Avoiding false discoveries in single-cell RNA-seq by revisiting the first Alzheimer’s disease dataset
Figures
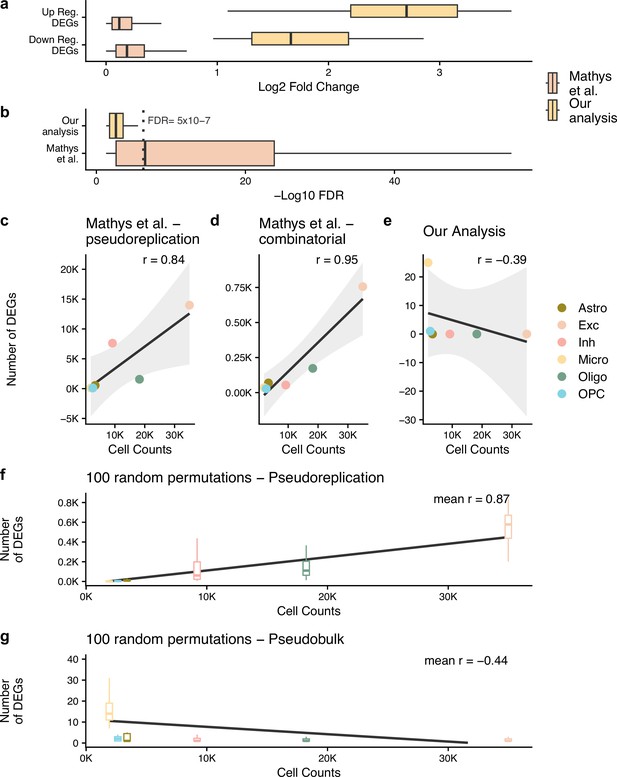
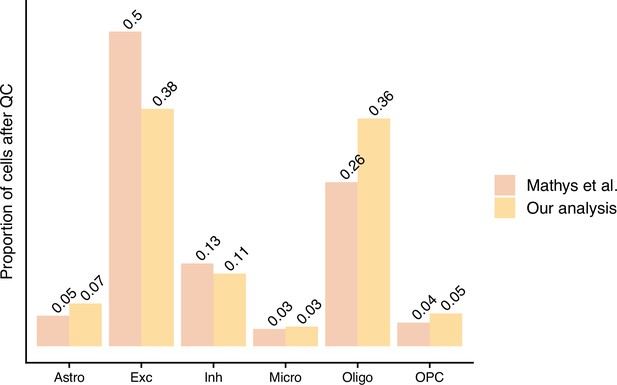
Pseudobulk differential expression results in far less dubious disease-related genes.
(a, b) The log2 fold change and -log10 false discovery rate (FDR) of the differentially expressed genes (DEGs) from the authors’ original work (Mathys et al.) and our reanalysis (Our analysis). In (b), we have marked an FDR of 5 × 10–7, dashed grey line, to highlight how small the p-values from Mathys et al.’s analysis are. For (a, b), n is based on the number of DEGs: 26 for our analysis and 23,923 for Mathys et al. (c–g) show the Pearson correlation between the cell counts after quality control (QC) and the number of DEGs identified - n is the 6 cell types tested. For (f, g) analysis, the samples have been randomly mixed between case and control patients - n = 100 random permutations. The different cell types are astrocytes (Astro), excitatory neurons (Exc), inhibitory neurons (Inh), microglia (Micro), oligodendrocytes (Oligo), and oligodendrocyte precursor cells (OPC).
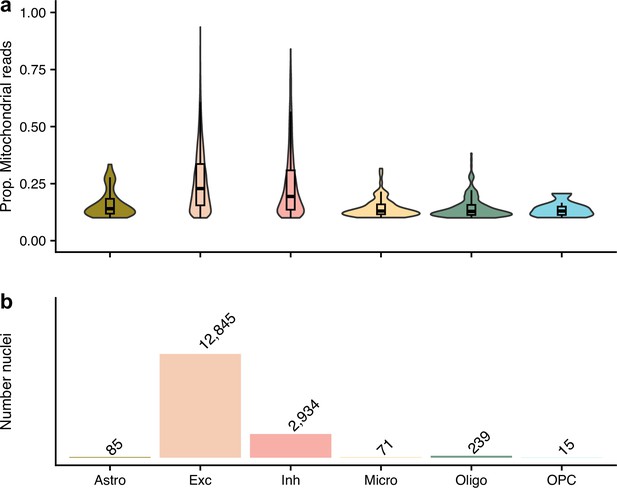
The nuclei that were removed from our quality control approach as their proportion of mitochondrial reads were ≥10%, but kept in the authors’.
