Plural molecular and cellular mechanisms of pore domain KCNQ2 encephalopathy
Figures
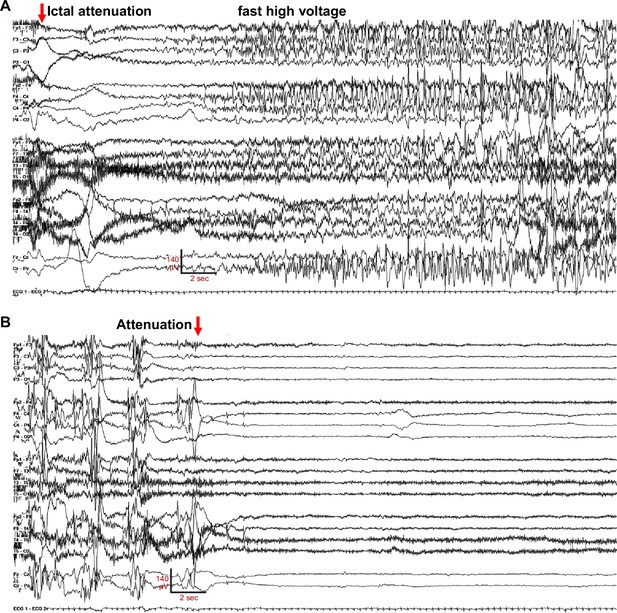
EEG of a bilateral onset seizure in G256W/+ individual 1, age 16 days.
The recording is continuous from (A) to (B). (A) Seizure onset with diffuse, bilateral amplitude EEG attenuation (red arrow), which is obscured in several electrodes by high-frequency muscle artifact (muscle artifact is better seen in Figure 1—video 1). (B) Seizure electrographic evolution to post-ictal voltage attenuation (red arrow). Settings: LFF 3 Hz, HFF 70 Hz, sensitivity 7uV/mm, 35 s/panel.
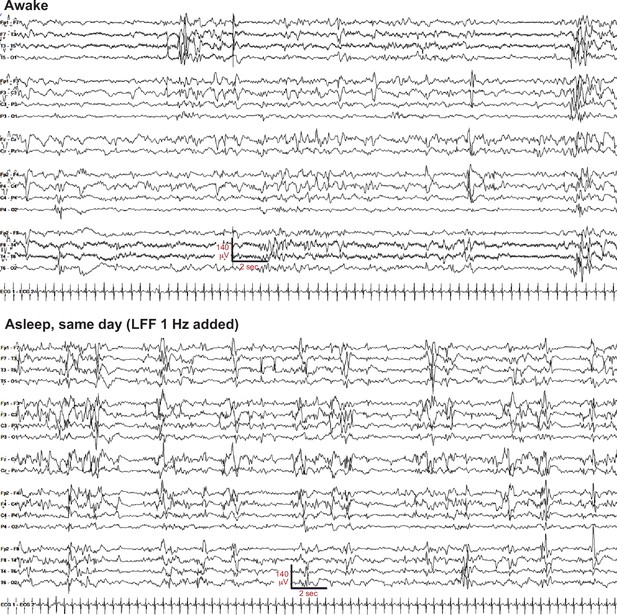
Examples of awake and sleep EEG background.
There is evidence of state change, with more discontinuity during sleep. The awake excerpt shows variable frequency composition, and excess multifocal sharps. Excess discontinuity and sharps indicate dysmaturity, but burst-suppression is not seen. Settings as in Figure 1, except LFF changed to 1 Hz.
EEG recording including pre-ictal, post-ictal attenuation and recovery of background of seizure excerpted in Figure 1.
As labeled, onset was preceded by eyeblink and muscle artifact. The interval of uninterrupted voltage attenuation between the end of the high voltage fast activity to the first epileptiform burst was 61 s. Interburst length progressively shortened in length, over about 3 min. Settings as in Figure 1.
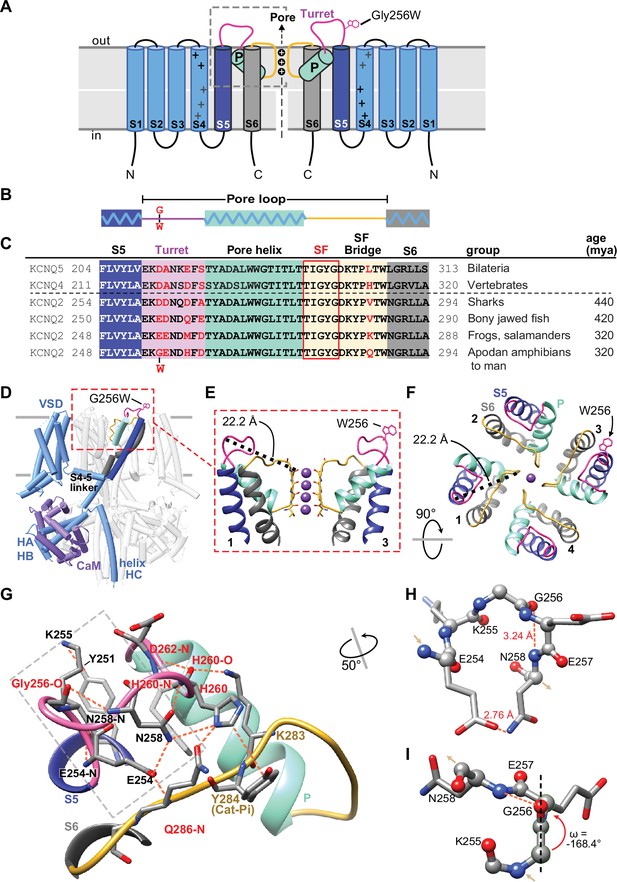
Gly256 is linked to the selectivity filter bridge segment via a hydrogen bond network among residues distinct to KCNQ2.
