Positive selection and relaxed purifying selection contribute to rapid evolution of male-biased genes in a dioecious flowering plant
Figures

Floral buds and flowers at anthesis of females (A, B) and males (C) in Trichosanthes pilosa.
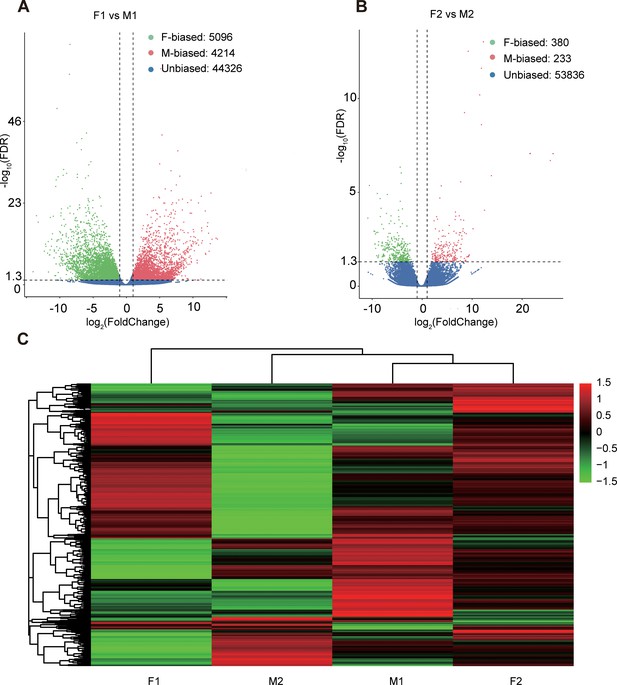
Sex-biased gene expression for floral buds and flowers at anthesis in males and females of Trichosanthes pilosa.
Volcano plots of average expression between female-biased, male-biased and unbiased genes in floral buds (A) and flowers at anthesis (B). M1 and F1 indicate male and female floral buds; M2 and F2 indicate male and female flowers at anthesis. The value of y coordinate represents -log10(FDR), and the value of x coordinate represents log2(Fold Change) identified by DESeq2. Heatmap of sex-biased gene expression (C) using hierarchical clustering analysis. Hierarchical gene clustering is based on Euclidean distance with an average of log2(FPKM) for differentially expressed genes. The color gradient represents from high to low (from red to green) gene expression.
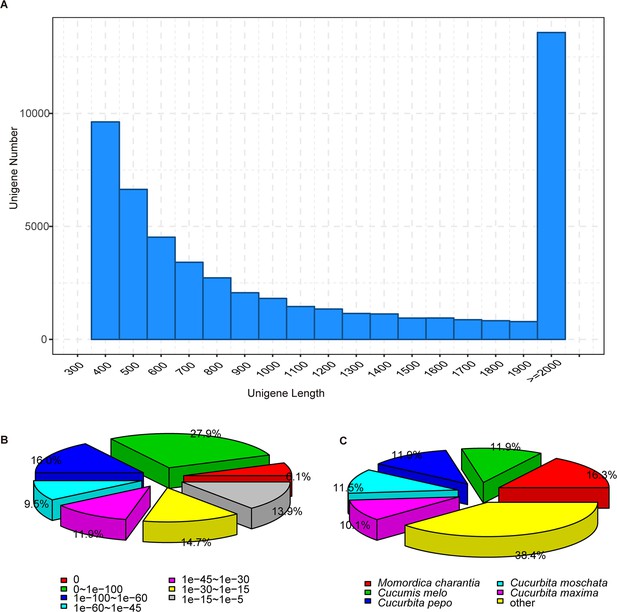
The length distribution of unigenes (A) and the e-value distribution (B) and the species distribution (C) of BLAST hits for each unigene.
