The Hippo kinase cascade regulates a contractile cell behavior and cell density in a close unicellular relative of animals
Figures
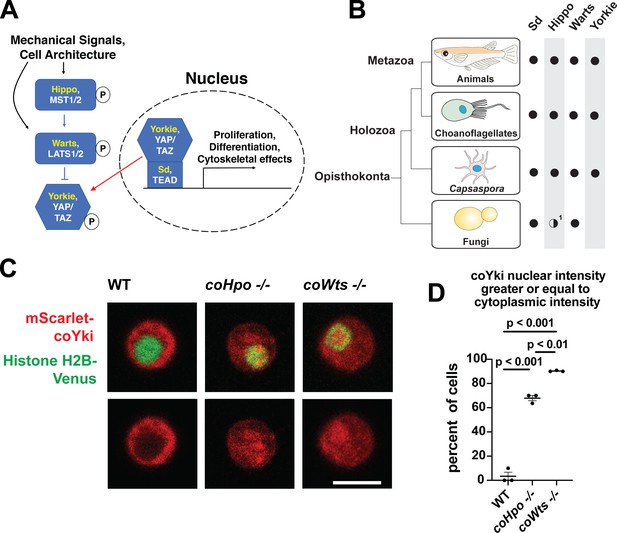
The Hippo pathway kinases regulate the subcellular localization of coYki.
(A) The Hippo pathway in Drosophila (yellow text) and mammals (white text). Figure from Phillips et al., 2022. (B) Conservation of Hippo pathway components in close unicellular relatives of animals. 1The Hippo kinase SARAH domain is absent from putative Hippo kinase orthologs in yeasts, but is present in the early-branching fungus Spizellomyces punctatus and in amoebozoans such as Dictyostelium, indicating SARAH domain loss in some fungal lineages. Data from Sebé-Pedrós et al., 2012. (C) Cells were transiently transfected with the indicated constructs and imaged by confocal microscopy. Scale bar is 5 microns. (D) Intensity of mScarlet-coYki signal in the nucleus and cytoplasm of cells was quantified. Where p-values are given, differences between conditions are significant (One-way ANOVA, Tukey’s test, n=3 with at least 10 cells measured for each independent experiment.) Values are mean ± SEM. Absence of error bars indicates that error is smaller than the plot symbol.
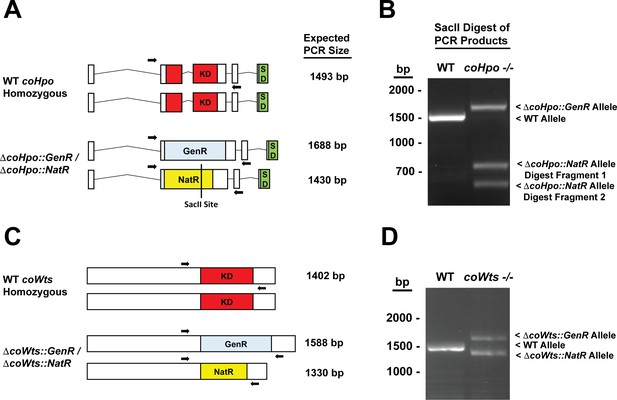
Knockout of coHpo and coWts.
(A) A diagram of the coHpo genomic loci in wild-type (WT) or a putative coHpo -/- knockout line is shown. ‘KD’ indicates kinase domain, ‘SD’ indicates the SARAH domain, ‘GenR’ indicates a geneticin resistance cassette, and ‘NatR’ indicates a nourseothricin resistance cassette. Arrows indicate a pair of primers used in PCR reactions. The same primer pair were used in all PCRs shown. Expected PCR product sizes for different alleles are given. A SacII site present only within the allele containing the NatR cassette is shown. (B) PCR products from WT and coHpo -/- cells using primers are shown in (A). Because the product sizes for the WT and ΔcoHpo::NatR alleles are close in size, we digested PCR products with SacII, which uniquely cuts the ΔcoHpo::NatR allele. The coHpo -/- cell line shows the expected products for a homozygous knockout. Sequencing of PCR products confirmed the identity of the WT, ΔcoHpo::GenR, and ΔcoHpo::NatR alleles. (C) A diagram of the coWts genomic loci in WT or a putative coWts -/- knockout line is shown. ‘KD’ indicates kinase domain, ‘GenR’ indicates a geneticin resistance cassette, and ‘NatR’ indicates a nourseothricin resistance cassette. Arrows indicate a pair of primers used in PCR reactions. The same primer pair were used in all PCRs shown. (D) PCR products from WT and coHpo -/- cells using primers are shown in (C). The coWts -/- cell line shows the expected products for a homozygous knockout. Sequencing of PCR products confirmed the identity of the WT, ΔcoWts::GenR, and ΔcoWts::NatR alleles.