(A, B) Cartoons showing KCNQ2 membrane topology, including transmembrane segments S1-S6 and the P-loop (turret segment, purple; H5 or P-helix, cyan; and selectivity filter segment, yellow). Positions of the K+ selective pore, and the G256W substitution within the turret are indicated. (C) Alignment of human KCNQ4 and KCNQ5 sequences with KCNQ2 sequences of major vertebrate groups. Background colors match panels A-B, and the five selectivity filter lining residues are boxed in red. At four aligned positions within the turret and one in the SFB, KCNQ2 substitutions have evolved in amphibians and tetrapods (residues highlighted in red). (D) Rendering of the WT KCNQ2-calmodulin tetrameric structure obtained by cryoEM (PDB 7cr3), highlighting one subunit and the position of the G256W substitution near the channel’s extracellular domain apex. The Trp256 sidechain is at scale but its rotamer is chosen arbitrarily. The subunit closest to the viewer is partially deleted to reveal the highlighted subunit more clearly. (E) Ribbon rendering of the extracellular part of the PGD. For clarity, only two opposing side subunits are shown (as schematically in A). A Trp side chain is added at one Gly256 α-carbon. The distance between the G256 α-carbon and Y280 carbonyl oxygen at the selectivity filter mouth is labeled. (F) Top down view of the KCNQ2 regions as in panel (E) but showing four subunits. The Trp rotamer is different from panels (D, E) The S5, S6, and P-helices are labeled. (G) Hydrogen bonding network of the KCNQ2 turret. All predicted bonds are shown as dashed orange lines. The network extends from the S5 helix (Y251) via the labelled turret residue atoms to bonds involving residues of the SFB. As in (C), KCNQ2 residues that diverge in vertebrates are colored red. (H, I) The turret peptide region near G256, which is boxed with a grey dashed line in (G). The main chain is shown as ball-and-stick; side chains as stick. A tight turn occurs at K255 to N258, stabilized by hydrogen bonding between the G256 carbonyl oxygen and N258 amide. (I) The G256-E257 peptide deviates from planarity (ω = +/-180°) by 11.6° (~2.6 sd). In and out arrows indicate N and C termini, respectively. Abbreviations: mya, million years ago; VSD, voltage-sensor domain; HA-HB, the cytoplasmic helices A and B; CaM, calmodulin.
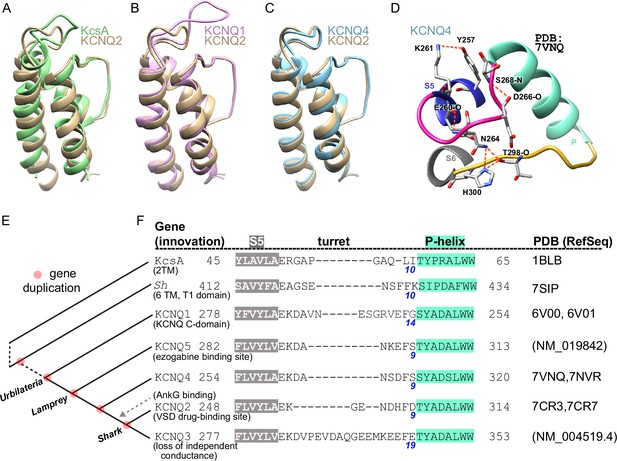
The G256W variant affects a divergent neuronal KCNQ turret structure enabling forming a bonding network linked to the ion selective pore.
(A–C) Aligned structural models of the extracellular portions of PGDs of KCNQ1, KCNQ4, and KcsA with that of KCNQ2. Single subunits are shown. (D) Cartoon of structural model of turret region of KCNQ4 highlighting the predicted hydrogen bonding network. Several bonds are conserved between KCNQ2 and KCNQ4, but the KCNQ4 network has fewer bonds (compare with Figure 2G). (E) Cladogram summarizing evolutionary relationships among several voltage-gated potassium channel genes. Gene duplication(s) are indicated by red circles, and are labeled by a common ancestor (or their extant descendant) possessing both duplicate genes. (F) Sequence alignments of KCNQ1-5 P loops and flanking S5 and S6 regions reveal relative conservation of KCNQ5, KCNQ4, and KCNQ2 (as in Figure 2C), and divergence of KCNQ1 and KCNQ3.
Movie illustrating position of the G256W substitution within the KCNQ2 channel pore turret and its distance to the selectivity filter.
Movie illustrating locations of residues contributing to a non-covalent bonding network extending from S5 to the selectivity filter.
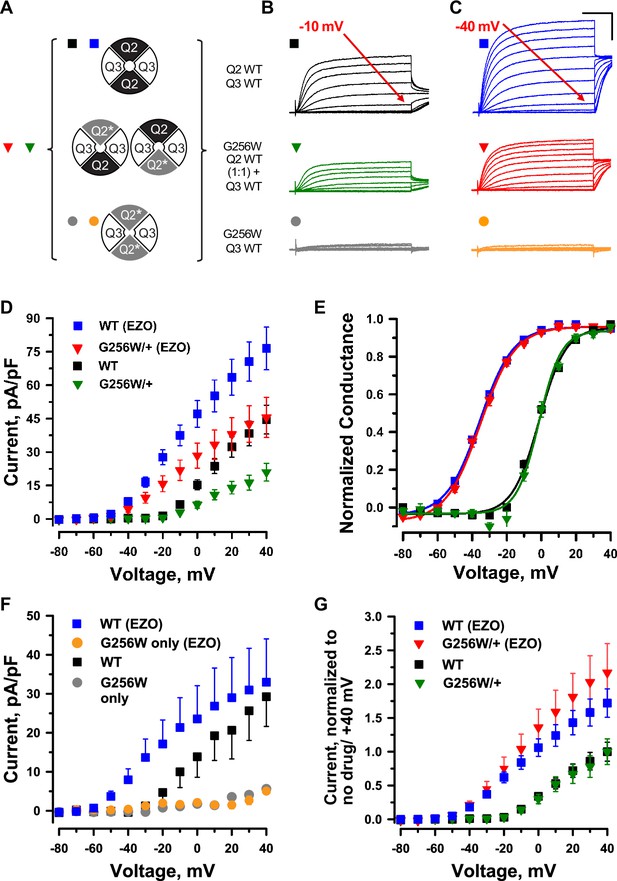
KCNQ2 G256W co-expression suppresses current in KCNQ2/KCNQ3 heteromeric channels.