(a) shows the proportion of mitochondrial reads across the different cell types. (b) gives the number of removed nuclei which were kept by the authors. The different cell types are astrocytes (Ast), excitatory neurons (Ex), inhibitory neurons (In), microglia (Mic), oligodendrocytes (Oli), and oligodendrocyte precursor cells (Opc).
Tables
Overview of the aggregated number of cells across samples removed at each step of the quality control (QC) as part of scFlow.
Note that cells can fail QC for more than one check, so only the total failed and total passed rows will sum to 100%.
| QC steps | Total cells | Percentage |
|---|---|---|
| Pre-QC | 35,389,440 | |
| Total failed | 35,337,874 | 99.85 |
| Minimum library size (n < 200) | 35,307,281 | 99.77 |
| Maximum library size | 4742 | 0.01 |
| Minimum expressed genes (n < 200) | 35,312,434 | 99.78 |
| Maximum library size/expressed genes (MAD> 4) | 2149 | 0.01 |
| Proportion of mitochondrial genes (≥ 0.1) | 1,097,738 | 3.10 |
| Multiplets (pK = 0.0054) | 581 | 0.00 |
| Total passed | 51,566 | 0.15 |
-
MAD, median absolute deviation.
The differentially expressed genes from our reanalysis using the same processed data the authors used and pseudobulk differential expression approach.
| Cell | logFC | logCPM | LR | p-Value | adj_pval | HGNC |
|---|---|---|---|---|---|---|
| Mic | 2.70178913 | 6.99794619 | 26.1418415 | 3.17E-07 | 0.00061349 | ACRBP |
| Mic | 1.48930071 | 8.06240877 | 28.6361217 | 8.73E-08 | 0.00019303 | APOC1 |
| Mic | 1.09327669 | 8.64199769 | 21.5323014 | 3.48E-06 | 0.00336416 | CD81 |
| Mic | –1.4157681 | 7.93884875 | 23.9955467 | 9.66E-07 | 0.00135806 | CD83 |
| Mic | 3.3782727 | 6.86183548 | 32.0804401 | 1.48E-08 | 4.58E-05 | CLEC1B |
| Mic | 2.84072452 | 6.74370542 | 21.7745509 | 3.07E-06 | 0.00316269 | EGF |
| Mic | 2.55769658 | 6.78345087 | 18.0468872 | 2.16E-05 | 0.01699007 | ELOVL7 |
| Mic | –1.2056098 | 8.33197499 | 22.6644045 | 1.93E-06 | 0.00229576 | IFI44L |
| Mic | –1.6616069 | 7.15366639 | 16.4801274 | 4.92E-05 | 0.03306938 | IFI6 |
| Mic | –1.9809425 | 7.00396289 | 17.9180823 | 2.31E-05 | 0.01699007 | IFIT3 |
| Mic | 2.76502672 | 6.72978805 | 20.6543637 | 5.50E-06 | 0.00472825 | ITGA2B |
| Mic | 1.90963403 | 7.01552233 | 16.3200189 | 5.35E-05 | 0.03448474 | MAP1A |
| Mic | –1.8194508 | 8.26208887 | 45.2221008 | 1.76E-11 | 1.36E-07 | NAMPT |
| Mic | 2.0945044 | 7.11048456 | 20.8068524 | 5.08E-06 | 0.00462318 | NEXN |
| Mic | –2.3789762 | 6.93896985 | 22.3912441 | 2.22E-06 | 0.00245752 | NR4A2 |
| Mic | –2.8553462 | 6.73713862 | 22.8029868 | 1.79E-06 | 0.00229576 | NR4A3 |
| Mic | 3.32873829 | 6.84942721 | 30.955327 | 2.64E-08 | 6.81E-05 | PF4 |
| Mic | 3.4213986 | 6.87326383 | 33.2621657 | 8.05E-09 | 3.11E-05 | PKHD1L1 |
| Mic | 3.64525677 | 6.93422174 | 38.661272 | 5.04E-10 | 2.60E-06 | PPBP |
| Mic | 2.30482679 | 8.10570443 | 60.7932697 | 6.34E-15 | 9.81E-11 | PTPRG |
| Mic | –1.0382468 | 8.11450266 | 15.5968273 | 7.84E-05 | 0.04850839 | RORA |
| Mic | 2.54636649 | 6.69202981 | 17.2532606 | 3.27E-05 | 0.02300507 | SDPR |
| Mic | –0.9629617 | 8.8434334 | 17.9319131 | 2.29E-05 | 0.01699007 | SYTL3 |
| Mic | –1.4215374 | 7.99629806 | 25.4736272 | 4.48E-07 | 0.00077092 | TMEM2 |
| Mic | 2.98901596 | 6.77276641 | 24.2100819 | 8.64E-07 | 0.00133637 | TUBB1 |
| Opc | –2.8274718 | 5.03371292 | 22.1334581 | 2.54E-06 | 0.04176231 | EGR1 |
-
CPM - Counts per Million, LR - fold change ratio, HGNC - HUGO Gene Nomenclature Committee.