-
Figure 1—figure supplement 1—source data 1
Original unedited file for the DNA gel image used to generate panel B.
- https://cdn.elifesciences.org/articles/90818/elife-90818-fig1-figsupp1-data1-v1.tif
-
Figure 1—figure supplement 1—source data 2
Uncropped file showing DNA gel image used to generate panel B including annotations of bands and sample labels from panel B.
- https://cdn.elifesciences.org/articles/90818/elife-90818-fig1-figsupp1-data2-v1.tif
-
Figure 1—figure supplement 1—source data 3
Original unedited file for the DNA gel image used to generate panel D.
- https://cdn.elifesciences.org/articles/90818/elife-90818-fig1-figsupp1-data3-v1.tif
-
Figure 1—figure supplement 1—source data 4
Uncropped file showing DNA gel image used to generate panel B including annotations of bands and sample labels from panel D.
- https://cdn.elifesciences.org/articles/90818/elife-90818-fig1-figsupp1-data4-v1.tif
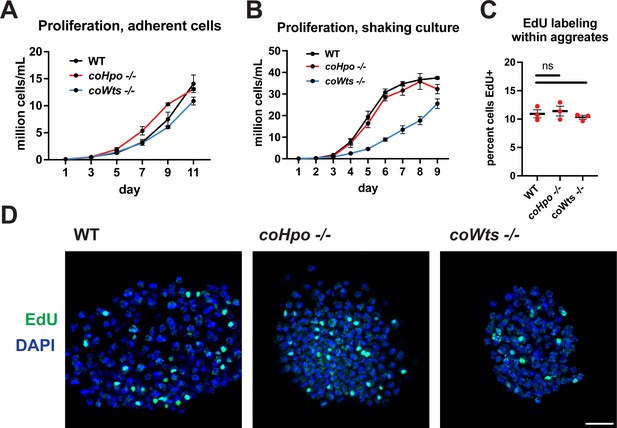
The Hippo pathway kinases do not restrict cell proliferation in Capsaspora.
Cells were grown in adherent culture in 96-well plates (A) or in shaking culture (B) and cell density was measured daily by hemocytometer. (C, D) Cells were inoculated under low-adherence conditions to induce aggregate formation, and 3 days after induction aggregates were incubated with EdU for 4 hr, fixed, and processed to detect EdU labeling. Scale bar is 10 microns. Differences in EdU labeling between WT and mutants were not significant (one-way ANOVA, Dunnett’s test, n=3 with three aggregates measured for each independent experiment). Black bars show mean ± SEM, and red dots show values for individual aggregates.
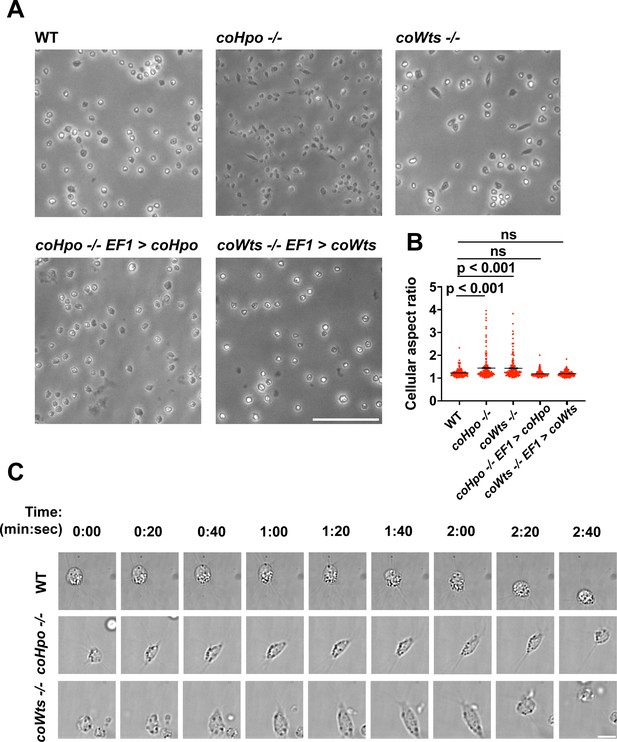
The Hippo pathway kinases regulate cell morphology and a contractile cell behavior in Capsaspora.