(A) Cartoon showing the expected combinations of WT and G256W subunits under heterozygosity based on a simple random association model and preferred 2:2 stoichiometry for KCNQ2 and KCNQ3. (B–G) In vitro dissection of effects of G256W heterozygosity on currents. (B, C) Mean current families are shown for the indicated combinations of expression of KCNQ2 and KCNQ3 prior to and after addition of 10 μM ezogabine (n=60, 50; 40, 31; 28, 24 for the upper, middle, and lower conditions). Scale: 250ms and 20 pA/pF. (D, E) Current/voltage and conductance/voltage relationships for the indicated WT only and G256W/WT electroporations into KCNQ3 stable expressing cells. (F) Current/voltage relationship for G256W (‘homozygous’) heteromeric channels, compared with the subset of WT control cells studied in parallel by automated patch recording. (G) Replot of data from panel (D) At each voltage, mean current is normalized to mean current at +40 mV in absence of ezogabine.
-
Figure 3—source data 1
Mean numerical data, n’s, and mutagenic primers used to produce Figure 3.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig3-data1-v2.xlsx
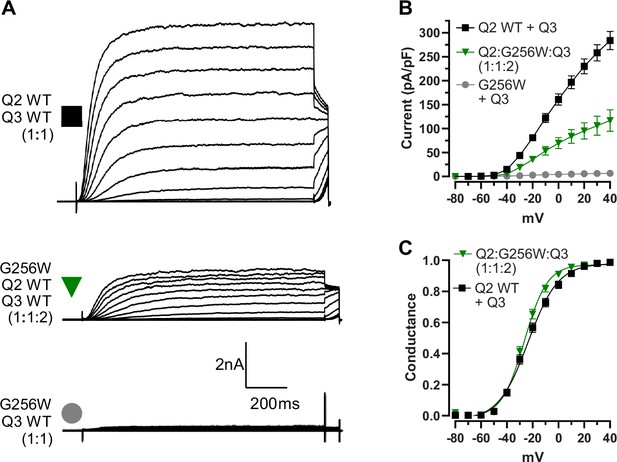
KCNQ2 G256W co-expression suppresses current in KCNQ2/KCNQ3 heteromeric channels recorded by manual patch-clamp.
(A) Representative current families for the indicated ratios of subunits. Note currents are larger than in Figure 3. (B, C) Current/voltage and conductance/voltage relationships for the indicated WT only and G256W/WT cells. WT: n=11 cells, Q2:G256W:Q3, n=15, G256WQ3: n=14.
-
Figure 3—figure supplement 1—source data 1
Numerical data and calculations used to produce Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig3-figsupp1-data1-v2.xlsx
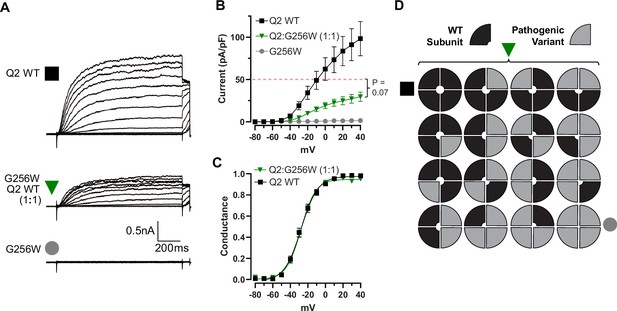
KCNQ2 G256W co-expression suppresses current in KCNQ2 homomeric channels recorded by manual patch-clamp.
(A) Representative current families for the indicated ratios of subunits. Homomeric currents are smaller than in Figure 3—figure supplement 1. (B, C) Current/voltage and conductance/voltage relationships for the indicated WT only and G256W/WT cells. (D) Cartoon showing the expected combinations of WT and G256W subunits under heterozygosity based on a simple random association model. Mutant subunits are included in 15/16 of channel tetramers. WT: n=10, Q2:G256W, n=19, G256W: n=11.
-
Figure 3—figure supplement 2—source data 1
Numerical data and calculations used to produce Figure 3—figure supplement 2.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig3-figsupp2-data1-v2.xlsx
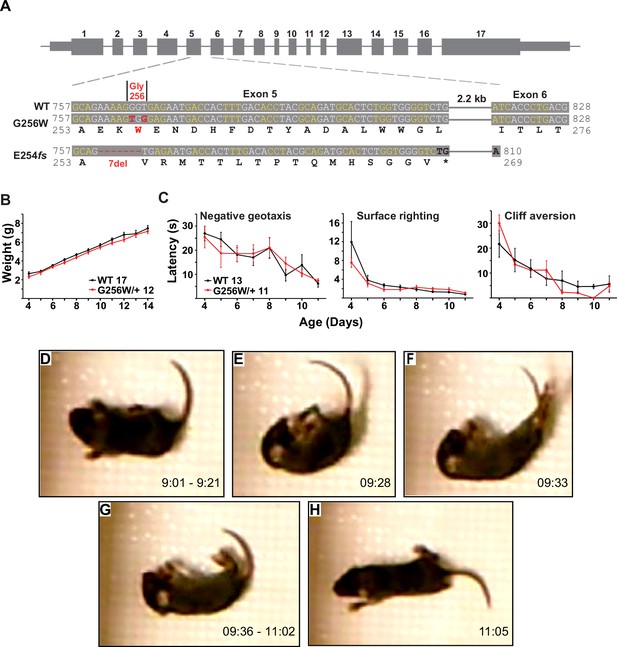
Immature heterozygous G256W mice exhibit normal development and have infrequent epileptic seizures.
(A) Upper, map of the Kcnq2 constructs. Lower, sequence alignments for the region between the middle of exon 5 and the beginning of exon 6. Although the human G256W variant is a single base substitution, Crispr/Cas9 editing introduced two substitutions, since the WT G256 codons differ between mouse (GGT) and human (GGG). Also aligned is the DNA and protein sequences of the frameshift mutation. (B) WT and G256W/+ mice showed no difference in weight gain during development. (C) WT and G256W/+ mice performed similarly in the developmental milestone assays for negative geotaxis, surface righting, and cliff aversion. (D–H) Screenshots of stages of a generalized seizure in a P10 G256W/+ mouse (see also: Figure 4—video 1). (D) Onset with immobility and myoclonic tail and forelimb shaking. (E) Abrupt fall to side with flexion posturing. (F) Evolution to hindlimb and tail extension posture. (G) Immobility with flaccid appearance, interrupted by brief episodes of tail, individual limb myoclonus or clonus. (H) Arouses, quickly regains upright posture, then normal mobility. Labels: time in 15 min source video.
