Chromatin and gene expression changes during female Drosophila germline stem cell development illuminate the biology of highly potent stem cells
Figures
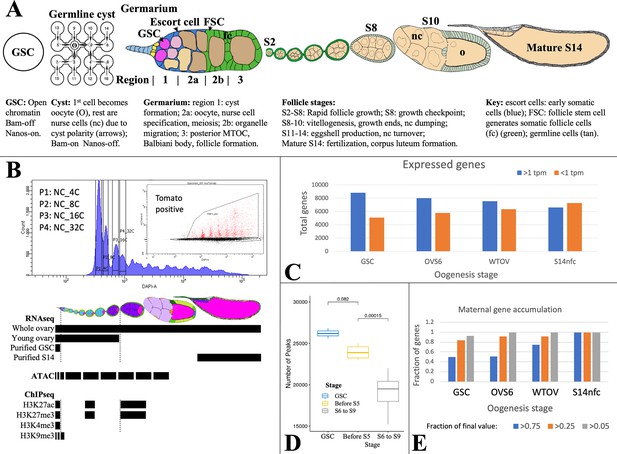
Analyzing Drosophila female germline chromatin.
(A) Stages of female gamete development in an ovariole. A single germline stem cell (GSC) and germline cyst are shown. To the right, a germarium is illustrated showing regions 1–3. Follicles in an ovariole are pictured at a lower magnification starting with stage 2 (S2). Below the diagrams, major events are summarized. (B) Fluorescence-activated cell sorting (FACS) purification (upper panel) of 4C-512C germ cells for ATAC after earlier separation of 2C and 4C GSCs, and 2C-16C follicle cells (see Figure 2). DAPI, DNA content; inset shows further purification based on germ cell marker expression (tomato). Below, follicle stages collected for RNA-seq and Chip-seq are shown by black bars. (C). The number of genes expressed (>1 tpm, blue) and off (<1 tpm, orange) in GSCs decreases as germ cell development proceeds downstream. (D) Box plot showing a decline in total ATAC peaks from GSCs to young ovaries (before S6) to whole ovaries (S6-9). (E) Stem cells already express a high fraction of 79 oocyte maternal effect genes defined in Drosophila genetic screens.
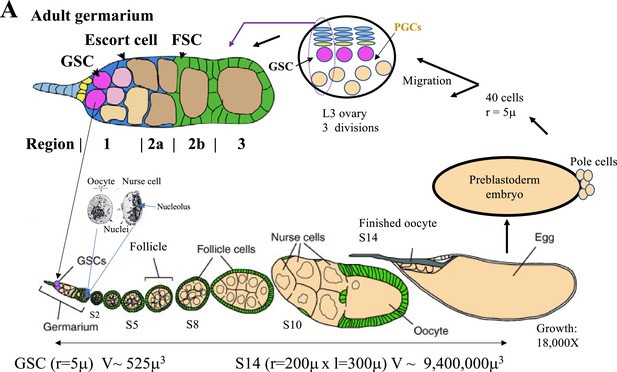
The Drosophila female germ cell cycle.
(A) The Drosophila female germ cell generational cycle. Starting with adult germline stem cells (GSCs, magenta, upper right) germ cells form germline cysts (tan) that move through germarium regions 1–3 and bud off as stage 2 follicles (S2). Mature S14 oocytes are fertilized and undergo 14 preblastoderm syncytial divisions, with pole cells budding off at division 9 to form primordial germ cells (PGCs). About 10 PGCs complete migration to the embryonic ovary, that develops new germaria prior to adulthood. The full cycle entails 19–20 germ cell divisions entailing an estimated >10,000-fold cytoplasmic volume increase.