(A) Cells were inoculated into glass-bottom chamber slides, and adherent cells were imaged 2 days after inoculation. Scale bar is 50 microns. (B) The aspect ratio of cells imaged as described in (A) was measured using ImageJ. The differences between wild-type (WT) and either coHpo -/- or coWts -/- cells are significant (one-way ANOVA, Dunnett’s test, n=3 with 50 cells measured for each independent experiment). Black bars show mean ± SEM, and red dots show values for 150 individual cells. (C) Cells were prepared as described in (A) and imaged by time-lapse microscopy. Scale bar is 10 microns.
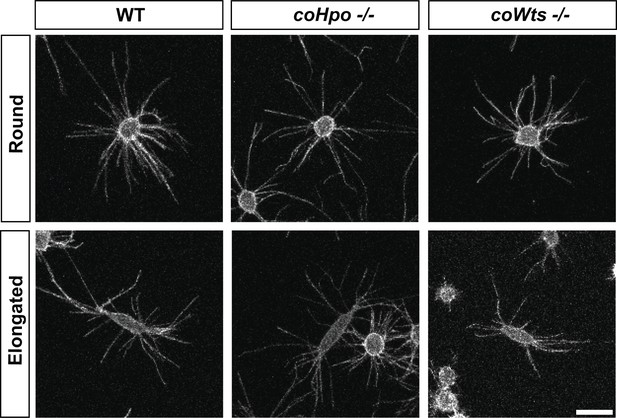
Filopodia are enriched at the poles of elongated cells.
Adherent cells stably expressing N-myristoylation motif (NMM)-Venus, a plasma membrane marker which allows for visualization of filopodia, were imaged live by confocal microscopy. Scale bar is 10 microns.
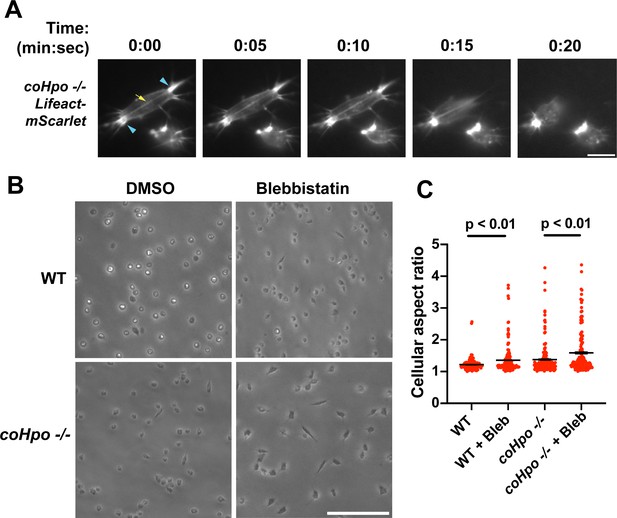
Characterization of actomyosin in contractile Capsaspora cells.
(A) Cells were inoculated on glass-bottom chamber slides, and at 2 days after inoculation adherent cells were imaged by time-lapse epifluorescence microscopy. Blue arrowheads indicate ventral F-actin-enriched foci, and the yellow arrow indicates the F-actin fiber that connects the foci. Scale bar is 10 microns. (B) Cells were inoculated on glass-bottom chamber slides. Two days after inoculation, DMSO or Blebbistatin at a concentration of 1 μM was added to cells. Cells were imaged 24 hr after the addition of blebbistatin. (C) The aspect ratio of cells prepared as in (B) was measured using ImageJ. For both wild-type (WT) and coHpo -/- cells, the difference between DMSO and blebbistatin treatment measurements are significant (t-test, n=3 with 50 cells measured for each independent experiment). Black bars indicate mean ± SEM, and red dots indicate measurements for 150 individual cells for each condition. Absence of error bars indicates that error is smaller than the plot symbol.
