Mapping the architecture of the initiating phosphoglycosyl transferase from S. enterica O-antigen biosynthesis in a liponanoparticle
Figures
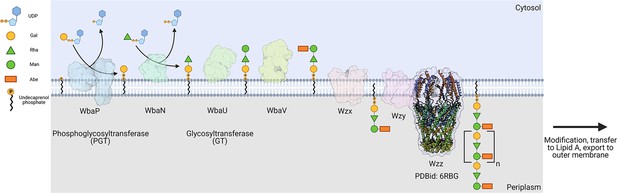
Biosynthesis of O-antigen repeat units in S.enterica serovar typhimurium.
The pathway is initiated by the transfer of a phospho-Gal onto undecaprenol phosphate (UndP), catalyzed by the LgPGT WbaP. A series of glycosyltransferases (GTs) add additional sugars to the nascent repeat unit (RU). The repeat unit is then flipped across the membrane by a Wzx-class flippase, and RUs are polymerized through the coordinated action of the Wzy polymerase and Wzz chain-length regulatory protein.
© 2023, Dodge et al. Figure 1 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
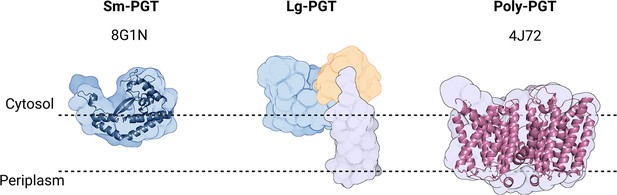
Overview of common phosphoglycosyl transferase (PGT) family members.
Sm-monoPGTs do occupy one leaflet of the membrane, comprise only the catalytic core of monotopic PGTs (monoPGTs) (blue surface), and are exemplified by the structurally characterized PglC from Campylobacter concisus (dark blue cartoon). Large monoPGTs (Lg-PGTs) feature the C-terminal conserved catalytic core domain (blue surface) and two uncharacterized N-terminal domains. These domains feature a predicted transmembrane helix bundle (purple surface) and a domain of unknown function (gold surface). Poly-PGTs such as MraY from Aquifex aeolicus (magenta cartoon) comprise a catalytic domain, which is structurally distinct Sm- or Lg-PGTs and have been demonstrated to dimerize in their active form.
© 2023, Dodge et al. Figure 2 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
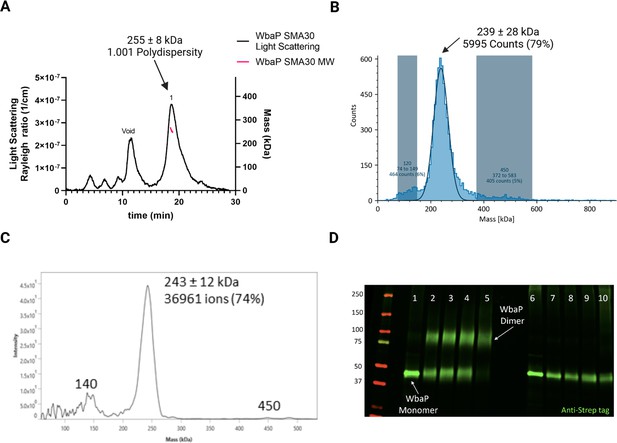
Characterization of S.enterica WbaP in SMALP.
(A) WbaP in styrene-maleic acid liponanoparticles (SMALP) analyzed by size exclusion chromatography with multi-angle light scattering (SEC-MALS). A small peak is observed at the void volume of the column, while the main peak has a calculated molecular weight of 255 ± 8 kDa, with a polydispersity value of 1.001. (B) WbaP in SMALP analyzed by mass photometry. The sample is monodisperse, and the main species has an apparent molecular weight of 239 ± 28 kDa. (C) Mass spectrum of WbaP in SMALP analyzed by Direct Mass Technology mode. (D) Western blot analysis of Lysine-reactive dithiobis(succinimidyl propionate) (DSP) crosslinker reacted with WbaP in SMALP. Lanes: 1: Control, 2: 0.1 mM DSP, 3: 0.25 mM DSP, 4: 1 mM DSP, 5: 5 mM DSP, 7: 0.1 mM DSP reduced, 8: 0.25 mM DSP reduced, 9: 1 mM DSP reduced, 10: 5 mM DSP reduced.