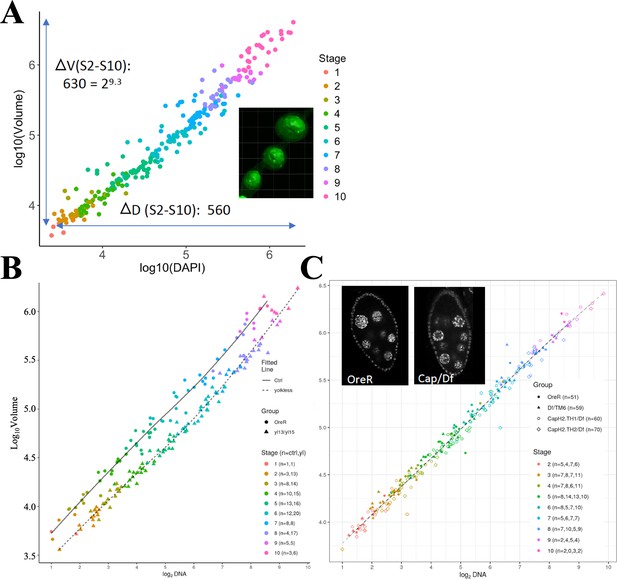
Growth profiles (total germ cell volume) of Drosophila ovarian follicles.
(A) Growth of wild type follicles shown by plotting volume (log10) determined as described in Materials and methods vs integrated DAPI (log10). An example of follicles with germline stem cell (GFP) labeled germ cells is shown at the right. Stages of the follicles are indicated by color key on the right. (B) Similar methods applied to reveal the growth of control vs yolkless follicles. (C) Similar methods used to determine the growth of control and Cap/Df follicles.
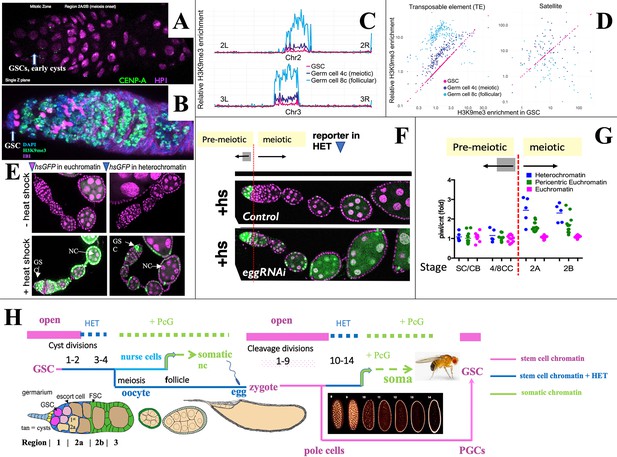
Heterochromatin forms in cysts downstream from the germline stem cell (GSC).
(A,B) Germaria stained for (A) H3K9me3-binding protein HP1a (pink) and CENP-A (green), or (B) H3K9me3 (green), fusome antibody 1B1 (pink) and DAPI (blue). (C) H3K9me3 Chip-seq of unambiguously mapped reads spanning chromosome 2 (lower) and 3 (upper) in 250 kb bins from GSC, 4C nurse cell (NC), or 8C NC. (D) Plots showing relative enrichment of reads mapping to transposable elements (TEs) or to satellite sequences in 4C, and 8C germ cells relative to GSCs. (E). A representative euchromatic hsGFP insertion after heat shock expresses GFP (green) in GSCs and nearly all downstream cyst and follicular NC (arrows). In contrast, a typical heterochromatic insertion after heat shock only expresses in GSCs and early cysts (upper arrow), but has become repressed in 16-cell cysts and NC in meiotic and later follicles (lower arrow; other labeled cells are somatic). (F) Repression of heterochromatic hsGFP expression after heat shock (+hs) (Control) requires the H3K9 methylase eggless/SETDB1 (eggRNAi). (G) Diagram shows the ratio of hsGFP expression in piwi(GLKD) or control (cnt) genetic backgrounds from individual hsGFP insertions (points) at the indicated developmental stages (Figure 1A) colored based on insert location in euchromatin (pink), pericentric chromatin (green), or centric heterochromatin (blue). Note: SC = GSC; CB = cystoblast (first cell of germline cyst). (H) Summary diagram of Drosophila female germ cell generational cycle showing two highly potent stem cell chromatin states (pink) beginning with the GSC or zygote and their downstream developmental trajectories. Both the GSC (maternal) and zygotic trajectories quickly undergo heterochromatin formation (blue) and then somatic cell production dependent on Polycomb repression (green). See text and Figure 1, Figure 1—figure supplement 1 for further details.