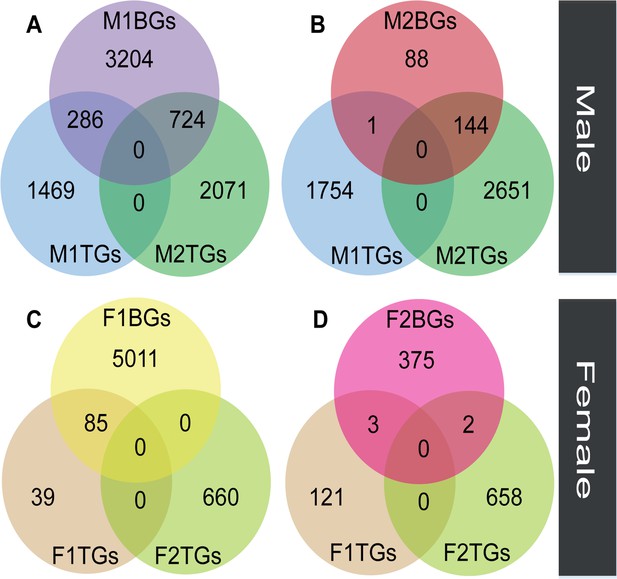
The overlap between sex-biased and tissue-biased genes in two types of sexes and tissues.
Male-biased genes in floral buds (M1BGs) (A) or flowers at anthesis (M2BGs) (B) overlapped with tissue-biased genes in floral buds (M1TGs) and flowers at anthesis (M2TGs). Female-biased genes in floral buds (F1BGs) (C) or flowers at anthesis (F2BGs) (D) overlapped with tissue-biased genes in floral buds (F1TGs) and flowers at anthesis (F2TGs).

The overlap between male-biased genes with faster evolutionary rates and tissue-biased genes in floral buds.
M1BGs indicate male-biased genes with faster evolutionary rates in male floral buds (M1-biased genes). M1TGs indicate tissue-biased genes in male floral buds (M1-tissue-biased genes). 343 M1BGs overlapped with 1755 M1TGs (A). 27 out of 343 M1BGs & M1TGs overlapped with M1BGs_98 (B). 27 out of 343 M1BGs & M1TGs overlapped with M1BGs_18 (C).
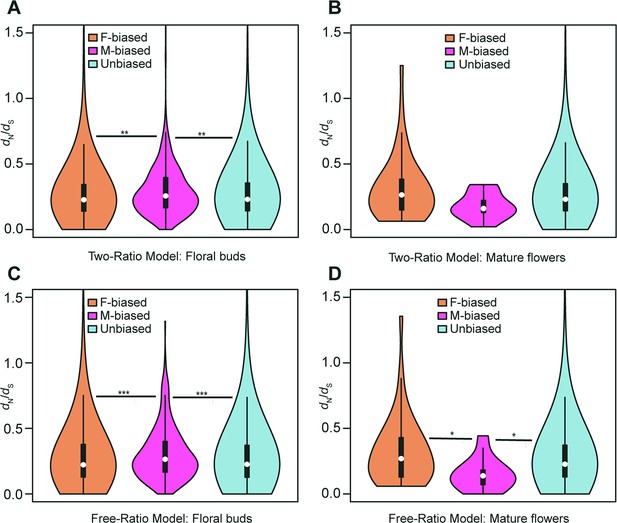
Violin plots of dN/dS values (0<ω<2) of female-biased, male-biased, and unbiased genes in floral buds and flowers at anthesis of Trichosanthes pilosa.
White dot indicates the median of dN/dS values for sex-biased and unbiased genes. Wilcoxon rank sum tests are used to test for significant differences (***p<0.0005, **p<0.005, and *p<0.05). The distributions of dN/dS values for female-biased, male-biased and unbiased genes in floral buds (A) and flowers at anthesis (B) using ‘two-ratio’ branch model. The distributions of dN/dS values for female-biased, male-biased and unbiased genes in floral buds (C) and flowers at anthesis (D) using ‘free-ratio’ branch model.
