BRAIDing receptors for cell-specific targeting
Figures
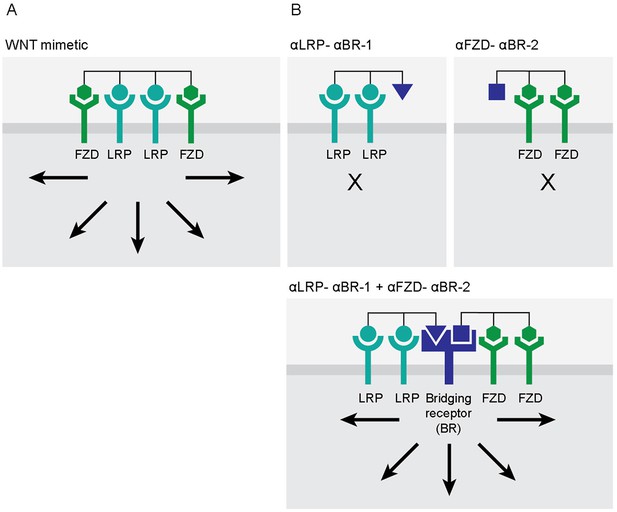
Activation of a signaling pathway by split molecules through binding to a bridging receptor on the target cell surface.
(A) Efficient WNT/β-catenin signaling requires two frizzled (FZD)-binding domains and two lipoprotein receptor-related protein (LRP)-binding domains in one WNT mimetic molecule. (B) WNT mimetic molecule is split into one molecule having two FZD-binding arms (2:0) and a second molecule having two LRP-binding arms (0:2) and then each tethered to a different ‘bridging element’ binding to a different epitope on the same bridging receptor (named αLRP-αBR-1 and αFZD-αBR-2) (top two panels). On target cells where the bridging receptor is expressed, FZD and LRP are then assembled by αLRP-αBR-1 and αFZD-αBR-2 via the bridging receptor, recreating the signaling-competent receptor complexes (bottom panel). Arrows indicate activation of WNT/β-catenin signaling.
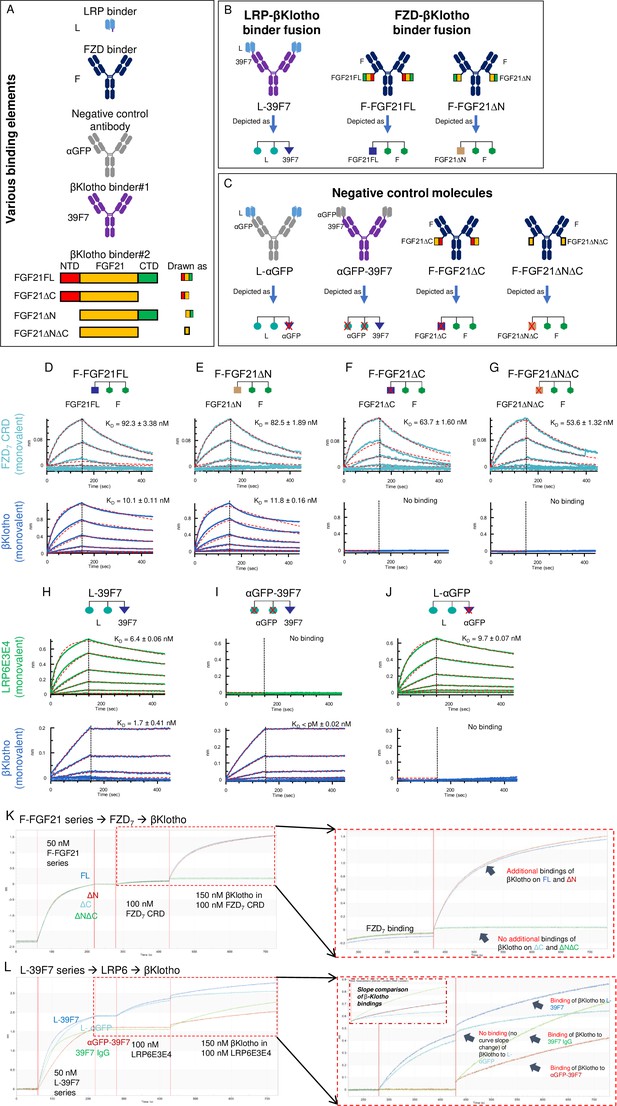
Diagrams of the molecules used in the experiment to provide proof of concept for SWIFT.