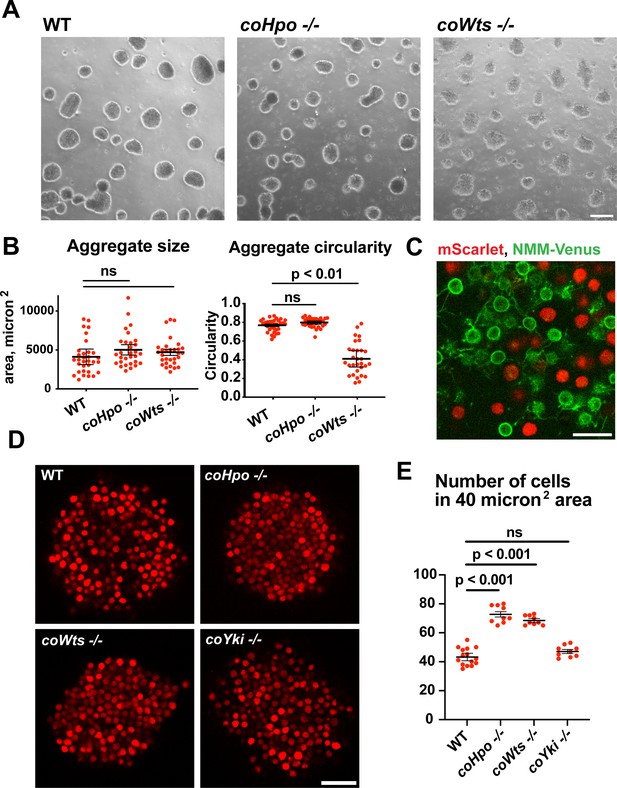
The Hippo pathway kinases regulate the density of cell packing in Capsaspora aggregates.
(A) Aggregates were induced by inoculating cells in low-adherence conditions, and aggregates were imaged 5 days after inoculation. Scale bar is 100 microns. (B) The size and circularity of aggregates imaged as in (A) was measured using ImageJ. Black bars indicate mean ± SEM (n=3 with 10 aggregates measured for each independent experiment), and red dots indicate values for 30 individual aggregates. For aggregate circularity, the difference between wild-type (WT) and coWts -/- aggregates are significant (one-way ANOVA, Dunnett’s test.) (C) Cells expressing either mScarlet or N-myristoylation motif (NMM)-Venus, a marker of the plasma membrane which allows visualization of filopodia, were mixed 1:1 and inoculated into low-adherence conditions to induce aggregation. After 5 days, aggregates were imaged by confocal microscopy. Scale bar is 10 microns. (D) Clonal populations of cells of the indicated genotype stably expressing mScarlet were inoculated into low-adherence conditions, and cell aggregates were imaged 5 days after inoculation by confocal microscopy. Scale bar is 20 microns. (E) The number of cells within a 40 micron2 area from aggregate optical sections imaged as in (D) was measured. Black bars indicate mean ± SEM (n=3 with three aggregates measured for each independent experiment), and red dots indicate values for nine individual aggregates for each condition. The difference between WT and either coHpo -/- or coWts -/- cells is significant (one-way ANOVA, Dunnett’s test.).
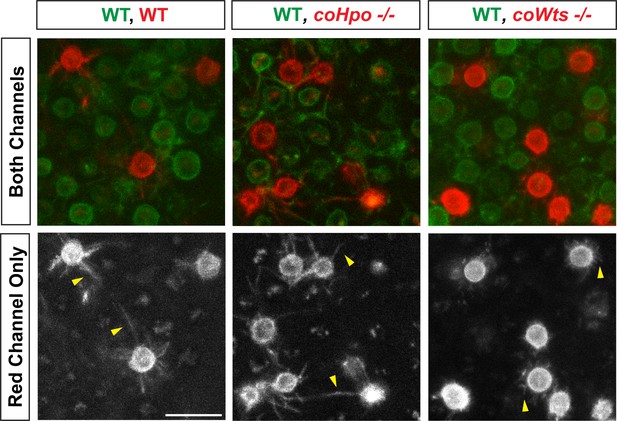
coWts -/- cells within multicellular aggregates show altered filopodial morphology and reduced filopodial length.
Wild-type (WT) cells stably expressing N-myristoylation motif (NMM)-Venus, which allows visualization of filopodia, were mixed with WT, coHpo -/-, or coWts -/- cells stably expressing NMM-mScarlet at a 9:1 cell ratio and were inoculated into low-adherence wells to induce aggregate formation. Cells within aggregates were then imaged 3 days after aggregate induction. Yellow arrowheads indicate examples of filopodia. Scale bar is 10 microns.