© 2023, Dodge et al. Figure 3 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
-
Figure 3—source data 1
Raw data for the blot shown in Figure 3D.
- https://cdn.elifesciences.org/articles/91125/elife-91125-fig3-data1-v1.zip
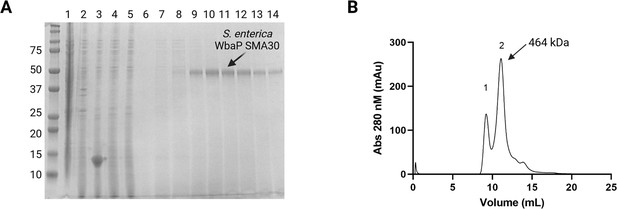
Purification and characterization of S.enterica WbaP in styrene-maleic acid liponanoparticle (SMALP).
(A) Coomassie-stained sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) of large-scale WbaP purification. Lanes: 1: Lysate, 2: Cell envelope fraction (CEF), 3: SMA30-solubilized CEF, 4: StreptactinXT flowthrough, 5: Wash 1, 6: Wash 2, 7–14: Biotin elution. (B) Purified WbaP separated on an Enrich SEC 650 column. Peak 1 corresponds to the void volume of the column, Peak 2 elutes at 11.09 mL, corresponding to a molecular weight of 464 kDa.
© 2023, Dodge et al. Figure 3—figure supplement 1 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
-
Figure 3—figure supplement 1—source data 1
Raw SDS-PAGE gel data relating to Figure 3—figure supplement 1A.
- https://cdn.elifesciences.org/articles/91125/elife-91125-fig3-figsupp1-data1-v1.zip
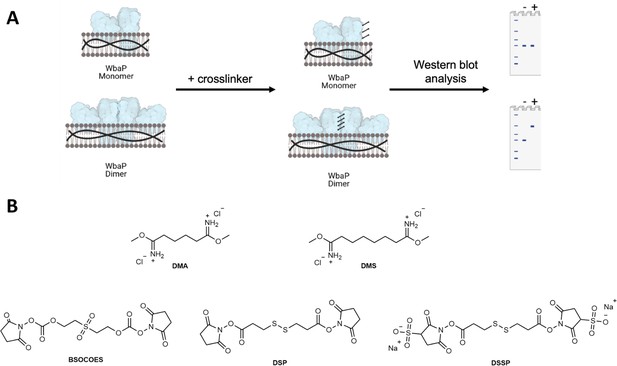
Crosslinking strategy for S.enterica WbaP in styrene-maleic acid liponanoparticle (SMALP).
(A) A cartoon scheme depicting the results of crosslinking for either a WbaP monomer, or a WbaP dimer. Crosslinking efficiency is read out by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) or western blot. (B) Panel of crosslinking compounds screened. DMA: dimethyl adipimidate, DMS: dimethyl suberimidate, BSOCOES: bis[2-(succinimidyloxycarbonyloxy)ethyl]sulfone, DSP: dithiobis(succinimidyl propionate), DTSSP: 3,3'-dithiobis(sulfosuccinimidyl propionate).