(A) Elements that are used in the split molecules. The LRP6 binder element is in scFv format; frizzled (FZD) binder is in IgG1 format; both αGFP IgG1 and αGFP scFv are used for assembly of the negative control molecules. Two types of βKlotho binders are used. Binder#1, 39F7 IgG1, a βKlotho monoclonal antibody; Binder#2, FGF21FL and different deletion variants. (B) Assembled SWIFT molecules. (C) Assembled negative control molecules. (D–G) Bindings of various F-FGF21 fusion proteins to FZD7 and βKlotho. Bindings of FZD7 and β-Klotho to F-FGF21FL (D), F-FGF21ΔN (E), F-FGF21ΔC (F), or F-FGF21ΔNΔC (G) were determined by Octet. (H–J) Binding of LRP6 and βKlotho to L-39F7 (H), αGFP-39F7 (I), or L-αGFP (J) were determined by Octet. Mean KD values were calculated for the binding curves with global fits (red dotted lines) using a 1:1 Langmuir binding model. (K) Step bindings of FZD7 and βKlotho to various F-FGF21 fusion proteins. Sequential binding of F-FGF21FL (blue sensorgram), F-FGF21ΔN (red sensorgram), F-FGF21ΔC (light-blue sensorgram), or F-FGF21ΔNΔC (green sensorgram), followed by FZD7 CRD, then followed by addition of βKlotho on Octet shows simultaneous engagement of both FZD7 and βKlotho to the indicated F-FGF21 proteins. Sensorgrams for FZD7 and βKlotho area (red dotted box) are enlarged at the right. (L) Step binding of LRP6 and βKlotho to L-39F7 and its control proteins. Sequential binding L-39F7 (blue sensorgram), L-αGFP (light-blue sensorgram), αGFP-39F7 (red sensorgram), or 39F7 IgG (green sensorgram), followed by LRP6E3E4, then followed by addition of βKlotho on Octet shows simultaneous engagement of both LRP6 and βKlotho to the indicated L-39F7 and its control proteins. Sensorgrams for LRP6 and βKlotho area (red dotted box) are enlarged on the right.
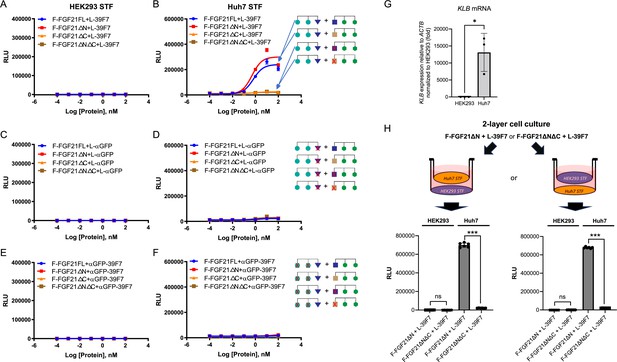
Dose-dependent Super TOP-FLASH (STF) assay of SWIFT molecules in HEK293 and Huh7 cells.
(A–F) Various F-FGF21 fusion proteins in the presence of L-39F7 in HEK293 STF cells (A) and Huh7 STF cells (B); various F-FGF21 fusion proteins in the presence of L-αGFP in HEK293 STF cells (C) and Huh7 STF cells (D); and various F-FGF21 fusion proteins in the presence of αGFP-39F7 in HEK293 STF cells (E) and Huh7 STF cells (F). (G) Expression of bridging receptor βKlotho (KLB) in HEK293 and Huh7 cells normalized to ACTB, relative to HEK293 expression. (H) STF responses in a two-layer cell culture system after 16 hr treatment with 10 nM of indicated molecules. Data are representative of three independent experiments performed in triplicate and are shown as mean ± SD. *p<0.05, ***p≤0.001 (unpaired t-test).