-
Figure 4—source data 1
Individual mouse and mean data used for Figure 4B.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig4-data1-v2.xlsx
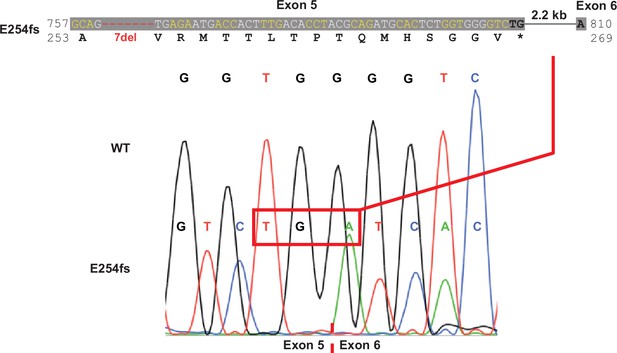
DNA, RNA, and predicted protein consequences of the G256W and E254fs*16 mutations.
Upper, DNA and predicted protein alignment of the frameshift mutation. Lower, Sanger sequence for cDNA from hippocampal mRNA of an E254fs/+ mouse. Splicing occurs at the WT junction, resulting in the predicted in-frame stop codon.
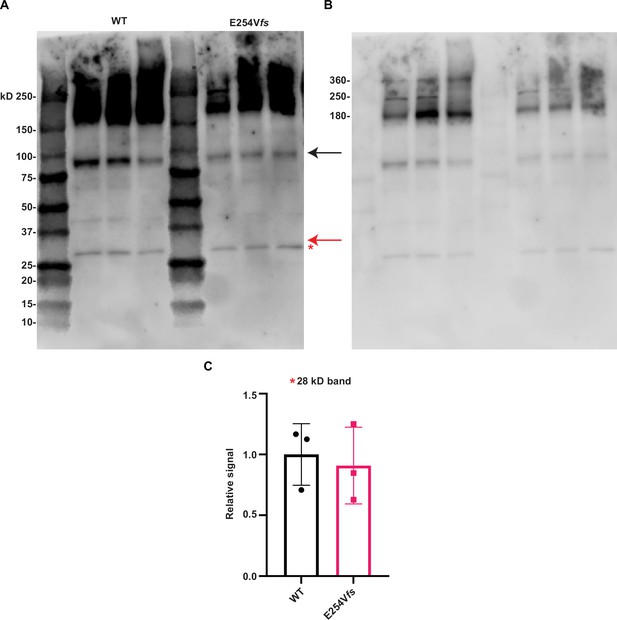
Western blotting reveals no evidence of the predicted E254fs truncated protein product.
(A) Western blot of WT and E254fs/+ cortical homogenates (3 biological replicates per genotype, all males), probed with KCNQ2 N-terminal antibody. Black arrow indicates the monomer (Mr ~85 kDa), red arrow indicates the estimated relative mobility (~29.7 kDa), of the truncated protein product made from the E254fs allele. Red asterisk indicates a~28 kDa band equally detected in both WT and E254fs/+. (B) Same blot as in (A) but windowed to show higher molecular weight bands. Bands at ~180 kDa and ~360 kDa consistent with predicted mobility of KCNQ2 dimers and tetramers, respectively. A band at ~250 kDa appears in all immunoblots of whole brain homogenates using our KCNQ2 N-terminal antibody. Nano-LC tandem mass spectrometry of peptides from an in-gel tryptic digest of this band showed high peptide counts for multiple abundant proteins and few KCNQ2 peptides. (C) Quantification of ~28 kDa band from (A).
-
Figure 4—figure supplement 2—source data 1
Band signal intensity data for the 28 kD band graphed in Figure 4—figure supplement 2C.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig4-figsupp2-data1-v2.xlsx
-
Figure 4—figure supplement 2—source data 2
This file is a zip folder containing an image of the uncropped blot with the relevant bands clearly labelled seen in Figure 4—figure supplement 2A, B.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig4-figsupp2-data2-v2.zip
-
Figure 4—figure supplement 2—source data 3
This file is a zip folder containing the original files of the full raw unedited blot seen in Figure 4—figure supplement 2A, B.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig4-figsupp2-data3-v2.zip
Generalized seizure in a P10 heterozygous G256W mouse.
This video includes from 9:01 to 11:05 of a 15:00 min period of open field observation. Animal recovers upright posture at 1:45 in the clip.
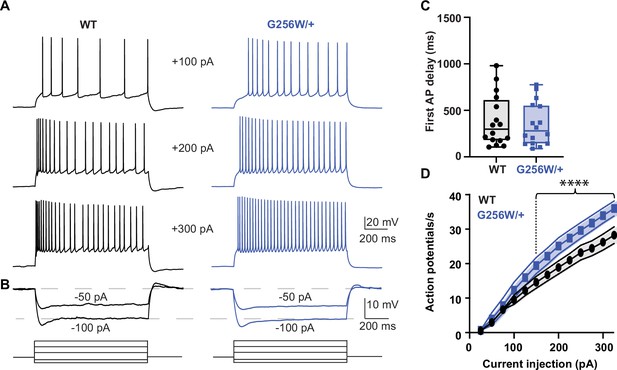
Heterozygous G256W mice have increased CA1 pyramidal cell excitability.
(A) Representative voltage responses to increasing current injection steps (step duration 1 s) in CA1 pyramidal neurons from WT and G256W/+ mice. The resting membrane potential was held at –65 mV. (B) Representative voltage responses to decreasing current injections steps (1 s) in CA1 pyramidal neurons from WT and G256W/+ mice. (C) Time to first action potential following step stimulus is not significantly different between groups (3 animals per group; WT and G256W/+, n=16 cells each). (D) Summary graph showing the effect of one copy of G256W on the action potential count (3 animals per group; WT and G256W/+, n=16 cells each, F(12,180)=5.8, **** is p<0.0001). Data are presented as mean and s.e.m.