© 2023, Dodge et al. Figure 3—figure supplement 2 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
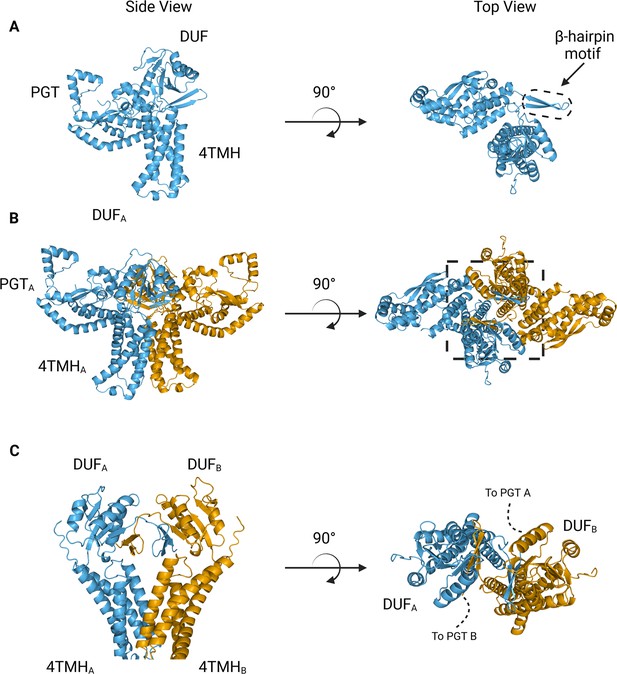
AlphaFold predictions of S.enterica WbaP monomer and dimer.
(A) WbaP monomer prediction. (B) WbaP dimer prediction. (C) Close-up view of predicted dimer interface showing the interdigitating β-hairpin motifs between DUFA and DUFB. Phosphoglycosyl transferase (PGT) domains hidden for clarity.
© 2023, Dodge et al. Figure 4 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
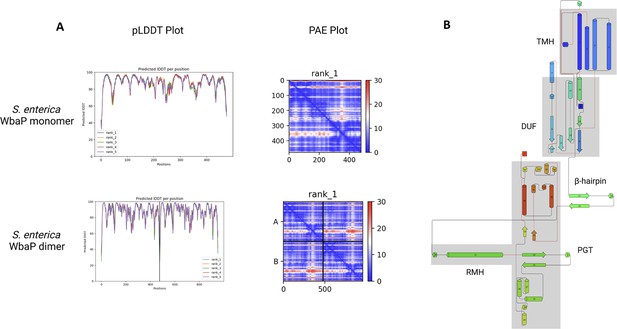
Analysis of S.enterica WbaP AlphaFold prediction.
(A) AlphaFold predicted local distance difference test (pLDDT) plots and predicted aligned error (PAE) plots for S. enterica WbaP monomer and dimer predictions. The overall confidence in both models is high. (B) Topology diagram of AlphaFold S. enterica WbaP prediction. Structural domains are shaded in gray. TMH: transmembrane helix, DUF: domain of unknown function, PGT: phosphoglycosyl transferase, RMH: re-entrant membrane helix.
© 2023, Dodge et al. Figure 4—figure supplement 1 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
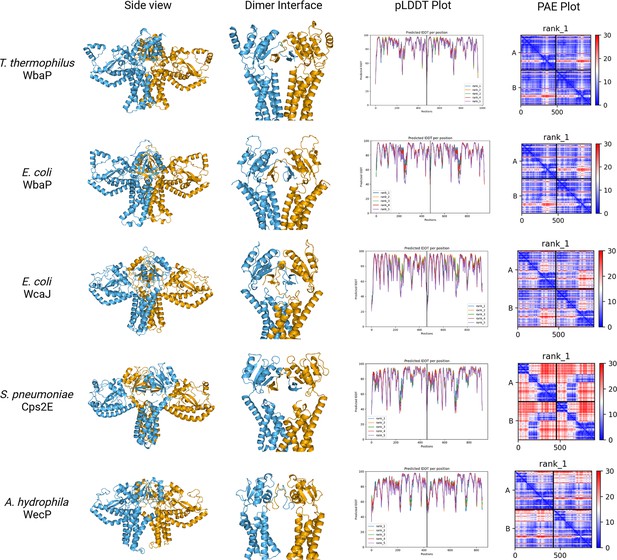
AlphaFold dimer predictions for various large monoPGTs (Lg-PGTs), along with predicted local distance difference test (pLDDT) plots and predicted aligned error (PAE) plots.