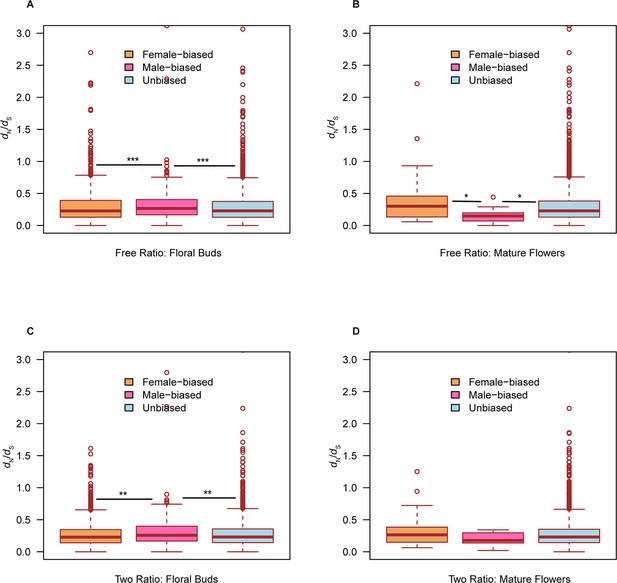
Boxplot of dN/dS values (including all ω values) of female-biased, male-biased, and unbiased genes in floral buds and flowers at anthesis of Trichosanthes pilosa.
White dot indicates the median of dN/dS values for sex-biased and unbiased genes. Wilcoxon rank sum tests are used to test for significant differences (***p<0.0005, **p<0.005, and *p<0.05). The distributions of dN/dS values for female-biased, male-biased, and unbiased genes in floral buds (A) and flowers at anthesis (B) using ‘free-ratio’ branch model. The distributions of dN/dS values for female-biased, male-biased, and unbiased genes in floral buds (C) and flowers at anthesis (D) using ‘two-ratio’ branch model.

Violin plots of effective number of codons (ENCs) values of female-biased, male-biased and unbiased genes in floral buds.
Significant differences using Wilcoxon rank sum tests are represented by * (***p<0.0005 and *p<0.05).
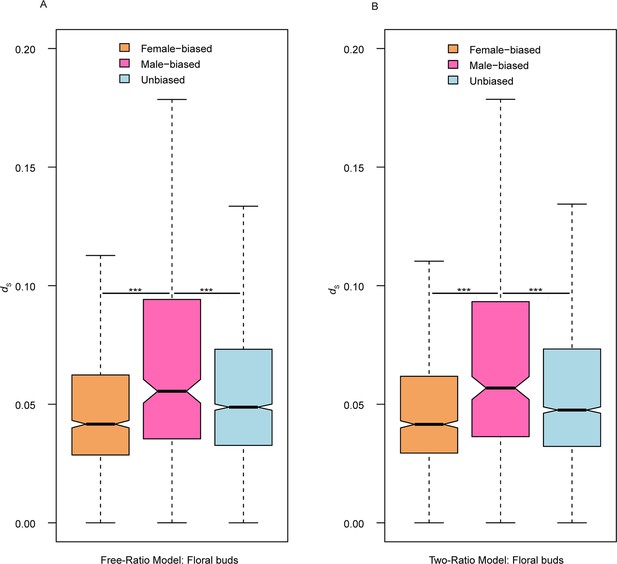
Boxplot of dS values of female-biased, male-biased, and unbiased genes in floral buds of dioecious Trichosanthes pilosa using ‘free-ratio’ (A) and ‘two-ratio’ (B) branch model.
Significant differences are represented by * in Wilcoxon rank sum tests (***p<0.0005).