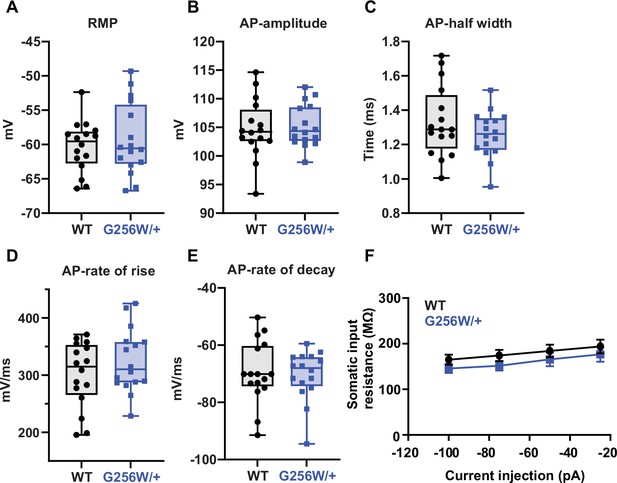
Several neuronal biophysical properties are unchanged.
(A) Resting membrane potential. (B) Action potential amplitude. (C) Action potential width. (D) Action potential rise slope. (E) Action potential decay. (F) Input resistance. For all panels, 3 animals per genotype, n=16 cells/genotype.
-
Figure 5—figure supplement 1—source data 1
Individual cell data used for Figure 5, Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig5-figsupp1-data1-v2.xlsx
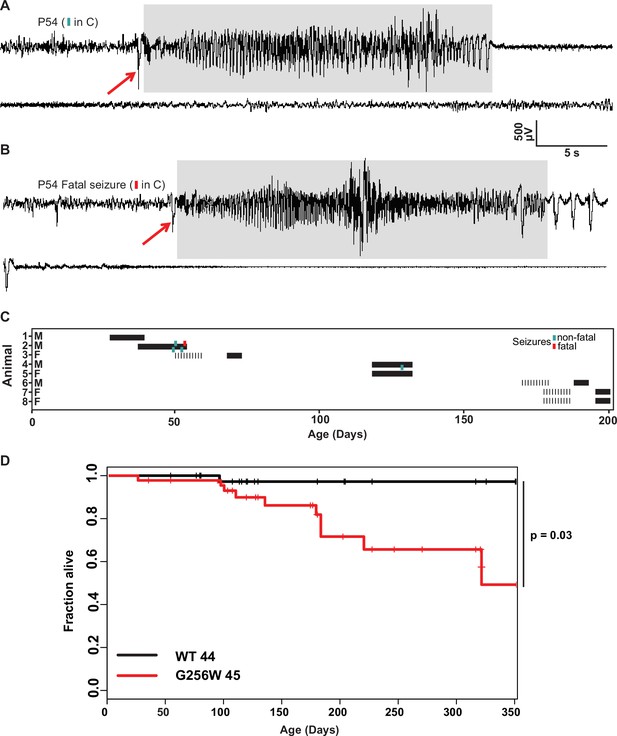
Convulsive seizures in adult heterozygous G256W mice show stereotyped electrographic features and reduced survival.
(A, B) EEGs of non-fatal and subsequent fatal seizure captured in a P54 male G256W/+ mouse (animal 2, C). Electrographic seizures were characterized by fast spiking, high amplitude activity lasting 15–20 s (highlighted in gray). Summary showing the sex, ages, duration of recordings and timing of seizures in 8 animals undergoing EEG. Turqoise hashmarks denote a survived seizure, red hashmark denote a fatal seizure. Black bars are periods on EEG; some recording were performed on a 6 hr/day schedule. (D) Survival curve of WT vs G256W/+ mice, hashmarks indicate censored mice. G256W/+ mice had signifcant mortality, p=0.0348 Cox propotional hazards model.
-
Figure 6—source data 1
Individual mouse survival data used for Figure 6D.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig6-data1-v2.csv
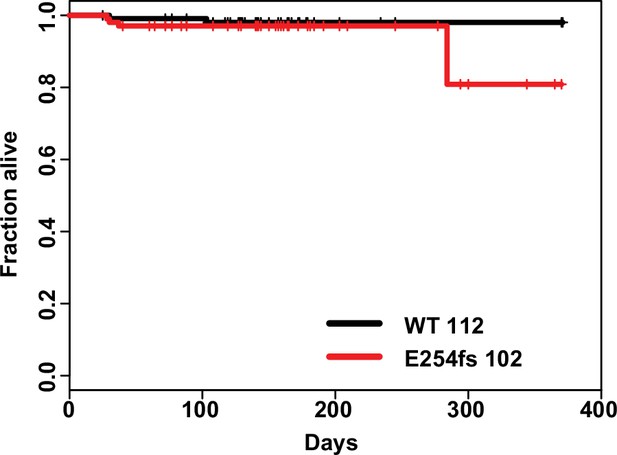
No significant mortality in heterozygous E254fs mice.
Survival curve of WT vs E254fs/+ mice, hashmarks indicate censored mice. E254fs/+ mice showed no significant mortality, p=0.452 Cox propotional hazards model.
-
Figure 6—figure supplement 1—source data 1
Individual mouse survival data used for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig6-figsupp1-data1-v2.csv
Video of a fatal convulsive seizure in a 4-month-old heterozygous G256W mouse.
Seizure onset (not shown) occurred 5–10 s prior to the start of recording with wild running and jumping, followed by arrest, then resumed (start of video). This was again followed by loss of postural control, followed by sustained forelimb flexor/hindlimb extensor posturing. Attempts to resuscitate the animal were begun immediately and were unsuccessful.