β-hairpin-mediated domain swaps are observed for each prediction. While the estimated quality of predictions varies from protein to protein, the overall confidence in the models is satisfactory, with the majority of the regions of the proteins having a pLDDT score ≥70.
© 2023, Dodge et al. Figure 4—figure supplement 2 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
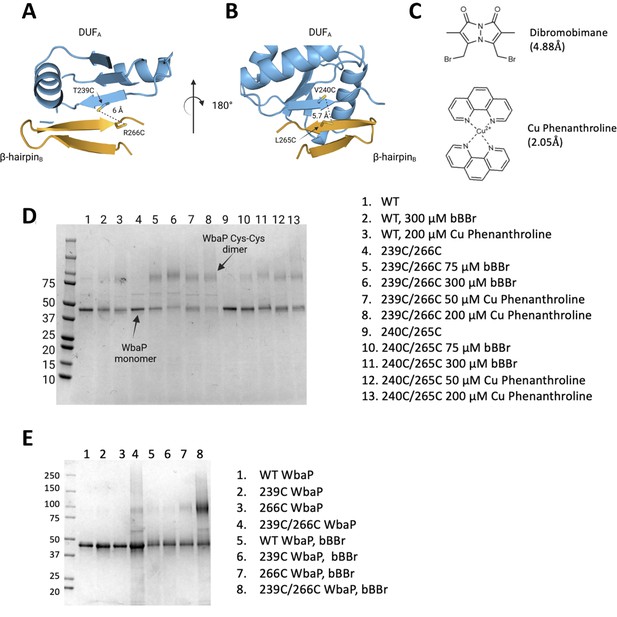
Probing the predicted interface of the WbaP dimer.
(A, B) AlphaFold prediction with Cys pair variants modeled as sticks. The β-hairpin of one monomer is shown in sky blue and the continuing β- strand of the other monomer is shown in orange. (C) Structure and crosslinking radii of thiol-reactive crosslinkers. (D) Crosslinking of Cys pair WbaP variants in styrene-maleic acid liponanoparticle (SMALP) using dibromobimane (bBBr) and Cu Phenanthroline. (E) bBBr crosslinking of single and double WbaP Cys variants in SMALP.
© 2023, Dodge et al. Figure 5 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
-
Figure 5—source data 1
Raw gel data for Figure 5D.
- https://cdn.elifesciences.org/articles/91125/elife-91125-fig5-data1-v1.zip
-
Figure 5—source data 2
Raw gel data for Figure 5E.
- https://cdn.elifesciences.org/articles/91125/elife-91125-fig5-data2-v1.zip
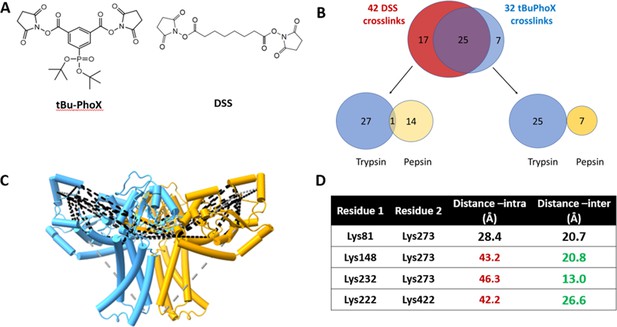
Crosslinking mass spectrometry (XLMS) of WbaP in styrene-maleic acid liponanoparticle (SMALP).