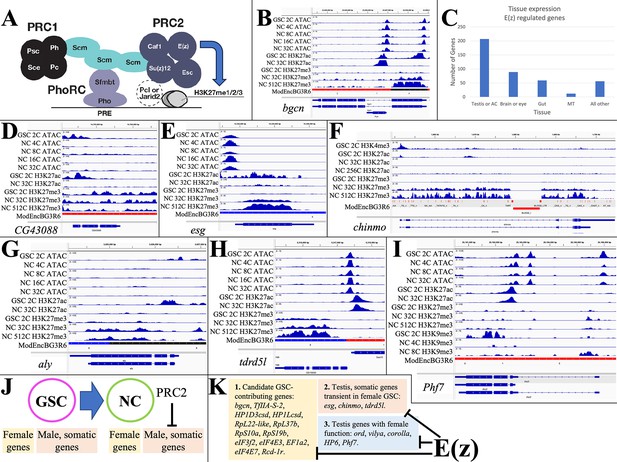
E(z)-dependent repression contributes to sex-differential gene expression.
(A) Diagram of Drosophila PRC1 and PRC2. (B) Chromatin changes around the germline stem cell (GSC) differentiation gene bgcn. The promoter-associated H3K27ac peak is lost by 32cNCs, when H3K27me3 has begun to accumulate. (C) The number of genes strongly downregulated by E(z) in young ovaries and their predominant tissue of expression. (D) Chromatin changes around the CG43088 testis gene related to a transposon. (E) The male GSC gene escargot (esg) downregulated by OVS6 shows increased H3K27me3. (F) The chinmo gene encoding a transcription regulator required in male and female germ cells is repressed as in (E). (G) The testis regulatory gene aly is repressed as in (E). (H) The tdrdl5 male germ cell regulator is repressed as in (E). (I) Phf7 gene regulation. (J) Summary of E(z)-dependent repression of male and somatic genes during early female gene expression. (K) Three subclasses of genes downregulated by E(z).
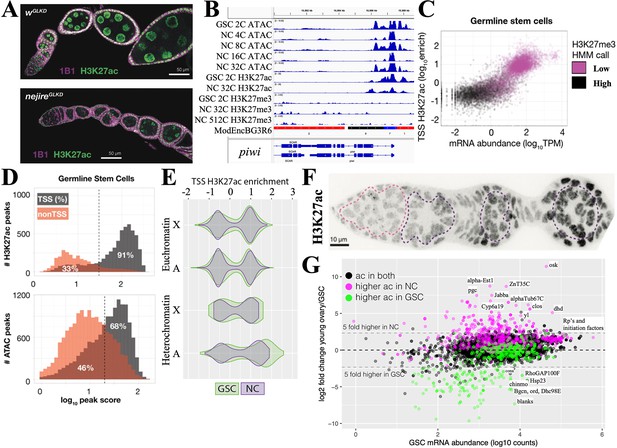
Maintenance and exit from the germline stem cell (GSC) state.