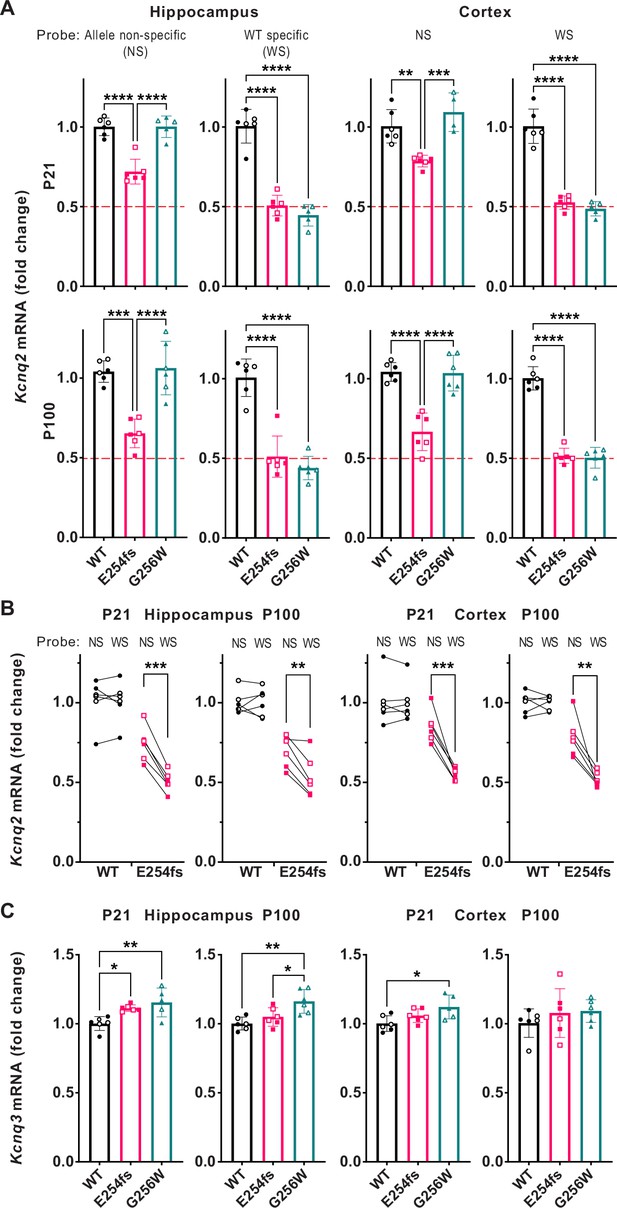
RT-qPCR shows incompletely efficient nonsense mediated decay of Kcnq2 E254fs/+ mRNA and increased Kcnq3 mRNA in E254fs/+ and G256W/+ mice.
(A) Upper, Kcnq2 mRNA levels in P21 hippocampus and neocortex. Levels were analyzed using TaqMan probes binding to both WT and variant Kcnq2 alleles (allele non-selective, NS), or binding only to WT only (WT selective, WS). Total Kcnq2 mRNA in E254fs/+ samples were significantly higher than 50% of WT (hippocampus: p=1.28 × 10–5, neocortex: p=3.5 × 10–7, one sample t-test). Lower, Kcnq2 mRNA levels in P100 hippocampus and neocortex, using the probes as above. Total Kcnq2 mRNA levels in E254fs/+ samples were significantly higher than the expected 50% of WT in hippocampus (p=0.0003) and neocortex: (p=0.0007). (B) Total and WT Kcnq2 mRNA, tested in parallel, by individual. Age, sex (male, open symbols), brain region, genotype, and probe are indicated. In all four tissues tested, E254fs/+ mice have greater total Kcnq2 mRNA than WT Kcnq2 mRNA (P21 hippocampus, p=0.0001; P100 hippocampus, p=0.005; P21 neocortex, p=0.0007; P100 neocortex, p=0.0053; pairwise t-test). (C) In P21 G256W/+ mice, Kcnq3 mRNA was significantly increased: 1.15-fold (+/-0.10, p=0.0043) in the hippocampus and 1.12-fold (+/-0.09, p=0.00213) in the neocortex. In P100 E254fs/+ mice, Kcnq3 mRNA significantly increased (1.11+/-0.02 fold, p=0.0245) in the hippocampus only. One way ANOVA, *=p < 0.05, **=p < 0.005, ***=p < 0.0005. (See Supplemental Data for statistical test calculations).
-
Figure 7—source data 1
The content of this file is used for the allele non-specific P21 Kcnq2 data seen in Figure 7A and P21 Kcnq3 data seen in Figure 7C.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig7-data1-v2.xlsx
-
Figure 7—source data 2
The content of this file is used for the allele non-specific P100 Kcnq2 data seen in Figure 7A Kcnq3 data seen in Figure 7C.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig7-data2-v2.xlsx
-
Figure 7—source data 3
The content of this file is used for the WT specific P21 Kcnq2 data seen in Figure 7A.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig7-data3-v2.xlsx
-
Figure 7—source data 4
The content of this file is used for the WT specific P100 Kcnq2 data seen in Figure 7A.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig7-data4-v2.xlsx
-
Figure 7—source data 5
The content of this file is used for Figure 7B.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig7-data5-v2.xlsx
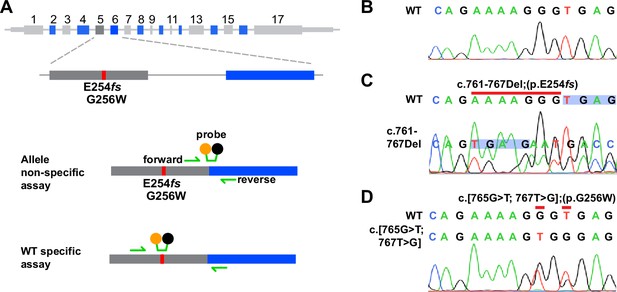
Measuring allele expression with allele-specific PCR amplification and cDNA Sanger sequencing.
(A) Probes used for allele non-selective and WT allele-selective RT-qPCR. (B) Upper, WT sequence (bases 758–771). Lower, Sanger trace of amplified Kcnq2 cDNA from WT hippocampus. (C) Upper, WT sequence; the red line indicates bases deleted in the E254fs allele. Lower, Sanger trace of cDNA from an E254fs/+ mouse. Blue shading highlights the shift following the deletion at positions 761–767. Peaks corresponding to E254fs transcripts are labeled and are smaller than WT peaks. (D) Upper, alignment of WT and missense variant DNA sequences, red lines highlight the two base substitutions at codon 256. Below, Sanger trace of amplified cDNA from G256W/+ hippocampus. Double peaks are visible at the bases mutated by Crispr.