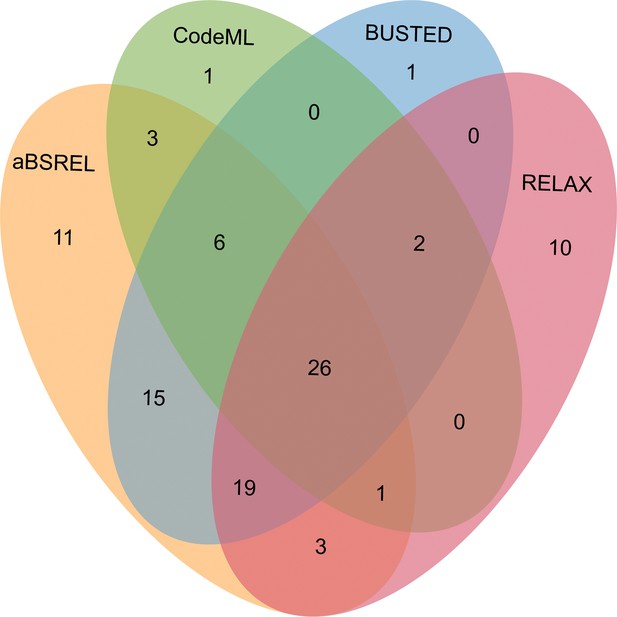
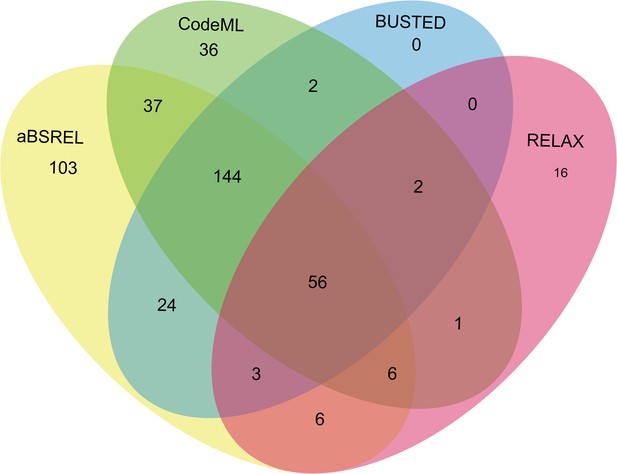
Venn diagrams of male-biased genes detected to be under positive selection using aBSREL, BUSTED, CodeML, and RELAX in floral buds.

Venn diagrams of female-biased genes under positive selection in floral buds.
Overlaps of female-biased genes were detected to be under positive selection using aBSREL, BUSTED, CodeML, and RELAX.
Venn diagrams of unbiased genes under positive selection in floral buds.
Overlaps of unbiased genes were identified to be under positive selection using aBSREL, BUSTED, CodeML, and RELAX.
Additional files
-
Supplementary file 1
Overview of sequencing reads from 12 samples of male and female plants in Trichosanthes pilosa.
- https://cdn.elifesciences.org/articles/89941/elife-89941-supp1-v1.xls
-
Supplementary file 2
Numbers of unigenes annotated in public databases.
- https://cdn.elifesciences.org/articles/89941/elife-89941-supp2-v1.xls
-
Supplementary file 3
The mapping rate of reads for each sample in floral buds and flowers at anthesis of Trichosanthes pilosa.
- https://cdn.elifesciences.org/articles/89941/elife-89941-supp3-v1.xls
-
Supplementary file 4
dN, dS, and ω values of each female-biased, male-biased, unbiased orthologous genes of floral buds and flowers at anthesis for each species using ‘two-ratio’ branch model of CodeML in PAML.
Two dioecious species Trichosanthes pilosa and T. kirilowii represent the foreground and two monoecious species T. anguina and Luffa cylindrica represent the background.
- https://cdn.elifesciences.org/articles/89941/elife-89941-supp4-v1.xls
-
Supplementary file 5
dN, dS, and ω values of each female-biased, male-biased, unbiased orthologous genes of floral buds and flowers at anthesis for each species using ‘free-ratio’ branch model of CodeML in PAML.
- https://cdn.elifesciences.org/articles/89941/elife-89941-supp5-v1.xls
-
Supplementary file 6
Genes under positive selection identified by branch-site model of CodeML in PAML and function in NR, KEGG, Swissport, and GO databases for male-biased orthologous genes in floral buds.
Two dioecious species Trichosanthes pilosa and T. kirilowii represent the foreground and two monoecious species, T. anguina and Luffa cylindrica represent the background.
- https://cdn.elifesciences.org/articles/89941/elife-89941-supp6-v1.xls
-
Supplementary file 7
Genes under episodic positive selection tested by aBSREL model in HyPhy and functions in NR, KEGG, Swissport, and GO databases for male-biased orthologous genes in floral buds.
The dioecious species Trichosanthes pilosa represents the foreground and other species represent the background.
- https://cdn.elifesciences.org/articles/89941/elife-89941-supp7-v1.xls
-
Supplementary file 8
Genes under episodic positive selection found by BUSTED model in HyPhy and functions in NR, KEGG, Swissport, and GO databases for male-biased orthologous genes in floral buds.
The dioecious species Trichosanthes pilosa represents the foreground (unconstrained branch) and other species represent the background (constrained branch).