(A) Structures of amino reactive crosslinkers used for XLMS analysis. (B) Overview of identified WbaP crosslinks from different protease treatments and crosslinkers. (C) Identified crosslinks mapped onto S. enterica WbaP AlphaFold dimer model. Disuccinimidyl suberate (DSS) crosslinks are shown in black, tert-butyl disuccinimidyl phenyl phosphonate (tBu-PhoX) crosslinks are shown in gray. (D) Residues linked by intermolecular crosslinks. The distance between these residues within a single monomer and between monomers is shown.
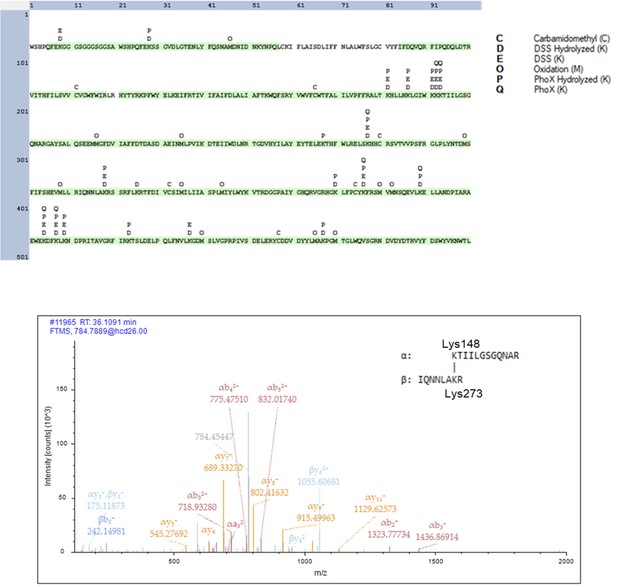
Crosslinking mass spectrometry analysis of S. enterica WbaP in styrene-maleic acid liponanoparticle (SMALP).
Top: Coverage and detected modifications across the WbaP construct a primary sequence. Residues highlighted in green were detected in fragments after proteolytic degradation. Bottom: Example of MS/MS spectrum of the disuccinimidyl suberate (DSS) inter-chain crosslinked fragment between Lys148 and Lys273.
© 2023, Dodge et al. Figure 6—figure supplement 1 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
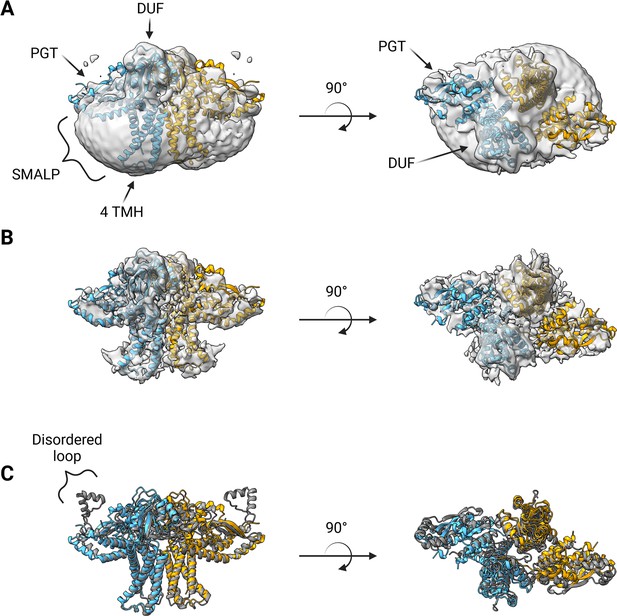
Structure of S.enterica WbaP in styrene-maleic acid liponanoparticle (SMALP).
WbaP dimer colored in blue and orange (A) Unsharpened map displaying clear density for stabilizing liponanoparticle. Unsharpened density colored light gray. (B) Sharpened map after masked local refinement. Sharpened density colored light gray. (C) Superposition of refined WbaP model and full-length AlphaFold prediction, RMSD: 2.83 Å. AlphaFold dimer colored gray. A predicted helix-turn-helix motif lacks density in the experimental map.
© 2023, Dodge et al. Figure 7 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.

EM processing workflow.