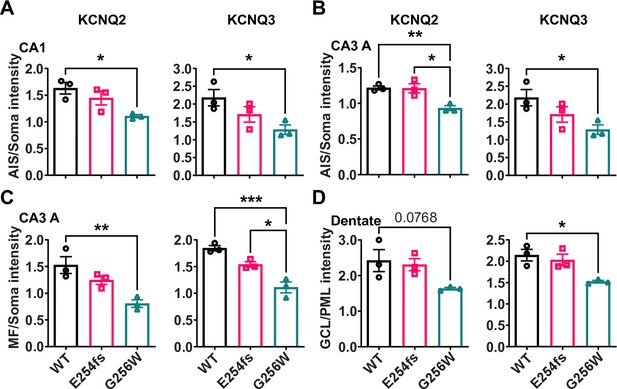
The ratios of axonal to somatic KCNQ2 and KCNQ3 labeling are reduced in CA1 and CA3 in heterozygous G256W mice.
(A, B) The ratios of AIS to somatic immunofluorescence intensity is significantly reduced for KCNQ2 and KCNQ3 in CA1 (A) and CA3 (B) for G256W/+ but not E254fs/+ mice. (C) The ratio of mossy fiber to somatic KCNQ2 and KCNQ3 immunofluorescence intensity is reduced in the CA3 for G256W/+ but not E254fs/+ mice. (D) In the dentate gyrus, the ratio between GCL and PML intensity is significantly reduced for KCNQ3 but not KCNQ2 in G256W/+ but not E254fs/+ mice. n=3 per genotype. One way ANOVA, *=p < 0.05, **=p < 0.005, ***=p < 0.0005.
-
Figure 8—source data 1
This file contains ROI data and calculations used for Figure 8.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig8-data1-v2.xlsx
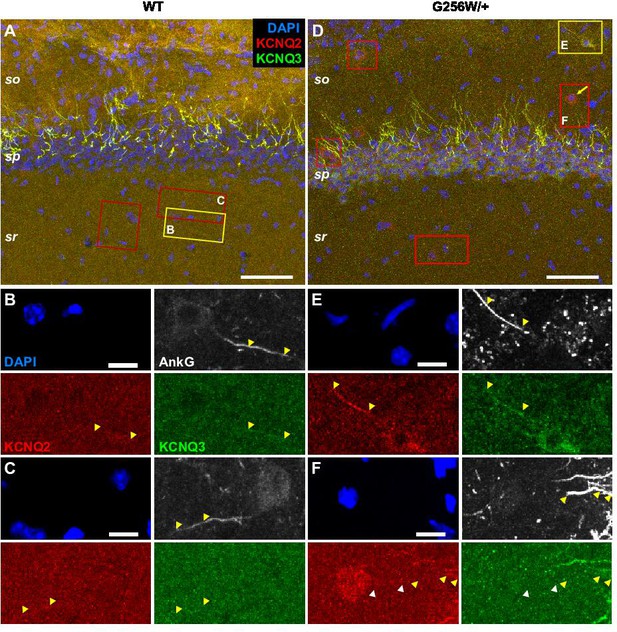
G256W/+ mouse interneurons in CA1 show somatic KCNQ2 labeling.
(A, D) Same source images of area CA1B as shown in Video 1, but including more of s. radiatum and s. oriens to show interneurons. In WT, positions of three S. radiatum interneurons are boxed, but comparison to higher magnification AnkG colabeling (e.g., B, C) shows these cells lack somatic labeling for KCNQ2 or KCNQ3. In G256W/+ image (D), four interneurons somatically labelled for KCNQ2 but not KCNQ3 are enclosed by red boxes. Yellow arrow indicates one interneuron highlighted with an arrow in Video 1 (KCNQ2 labeled, KCNQ3 unlabeled). The yellow box encloses the one interneuron found with somatically co-labeling for KCNQ2 and KCNQ3. (E) Individual laser channels for the interneuron enclosed by yellow box in (D). The soma is labeled for both KCNQ2 and KCNQ3. The nearby AIS, showing AnkG, KCNQ2, and KCNQ3 labeling, may arise from this or a different cell, as its origin was not verified by higher resolution re-imaging. (F) Interneuron somatically labeled for KCNQ2, not KCNQ3. Its AIS appears to arise from a weakly KCNQ2 labeled process (white arrowheads). In B, C and E, F, yellow arrowheads show AISs that are labeled strongly for AnkG and weakly and only distally for KCNQ2 and KCNQ3. Scales: A, 50 µm; B, 10 µm.
-
Figure 8—figure supplement 1—source data 1
This file contains interneuron labeling data cited in results related to Figure 8—figure supplement 1.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig8-figsupp1-data1-v2.xlsx
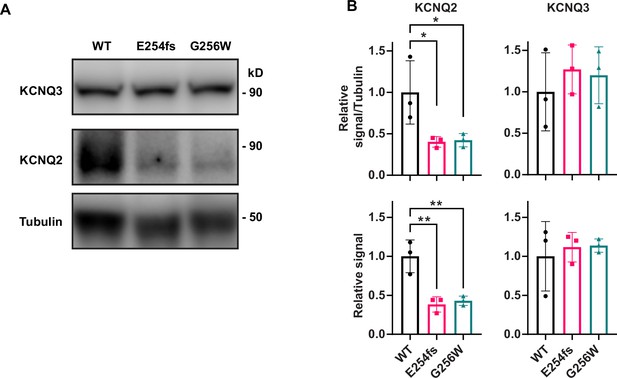
KCNQ2 protein is reduced in neocortex of P21 heterozygous E254fs and G256W mice.
(A) Representative western blots for all three genotypes probed for tubulin, KCNQ2 and KCNQ3. (B) Quantified KCNQ2 monomer and KCNQ3 signal relative to WT, normalized to tubulin and by protein loaded as assayed by BCA. n=3 per genotype, all males. One way ANOVA, *=p < 0.05, **=p < 0.005.
-
Figure 9—source data 1
This file has the quantification of signal intensities that appear in Figure 9B, Figure 9—figure supplement 1C.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig9-data1-v2.xlsx
-
Figure 9—source data 2
This file is a zip folder containing uncropped images of the blots with relevant bands clearly labelled, the source files for Figure 9A and Figure 9—figure supplement 1A, B.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig9-data2-v2.zip
-
Figure 9—source data 3
This zip folder contains raw blot image files (requires Image Studio Lite software) and unlabeled, intensity-windowed 8-bit tiff files used for blot figures.