- https://cdn.elifesciences.org/articles/89941/elife-89941-supp8-v1.xls
-
Supplementary file 9
Genes under relaxed selection detected by RELAX model in HyPhy and functions in NR, KEGG, Swissport, and GO databases for male-biased orthologous genes in floral buds.
The dioecious species Trichosanthes pilosa represents the foreground (test) and other species represent the background (reference).
- https://cdn.elifesciences.org/articles/89941/elife-89941-supp9-v1.xls
-
Supplementary file 10
Genes under intensified positive selection identified by RELAX model in HyPhy and functions in NR, KEGG, Swissport, and GO databases for male-biased orthologous genes in floral buds.
The dioecious species Trichosanthes pilosa represents the foreground (test) and other species represent the background (reference).
- https://cdn.elifesciences.org/articles/89941/elife-89941-supp10-v1.xls
-
Supplementary file 11
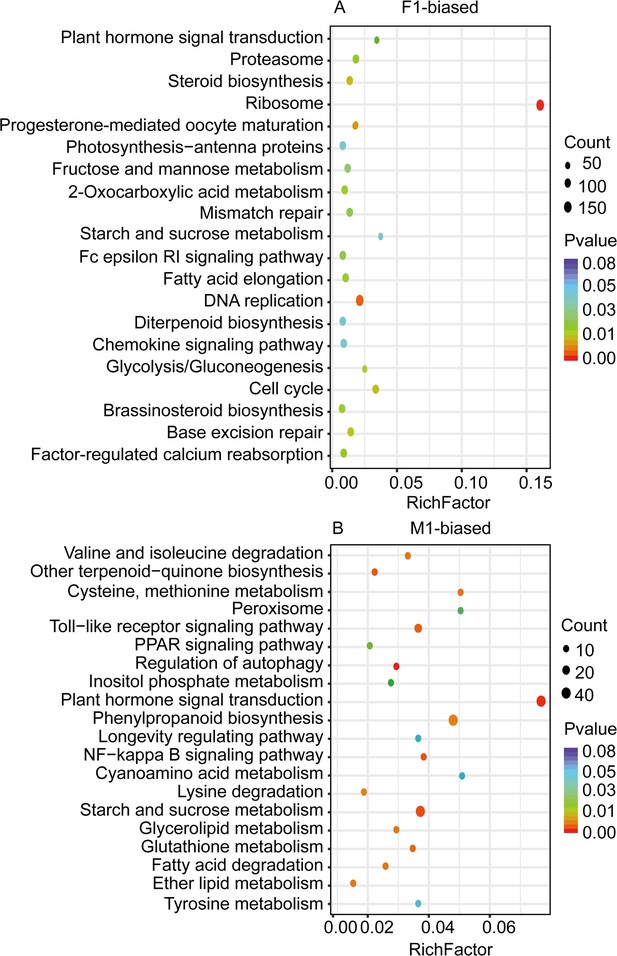
KEGG pathway enrichment analysis of female-biased and male-biased genes in floral buds of the dioecious Trichosanthes pilosa.
- https://cdn.elifesciences.org/articles/89941/elife-89941-supp11-v1.xls
-
Supplementary file 12
Functions and references associated with abiotic stress and immune responses, organ developments of male-biased genes under significant positive selection (p<0.05) in floral buds.
- https://cdn.elifesciences.org/articles/89941/elife-89941-supp12-v1.docx
-
Supplementary file 13
Functions and references associated with abiotic stress and immune responses, organ developments of male-biased genes under significant relaxed selection (p<0.05) in floral buds.
- https://cdn.elifesciences.org/articles/89941/elife-89941-supp13-v1.docx
-
Supplementary file 14
The expressions and functions of some male-biased genes associated with senescence, raceme inflorescence development and early flowering in floral buds.
- https://cdn.elifesciences.org/articles/89941/elife-89941-supp14-v1.xls
-
MDAR checklist
- https://cdn.elifesciences.org/articles/89941/elife-89941-mdarchecklist1-v1.docx
-
Source data 1
Reference transcriptome, orthology data, and alignments.
- https://cdn.elifesciences.org/articles/89941/elife-89941-data1-v1.zip