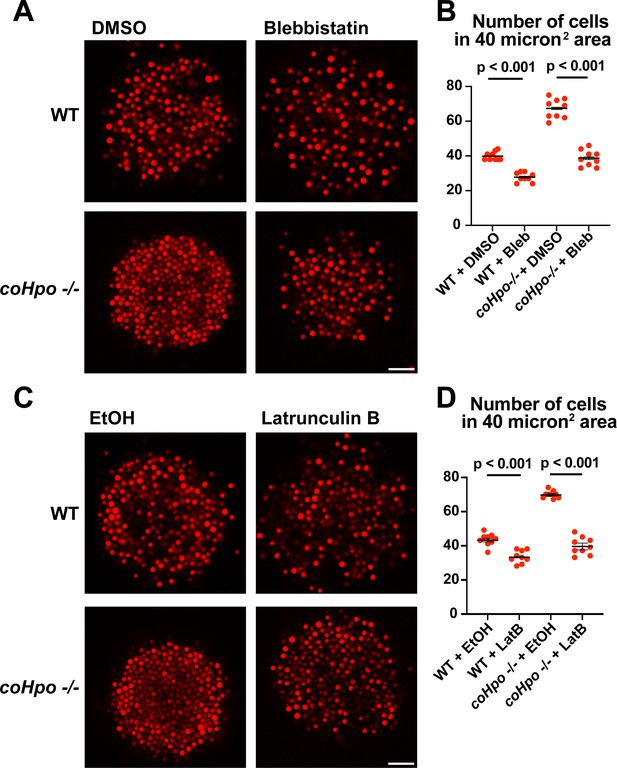
Perturbation of the actomyosin network affects cell packing in multicellular Capsaspora aggregates.
(A) Aggregates were treated with 20 μM blebbistatin or DMSO as a vehicle control for 20 minand then imaged by confocal microscopy. Scale bar is 20 microns. (B) Cells per area were quantified from images obtained as in (A). Black bars indicate means ± SEM (n=3 with three aggregates measured for each independent experiment), and red dots indicate values for nine individual aggregates for each condition. Absence of error bars indicates that error is smaller than the plot symbol. For both wild-type (WT) and coHpo -/- aggregates, the difference between DMSO and blebbistatin treatment is significant (t-test). (C) Aggregates were treated with 5 μg/ml Latrunculin B or ethanol as a vehicle control for 1 hr and then imaged by confocal microscopy. Scale bar is 20 microns. (D) Cells per area were quantified from images obtained as in (C). For both WT and coHpo -/- aggregates, the difference between ethanol and Latrunculin B treatment is significant (n=3, t-test.).
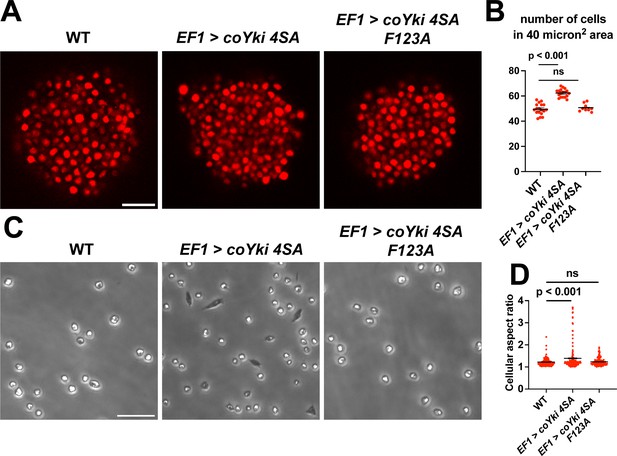
Expression of a constitutively active form of coYki phenocopies loss of coHpo or coWts in Capsaspora.