© 2023, Dodge et al. Figure 7—figure supplement 1 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
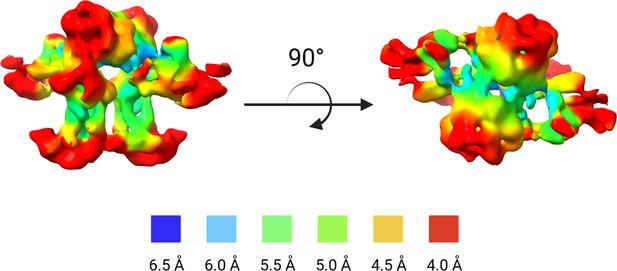
Local resolution estimation of masked, un-sharpened WbaP reconstruction.
Areas buried within the membrane tend to exhibit an overall lower local resolution than more surface-exposed regions.
© 2023, Dodge et al. Figure 7—figure supplement 2 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
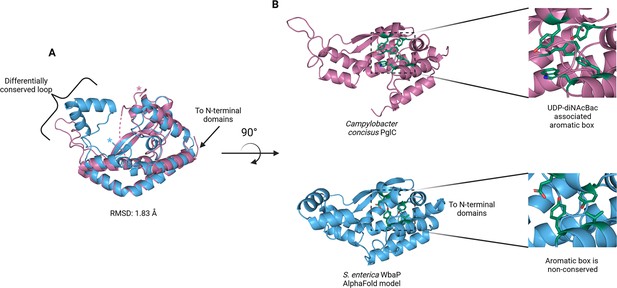
Comparison of S. enterica WbaP phosphoglycosyl transferase (PGT) domain and PglC from Campylobacter concisus (PDBid 8G1N).
WbaP is shown in light blue, PglC is in magenta. (A) Superposition of WbaP and PglC. While the overall RMSD is 1.83 Å, regions in PglC which can be used to computationally assign substrate are significantly different in WbaP. A loop region in PglC is replaced with a helix-turn-helix motif in WbaP, and the C-terminus of WbaP is significantly shorter than that of PglC. (B) Top: an aromatic box motif in PglC is conserved among PGTs that utilize UDP-diNAcBac. Aromatic box residues are shown as dark green sticks. Bottom: Residues found in S. enterica WbaP at homologous positions to aromatic box residues in PglC are shown as dark green sticks. Aromatic box residues are non-conserved in WbaP.
© 2023, Dodge et al. Figure 8 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
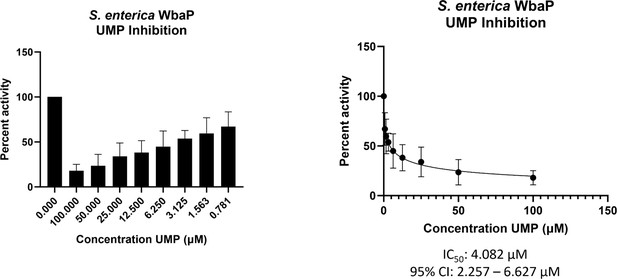
WbaP UMP titration assay.
Left: Percent activity vs UMP concentration. A UMP-dependent decrease in WbaP activity is observed. Right: Curve-fitting of UMP inhibition data yields an IC50 apparent of 4.1 μM. Data were collected in triplicate with error bars representing standard deviation.
