
Supplementary file 2
Significant SNPs in the GWA analysis of the ancestral and evolved TuMV isolates and posterior conditional GWA analysis on the significant SNP on the chromosome 2.
- https://cdn.elifesciences.org/articles/89749/elife-89749-supp2-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/89749/elife-89749-mdarchecklist1-v1.pdf