(A) Ovarioles subjected to white gene (wGLKD) or nejire gene (nejireGLKD) germline knockdown. Levels of H3K27ac (green) are greatly reduced in nejGLKD and germ cell development is arrested at stages 1–2. Hts staining (1B1, purple) highlights cell membranes and fusomes. (B) Chromatin tracks around the conserved germline gene (CGG) piwi which is expressed throughout oogenesis. H3K27me3 does not increase. (C) mRNA abundance vs the transcription start site (TSS) levels of H3K27ac are plotted versus GSC gene expression. Genes are colored according to hidden Markov model (HMM) calls of H3K27me3 levels. (D) The number of gene-associated H3K27ac peaks (upper) or ATAC peaks (lower) in GSCs is plotted versus log10 peak score. 81% of H3K27ac peaks and 68% of ATAC peaks are associated with TSSs; the remaining peaks may be associated with enhancer regions. (E) Violin plots showing the distribution of H3K27ac enrichment at transcription start sites (TSS) of genes in GSC (green) and nurse cells (gray). Plots are subdivided by gene location in euchromatin (upper) or heterochromatin (lower), and chromosome ( X or autosome (A)). (F) Germarium immunostained for H3K27ac showing low levels in early germ cells (dashed pink circle) and increase in S1-S3 nurse cells (purple dashed circles). (G) Plot of GSC mRNA abundance vs expression change in young ovary compared to GSCs. A strong correlation can be seen between genes with changes in mRNA levels and changes in H3K27ac levels. Genes (dots) are colored according to changes in ac = H3K27ac levels.
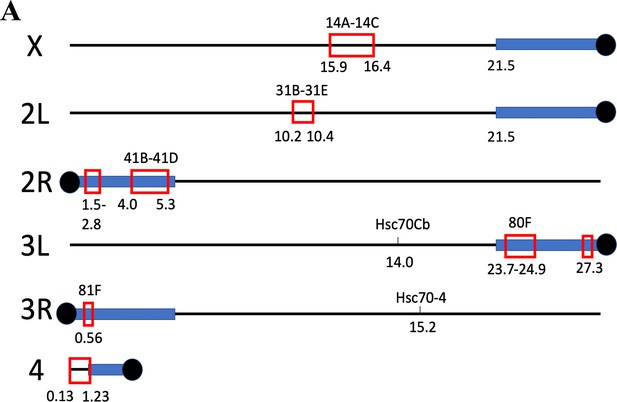
Genes with high H3K27ac levels in germline stem cells (GSCs) cluster in heterochromatin and two euchromatic domains.
(A) Diagram of the six Drosophila chromosome arms showing zones of clustered genes with high H3K27ac that are expressed in GSCs and downstream germ cells. Heterochromatin = blue boxes. Numbers below line are coordinates in Mb. Numbers above line are cytogenetic regions. Black circles are centromeres.

Genes induced in germ cells downstream from germline stem cells (GSCs) mediate a metabolic and growth transition.
(A) Germ cell-expressed transcription regulators with the largest fold change increases in young nurse cell (NC) (OVS6) compared to GSCs. (B) Gene ontology (GO) terms for genes upregulated in young ovaries vs GSCs. The GO categories reveal that a dramatic metabolic shift occurs between GSCs and OVS6 NCs. (C) Expression values (tpm) in GSCs and OVS6 NCs of glycolytic genes showing mostly upregulation but a shutoff of Ldh. (D) Diagram showing upregulated transmembrane transport genes (first GO category in B). Sub-cellular localization of a sample (20) of membrane transporters (Dros name/Mouse name) is indicated (arrows) on the cell drawing showing plasma membrane (red), mitochondrion (blue), or Golgi (orange) along with their fold change (increase) in NC. Many are key regulators of cellular import or mitochondrial metabolism that help generate the precursor nucleotides, lipids, and carbohydrates needed for rapid cellular growth. For complete list see Supplementary file 1e.