(A) Aggregates were imaged 5 days after inoculation. Scale bar is 20 microns. (B) The number of cells within a 40 micron2 area within an aggregate was measured. Black bars indicate mean ± SEM (n=3 with three aggregates measured for each independent experiment), and red dots indicate values for 9 individual aggregates for each condition. The difference between wild-type (WT) and EF1 >coYki 4SA is significant (one-way ANOVA, Dunnett’s test). (C) Cells were inoculated into glass-bottom chamber slides and imaged 2 days after inoculation. Scale bar is 25 microns. (D) The aspect ratio of cells imaged as in (C) was measured using ImageJ. Black bars show mean ± SEM (n=3 with 40 cells measured for each independent experiment), and red dots show values for 150 individual cells. The difference between WT and EF1 >coYki 4SA is significant (one-way ANOVA, Dunnett’s test).
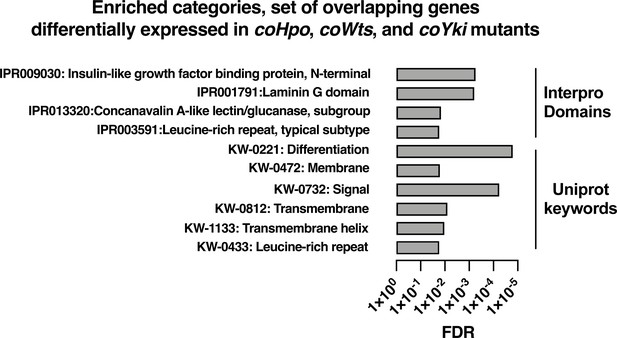
Enrichment analysis of the overlapping set of genes differentially expressed in coHpo, coWts, and coYki mutant cells.
Categories with FDR <0.05 are shown. Interpro or Uniprot accession numbers are given for each enriched category. FDR: false discovery rate.
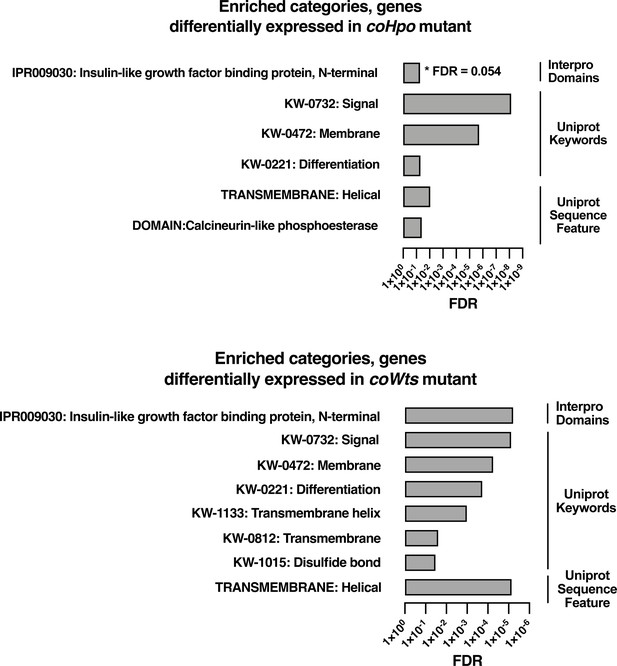
Enrichment analysis for genes differentially expressed in coHpo or coWts mutant cells.
Categories with FDR <0.05 are shown, except where FDR is indicated. Interpro or Uniprot accession numbers are given for enriched categories where appropriate. FDR: false discovery rate.
Videos
Time-lapse microscopy of adherent wld-type (WT) Capsaspora cells.
Time-lapse microscopy of adherent coHpo -/- cells.
Time-lapse microscopy of adherent coWts -/- cells.
Time-lapse confocal microscopy of adherent wild-type (WT) cells expressing Lifeact-mScarlet.
This video serves as a control for Video 5.