Pearson correlation between our pseudobulk differential expression analysis and the authors’ pseudoreplication analysis on all genes found to be significant at different adjusted p-value cut-offs from the authors’ pseudoreplication analysis.
| Pseudoreplication adjusted p-value cut-off | Number of genes compared | Pearson correlation |
|---|---|---|
| 0.01 | 20,152 | 0.8646269 |
| 0.05 | 23,903 | 0.8708275 |
| 0.1 | 26,382 | 0.8721126 |
| 0.25 | 32,117 | 0.8764692 |
| 0.5 | 42,022 | 0.8751554 |
| 1 | 84,467 | 0.826248 |
The differentially expressed genes from our reanalysis using the reprocessed data and pseudobulk differential expression approach.
| Cell | logFC | logCPM | LR | p-Value | adj_pval | ensembl_id | HGNC |
|---|---|---|---|---|---|---|---|
| OPC | –4.1544663 | 4.92100803 | 21.6911445 | 3.20E-06 | 0.04985906 | ENSG00000166573 | GALR1 |
| Astro | –4.5845276 | 4.7965143 | 22.2367847 | 2.41E-06 | 0.037634 | ENSG00000137959 | IFI44L |
| Micro | –3.7616619 | 7.32875316 | 26.8149688 | 2.24E-07 | 0.00077905 | ENSG00000077238 | IL4R |
| Micro | –2.0681446 | 7.88736441 | 17.5929095 | 2.74E-05 | 0.0346187 | ENSG00000105835 | NAMPT |
| Micro | –1.6757556 | 7.58472506 | 19.1736829 | 1.19E-05 | 0.02076348 | ENSG00000118257 | NRP2 |
| Micro | –3.1556403 | 6.85232653 | 19.2064627 | 1.17E-05 | 0.02076348 | ENSG00000135363 | LMO2 |
| Micro | –3.4339265 | 6.9290472 | 19.5975589 | 9.56E-06 | 0.02076348 | ENSG00000138135 | CH25H |
| Micro | –2.8183109 | 6.77500676 | 16.907959 | 3.92E-05 | 0.04550806 | ENSG00000142408 | CACNG8 |
| Micro | 2.90076647 | 8.34560617 | 45.5144266 | 1.52E-11 | 2.11E-07 | ENSG00000144724 | PTPRG |
| Micro | 3.25867589 | 6.91671013 | 16.5519147 | 4.73E-05 | 0.0490155 | ENSG00000163106 | HPGDS |
| Micro | –2.0290905 | 7.12321166 | 16.4746746 | 4.93E-05 | 0.0490155 | ENSG00000171612 | SLC25A33 |
| Micro | –3.4657301 | 6.93307221 | 19.7883301 | 8.65E-06 | 0.02076348 | ENSG00000172243 | CLEC7A |
| Micro | –4.172807 | 7.16813583 | 34.3515807 | 4.60E-09 | 3.20E-05 | ENSG00000174600 | CMKLR1 |
| Micro | –3.1984588 | 6.87310555 | 18.5335889 | 1.67E-05 | 0.0232342 | ENSG00000227531 | RP11-202G18.1 |
| Micro | 3.40562887 | 6.9381703 | 18.5526502 | 1.65E-05 | 0.0232342 | ENSG00000228058 | RP11-552D4.1 |
| Micro | 4.46073301 | 7.66559163 | 29.7716679 | 4.86E-08 | 0.00022549 | ENSG00000253496 | RP11-13N12.1 |