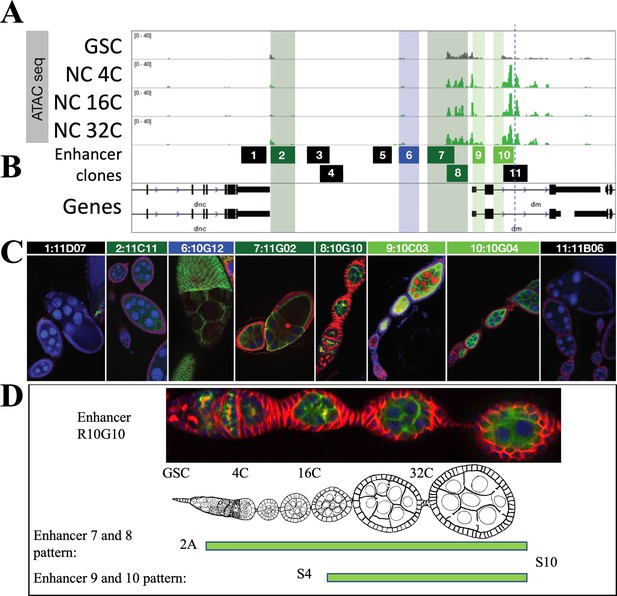
Analysis of chromatin changes at enhancers of Myc during oogenesis.
(A) Myc region chromatin accessibility assayed by ATACseq in germline stem cells (GSCs) and 4C-32C nurse cells (NCs). (B) Genomic DNA fragments from this region (1–11, indicated) cloned and integrated within Janelia GAL4 lines (Jenett et al., 2012) are shown. (C) A summary of the GFP expression driven by eight tested fragments in Janelia lines with indicated names. (D) Enlarged view of GFP expression from clone 8 (R10G10), which is similar to clone 7 and is shown below. The pattern of expression is indicated to show low level (clones 7 and 8) and higher-level (clones 9 and 10) GFP expression starting about stage 4.
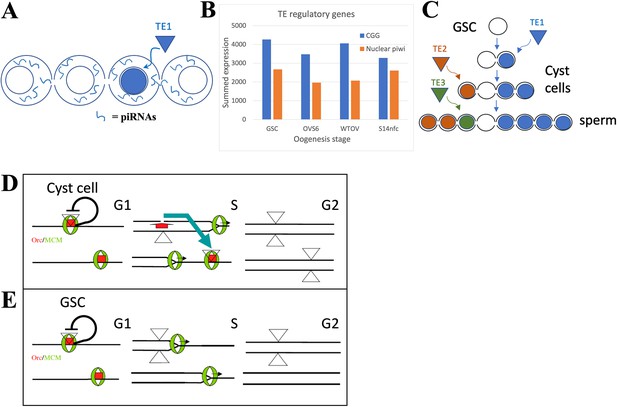
The germline stem cell (GSC) state provides high immunity to transposable elements (TEs).
(A) Proposed amplification of TE resistance by a germline cyst or syncytial stage downstream from a highly potent stem cell by amplifying and sharing piRNA (blue lines) between all connected cells following TE1 movement in one cell. (B) Summed expression (tpm) of conserved germline genes (CGGs) (blue) involved in transposon regulation and nuclear piwi genes (orange). (C) Illustrative diagram of cluster analysis in a male GSC lineage with two cyst divisions and a single meiotic division leading to eight sperm. The number of sperm with identical TE insertions depends of the time of insertion during cyst formation, as shown by three different TE examples that insert at different stages. (D) Model of P element (triangles) copy number increase on chromosomes (horizontal lines) during a cyst cell or meiotic germ cell cycle by ‘replication timing’. Red = ORC proteins, green = MCM proteins, black line shows postulated repression prior to loss of pre-replication complex in S phase by origin activation or fork passage. Blue arrow shows transposition from a replicated to an unreplicated region recognized by unfired pre-replication complex during later S phase (modified from Spradling et al., 2011). (E) Like (D), but in a GSC whose short G1 is proposed activate all origins simultaneously.
Additional files
-
Supplementary file 1
The Drosophila female germ cell cycle.
(a) All processed data. (b) E(z)-dependent gene downregulation. (c) Mostly heterochromatic germline stem cell (GSC) genes with high H3K27ac. (d) Genes upregulated in young follicles relative to GSCs. (e) 41 membrane transporters upregulated more than 10-fold in young follicles.
- https://cdn.elifesciences.org/articles/90509/elife-90509-supp1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/90509/elife-90509-mdarchecklist1-v1.docx