Time-lapse confocal microscopy of adherent coHpo -/- cells expressing Lifeact-mScarlet.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Capsaspora owczarzaki) | coHpo | Genbank | CAOG_01932 | |
| Gene (Capsaspora owczarzaki) | coWts | Genbank | CAOG_00619 | |
| Gene (Capsaspora owczarzaki) | coYki | Genbank | CAOG_07866 | |
| Cell line (Capsaspora owczarzaki) | WT | ATCC | 30864 | |
| Recombinant DNA reagent | pJP80 | Phillips et al., 2022 | Capsaspora expression vector: mScarlet-coYki fusion, Geneticin resistance | |
| Recombinant DNA reagent | pONSY-coH2B:Venus | Addgene | Addgene Plasmid #111877 | Capsaspora expression vector: Histone H2B-Venus fusion |
| Recombinant DNA reagent | pJP122 | This paper | Capsaspora expression vector: coHpo transgene, Hygromycin B Resistance | |
| Recombinant DNA reagent | pJP125 | This paper | Capsaspora expression vector: coWts transgene, Hygromycin B Resistance | |
| Recombinant DNA reagent | pJP145 | This paper | Capsaspora expression vector: NMM-Venus, Hygromycin B Resistance | |
| Recombinant DNA reagent | pJP118 | Phillips et al., 2022 | Capsaspora expression vector: Lifeact-mScarlet, Hygromycin B Resistance | |
| Recombinant DNA reagent | pJP103 | Phillips et al., 2022 | Capsaspora expression vector: mScarlet, Hygromycin B Resistance | |
| Recombinant DNA reagent | pJP90 | Phillips et al., 2022 | Capsaspora expression vector: coYki 4SA, Geneticin Resistance | |
| Sequence-based reagent | JP201: coHpo KO Forward: homologous arm +actin promoter | This paper | PCR primers | CCCGCCTCGTCCCAACAACAGCTCGTACGGCTCGGTCTTCAAGGCTCGCCACAAGGACACCCAGTCCATCCTCGCCGTCAAGCAGGTGCCCCTTGAGAACACAAAAATGCTGATTGTTTG |
| Sequence-based reagent | JP202: coHpo KO Reverse: homologous arm +actin terminator | This paper | PCR primers | AAGCAACATTGCTTCGTTGACTAACGCTTCATGGTGCCGTCCTCGCCCATGACCATCGTGCCGGAGTCGCCAGTGACCATCGTTCCGCTATCAACAAGGGTTTTTTCTTTGTACAAGATCAC |
| Sequence-based reagent | JP203: coWts KO Forward: homologous arm +actin promoter | This paper | PCR primers | CAAACGGCGAGACCAGCTCGAGATTGAGATGGCCAAGATGGACCTGACCGATGTTCAAAAGACCCAGTTGCGTCGCATCCTCCGCATGAAGGAATCAGAGACAAAAATGCTGATTGTTTG |
| Sequence-based reagent | JP204: coWts KO Reverse: homologous arm +actin terminator | This paper | PCR primers | GCATTTTAATGTGCTCGACCGTCTCCCGGCCGAGACGATCGGAGGATTCACAGCAGAGCCGGGAGATGAGGTCTTTGGACTCGCGCGAAATCTTTGCCCGTTTTTTCTTTGTACAAGATCAC |
| Sequence-based reagent | JP205: coHpo Diagnostic Forward | This paper | PCR primers | CAAAAACAACAGCGAAAACG |
| Sequence-based reagent | JP206: coHpo Diagnostic Reverse | This paper | PCR primers | CCATCGGAGGAAACTAAAGG |
| Sequence-based reagent | JP207: coWts Diagnostic Forward | This paper | PCR primers | TGCTGCTGAAAATGAAAACG |
| Sequence-based reagent | JP208: coWts Diagnostic Reverse | This paper | PCR primers | GAACCTCACCAGGATTTTGC |
| Commercial assay or kit | TransIT-X2 Dynamic Delivery System | Mirus Bio | MIR 6003 | |
| Commercial assay or kit | Click-iT Plus EdU Cell Proliferation Kit for Imaging, Alexa Fluor 488 dye | ThermoFisher | C10637 | |
| Chemical compound, drug | Blebbistatin | Sigma-Aldrich | B0560 | |
| Chemical compound, drug | Latrunculin B | Sigma-Aldrich | L5288 |
Additional files
-
Supplementary file 1
Genes with actin-binding functional annotation showing differential expression in coYki mutant cells and also exhibiting differential expression in coHpo and/or coWts mutant cells.
- https://cdn.elifesciences.org/articles/90818/elife-90818-supp1-v1.xlsx
-
Supplementary file 2
Lists of genes differentially expressed in coHpo mutant cells, coWts mutant cells, and the overlapping set of genes differentially expressed in coHpo, coWts, and coYki mutant cells.
- https://cdn.elifesciences.org/articles/90818/elife-90818-supp2-v1.xlsx
-
Supplementary file 3
Sequences of synthesized gene fragments used in this study.
- https://cdn.elifesciences.org/articles/90818/elife-90818-supp3-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/90818/elife-90818-mdarchecklist1-v1.docx