© 2023, Dodge et al. Figure 9 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
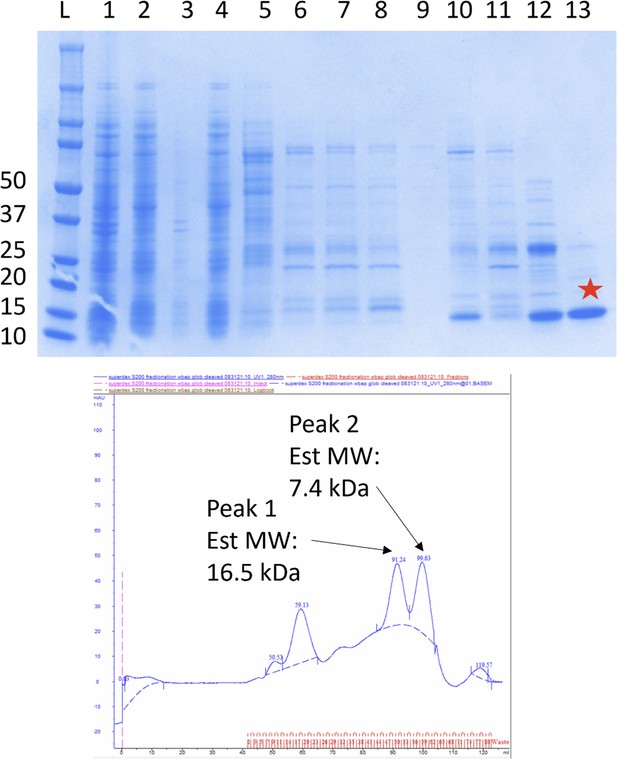
Purification of S.enterica WbaP soluble domain of unknown function domain of unknown function (DUF) truncation.
Top: Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel monitoring purification. The final purified material is indicated by a red star. Predicted molecular weight: 14.83 kDa. Lanes: 1: Lysate, 2: Soluble, 3: Pellet, 4: flowthrough, 5: wash, 6–8: Elution, 9: Post-TEV flowthrough, 10: low imidazole Elution, 11: high imidazole elution, 12: S200 peak 1, 13: S200 peak 2. Bottom: FPLC chromatogram of S200 separation.
© 2023, Dodge et al. Figure 9—figure supplement 1 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
-
Figure 9—figure supplement 1—source data 1
Raw SDS-PAGE gel data for Figure 9—figure supplement 1A.
- https://cdn.elifesciences.org/articles/91125/elife-91125-fig9-figsupp1-data1-v1.zip
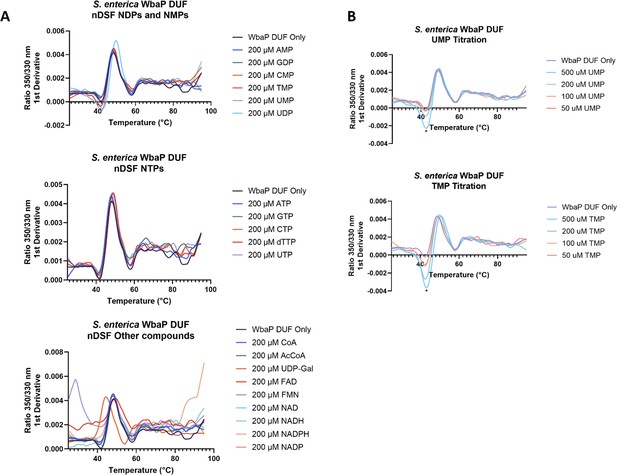
Nano differential scanning fluorimetry (nDSF) of purified soluble WbaP DUF truncation.
(A) Top: Incubation of DUF with selected nucleotide triphosphates (NTPs). Middle: Incubation of DUF with selected nucleotide diphosphates (NDPs) and nucleotide monophosphates (NMPs). Bottom: Incubation of DUF with additional putative small molecule ligands. (B) Top: UMP titration, change in Trp/Tyr fluorescence 1st derivative indicated with an asterisk. Bottom: TMP titration, change in Trp/Tyr fluorescence 1st derivative indicated with an asterisk. See Supplementary file 1C for tabulated shifts in melting temperature and full ligand names.