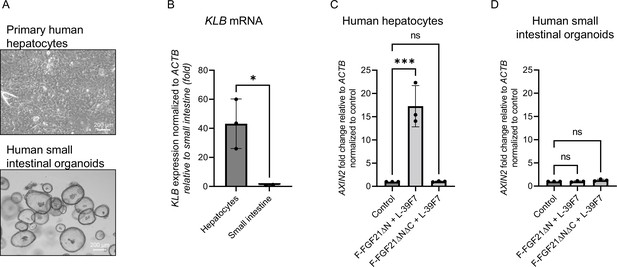
Activity of targeted molecules in primary human cells.
(A) Representative images of primary human hepatocyte cultures in 2D or human small intestinal organoids. Scale bar: 200 µm. (B) Expression of bridging receptor βKlotho (KLB) in hepatocytes or small intestinal cells. (C) WNT target gene AXIN2 expression normalized to control treatment after 24 hr treatment with 10 nM of the indicated molecules in human hepatocytes. (D) WNT target gene AXIN2 expression normalized to control treatment after 24 hr treatment with 10 nM of the indicated molecules in human small intestinal organoids. *p<0.05 (unpaired t-test), ***p≤0.001 (one-way ANOVA), each data point represents an independent experiment performed in duplicate.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | Expi293F cells | Thermo Fisher Scientific | A14527 | |
| Cell line (H. sapiens) | HEK293 STF | https://doi.org/10.1371/journal.pone.0009370 | Cells containing a luciferase gene controlled by a WNT-responsive promoter | |
| Cell line (H. sapiens) | Huh7 STF | https://doi.org/10.1038/s41598-020-70912-3 | Cells containing a luciferase gene controlled by a WNT-responsive promoter | |
| Biological sample (H. sapiens) | Primary hepatocytes | BioIVT | X008001-P | 10-Donor Human Cryoplateable |
| Biological sample (H. sapiens) | Human small intestinal organoids | Kuo Lab Stanford | NA | Primary cell derived 3D organoid culture |
| Peptide, recombinant protein | Human βKlotho | Fisher Scientific | Cat# 5889KB-050 | |
| Peptide, recombinant protein | Human FZD7 CRD | https://doi.org/10.1038/s41467-021-23374-8 | Produced in Expi293F cells | |
| Peptide, recombinant protein | Human LRP6 E3E4 | https://doi.org/10.1016/j.chembiol.2020.02.009 | Produced in Expi293F cells | |
| Peptide, recombinant protein | Fc-R-spondin 2 | https://doi.org/10.1038/s41598-020-70912-3 | Produced in Expi293F cells | |
| Peptide, recombinant protein | Recombinant Human EGF | PeproTech | Cat# AF-100-15 | |
| Peptide, recombinant protein | Recombinant Human Noggin | PeproTech | Cat# 120-10C | |
| Peptide, recombinant protein | Human Gastrin I | Tocris | Cat# 30061 | |
| Commercial assay or kit | Luciferase Assay System | Promega | E1501 | |
| Chemical compound, drug | IWP2 | Tocris Bioscience | Cat# 3533 | Porcupine inhibitor |
| Chemical compound, drug | B27 | Thermo Scientific | Cat# 17504044 | |
| Chemical compound, drug | N2 | Thermo Scientific | Cat# 17502048 | |
| Chemical compound, drug | N-Acetylcysteine | Sigma-Aldrich | Cat# A9165 | |
| Chemical compound, drug | Nicotinamide | Sigma-Aldrich | Cat# N0636 | |
| Chemical compound, drug | A83-01 | Tocris | Cat# 2939 | |
| Chemical compound, drug | SB202190 | Tocris | Cat# 126410 | |
| Software, algorithm | Octet Data Analysis 9.0 | Sartorius | https://www.sartorius.com/en/products/protein-analysis/octet-bli-detection/octet-systems-software | |
| Software, algorithm | Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ | |
| Other | Matrigel Matrix | Corning | CB40230C | Extracellular matrix for 3D organoid growth and plate coating |





