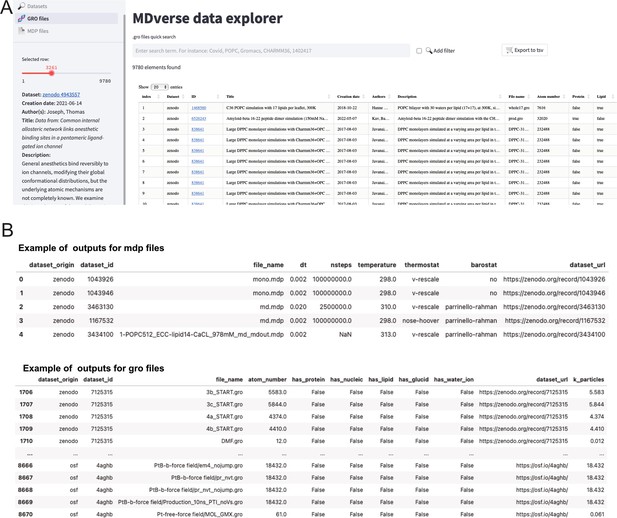
MDverse, shedding light on the dark matter of molecular dynamics simulations
Figures
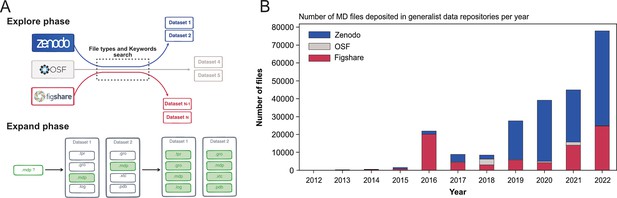
Explore and Expand ($Ex^2$) strategy used to index MD-related files and number of deposited files in generalist data repositories, identified by this strategy.
(A) Explore and Expand () strategy used to index and collect MD-related files. Within the explore phase, we search in the respective data repositories for datasets that contain specific keywords (e.g. ‘molecular dynamics’, ‘md simulation’, ‘namd’, ‘martini’...) in conjunction with specific file extensions (e.g. ‘mdp’, ‘psf’, ‘parm7’...), depending on their uniqueness and level of trust to not report false-positives (i.e. not MD related). In the expand phase, the content of the identified datasets is fully cataloged, including files that individually could result in false positives (such as e.g. ‘.log’ files). (B) Number of deposited files in generalist data repositories, identified by our strategy.
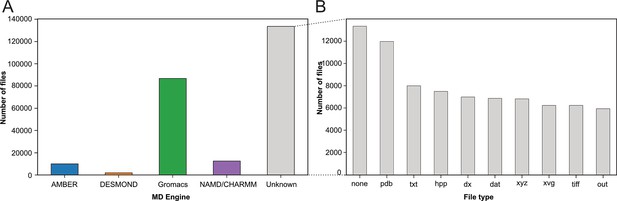
Categorization of index files based on their file types and assigned MD engine.
(A) Distribution of files among MD simulation engines (B) Expansion of (A) MD Engine category ‘Unknown’ into the 10 most observed file types.
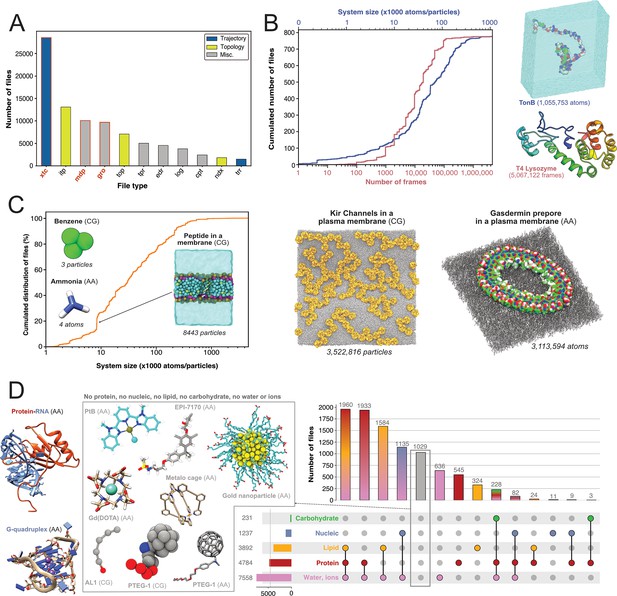
Content analysis of .xtc and .gro files.
(A) Number of Gromacs-related files available in searched data repositories. In red, files used for further analyses. (B) Simple analyze of a subset of .xtc files with the cumulative distribution of the number of frames (in green) and the system size (in orange). (C) Cumulative distribution of the system sizes extracted from .gro files. (D) Upset plot of systems grouped by molecular composition, inferred from the analysis of .gro files. For this figure, 3D structures of representative systems were displayed, including soluble proteins such as TonB and T4 Lysozyme, membrane proteins such as Kir Channels and the Gasdermin prepore, Protein-/RNA and G-quadruplex and other non-protein molecules.
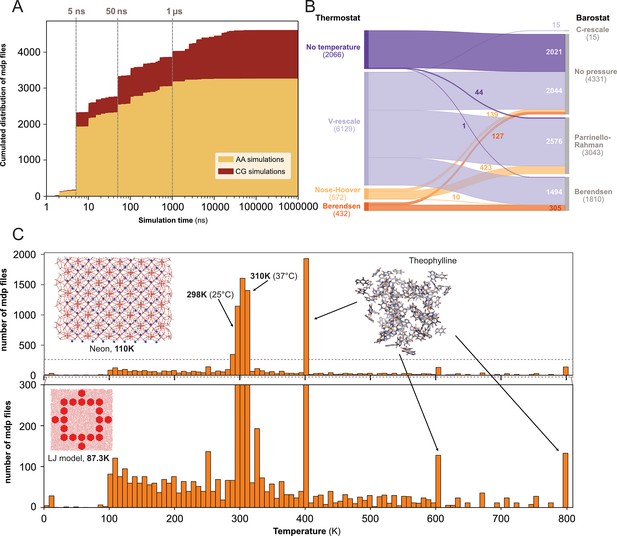
Content analysis of .mdp files.
(A) Cumulative distribution of .mdp files versus the simulation time for all-atom and coarse-grain simulations. (B) Sankey graph of the repartition between different values for thermostat and barostat. (C) Temperature distribution, full scale in upper panel and zoom-in in lower panel.
Tables
Statistics of the MD-related datasets and files found in the data repositories Figshare, OSF, and Zenodo.
| Data repository | datasets | first dataset | latest dataset | files | total size (GB) | zip files | files within zip | total files |
|---|---|---|---|---|---|---|---|---|
| Zenodo | 1011 | 19/11/2014 | 05/03/2023 | 20,250 | 12,851 | 1780 | 141,304 | 161,554 |
| Figshare | 913 | 20/08/2012 | 03/03/2023 | 3336 | 736 | 590 | 74,720 | 78,056 |
| OSF | 55 | 24/05/2017 | 05/02/2023 | 6146 | 495 | 14 | 0 | 6146 |
| Total | 1979 | – | – | 29,732 | 14,082 | 2384 | 216,024 | 245,756 |