© 2023, Dodge et al. Figure 9—figure supplement 2 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
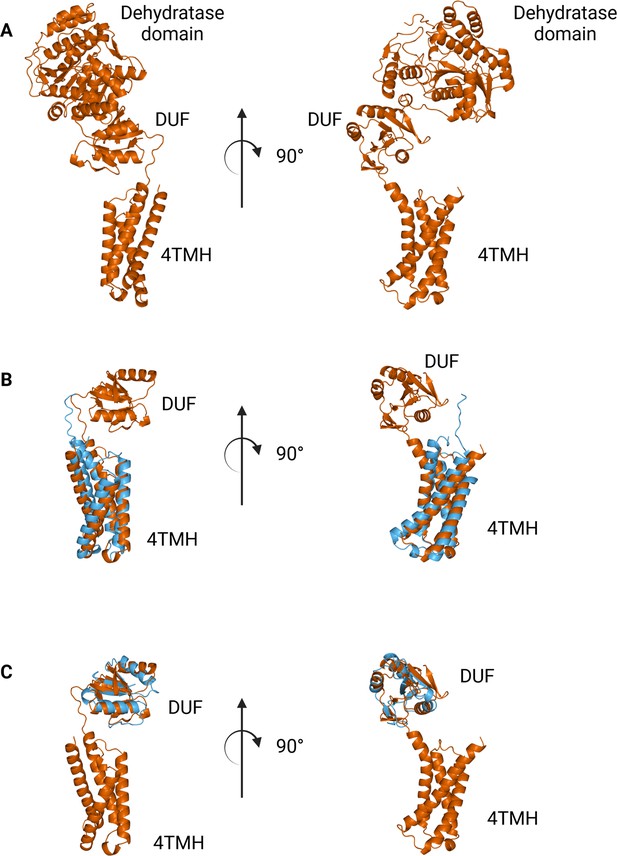
Comparison between N-terminal domains from C.jejuni PglF (Uniprot: Q0P9D4), a nucleotide sugar dehydratase, and S. enterica WbaP.
(A) AlphaFold prediction of full-length PglF. (B) Superposition of C. jejuni PglF and S. enterica WbaP 4TMH domain. RMSD: 4.3 Å. Dehydratase domain is omitted for clarity. (C) Superposition of C. jejuni PglF and S. enterica WbaP domain of unknown function (DUF) domain. RMSD: 2.3 Å. Dehydratase domain is omitted for clarity.
© 2023, Dodge et al. Figure 10 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
Tables
Summary of the differences between phosphoglycosyl transferase (PGT) superfamilies.
| Sm-PGT | Lg-PGT | Poly-PGT | |
|---|---|---|---|
| Catalytic Domain Topology | Monotopic | Monotopic | Polytopic |
| Domains | PGT only | PGT & uncharacterized accessory domains | PGT only |
| Evolutionary Conservation | Prokaryotic | Prokaryotic | Prokaryotic & eukaryotic |
| Oligomeric State | Monomer Ray et al., 2018; Anderson et al., 2023 | Unknown | Dimer Chung et al., 2013; Oluwole et al., 2022 |
| monoPGT SSN Abundance in superfamily O’Toole et al., 2021b | 38% | 47% | N/A |
Additional files
-
Supplementary file 1
Additional tables relating to the initial crosslinking screen, EM structure data and processing, and NanoDSF ligand screening.
(A) Screening chemical crosslinkers for the detection of S. enterica WbaP oligomers. S. enterica WbaP in styrene-maleic acid liponanoparticle (SMALP) was reacted with a panel of lysine-reactive crosslinkers with variable chemistries, solubilities, and lengths. Samples were analyzed by Western blot to detect the presence of crosslinked oligomers. Crosslinking efficiency was annotated as high (>30% total protein crosslinked to dimer), moderate (<30% total protein crosslinked to dimer), or none (no protein crosslinked to dimer). (B). Cryo-EM data collection, refinement, and validation statistics. (C) NanoDSF nucleotide ligand screen for soluble WbaP DUF truncation.
- https://cdn.elifesciences.org/articles/91125/elife-91125-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91125/elife-91125-mdarchecklist1-v1.docx