- https://cdn.elifesciences.org/articles/91204/elife-91204-fig9-data3-v2.zip
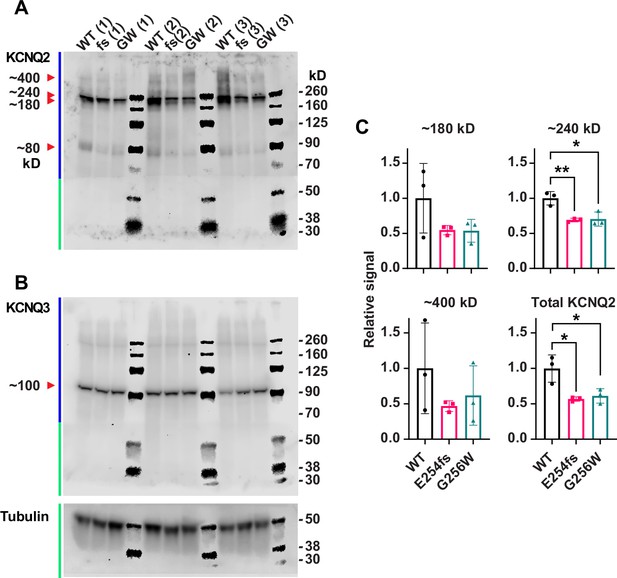
KCNQ2 antibodies, unlike KCNQ3, show complex electrophoretic banding pattern, and reduced levels in E254fs/+ and G256W/+ mice.
PVDF filter with electrotransferred brain proteins was cut between the 70 and 50 kDa markers. Each lane contains homogenate from an individual animal. Upper portion of the filter (blue bar at L) was initially probed for KCNQ2; lower portion (green bar at L) was initially probed for tubulin, then stripped and reprobed for KCNQ2. Next, the entire filter was stripped and probed again for KCNQ3. (A) KCNQ2 blots. Arrowheads point to KCNQ2 monomer band (Mr ~80 kDa), and candidate dimeric and oligomeric bands of Mr ~180 to~400 kDa. (B) Sequential probe of same filter from A, using KCNQ3 and tubulin antibodies. Arrowhead points to predicted KCNQ3 monomer with a Mr ~100 kDa. (C) Quantification of the indicated bands from the KCNQ2 blots, and the sum of the 4 KCNQ2 band intensities. Symbols are individual animals (n=3 per genotype). Means and SEMs are shown. One way ANOVA, *=p < 0.05.
Videos
Heterozygous G256W mice show reduced KCNQ2 and KCNQ3 labeling of CA1 pyramidal cell AISs and increased labeling of neuronal somata.
Identically processed age P21 tissue sections of WT (upper) and G256W/+ (lower) mice; area CA1B was imaged under identical settings. Confocal image stacks are shown as maximal intensity projections. In the animation, channels for the indicated markers are allowed to fade into the next, enabling evaluation of colabeling. DAPI marks cell nuclei. AnkG strongly marks AISs and lightly labels somata and proximal apical dendrites. An arrow highlights one stratum oriens interneuron somatically labeled for KCNQ2 only. Labels: DAPI, 4',6-diamidine-2'-phenylindole; so, stratum oriens; sp, stratum pyramidale, sr, stratum radiatum. Scale: 50 μm.
In CA1, the KCNQ2 and KCNQ3 cellular and subcellular immunolabeling patterns appear similar for WT and heterozygous E254fs mice.
Ankyrin-G marks position of AISs. KCNQ2 and KCNQ3 strongly label CA1 AISs in E254fs/+ mice, and do not show increased somatic labeling compared to WT. Highlighted by an arrow is one interneuron in stratum pyramidale that was somatically labeled for KCNQ2 only. Scale: 50 μm.
Heterozygous G256W mice show increased CA3 pyramidal cell somatic labeling and reduced mossy fiber labeling for KCNQ2 and KCNQ3.
Yellow lines demarcate the borders of sp; the sp-sl border is cut obliquely through the tissue section in the G256W/+ sample. PanNav strongly labels the unmylenated axons of the mossy fibers in stratum lucidum of both samples. PanNav also labels the obliquely cut AISs of pyramidal cell neurons, which are mostly located within sp. Scale: 50 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Rabbit anti-KCNQ2N1 | PMID: 11739564, PMID: 14762142 | RRID:AB_2314688 | IHC (1:200) WB (1:400) |
| Antibody | Guinea pig anti-KCNQ3N1 | PMID: 16525039, PMID: 18827480 | RRID:AB_3303619 | IHC (1:500) WB (1:1000) |
| Antibody | Mouse anti-PanNav igG1 | Sigma Aldrich | Catalog #: S8809 RRID:AB_477552 | IHC (1:200) |
| Antibody | Anti-α-Tubulin antibody, Mouse monoclonal | Sigma Aldrich | Catalog #: T6199 RRID:AB_477583 | WB (1:10,000) |
| Antibody | Mouse anti-Ankyrin G IgG2a | Neuromabs | Clone N106/36.1 RRID:AB_10697718 | IHC (1:1000) |
| Antibody | Goat anti-Rabbit IgG Alexa Fluor Plus 555 | Invitrogen | Catalog #: A32732 UH287772 RRID:AB_2633281 | IHC (1:500) |
| Antibody | Cy5 AffiniPure Donkey Anti-Guinea Pig IgG (H+L) | Jackson ImmunoResearch | Code: 706-175-148 Lot #: 144177 RRID:AB_2340462 | IHC (1:500) |
| Antibody | Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Catalog #: A-21121 Lot #: 2083196 RRID:AB_2535764 | IHC (1:500) |
| Antibody | Goat anti-Mouse IgG2a Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Catalog #: A-21131 Lot #: 1145167 RRID:AB_2535771 | IHC (1:500) |
| Antibody | Peroxidase IgG Fraction Monoclonal Mouse Anti-Rabbit IgG, light chain specific | Jackson ImmunoResearch | Code: 211-032-171 Lot #: 97224 RRID:AB_2339149 | WB (1:5000) |
| Antibody | Peroxidase AffiniPure F(ab')₂ Fragment Goat Anti-Guinea Pig IgG (H+L) | Jackson ImmunoResearch | Code: 106-036-003 Lot #: 121003 RRID:AB_2337405 | WB (1:5000) |
| Antibody | Peroxidase AffiniPure Goat Anti-Mouse IgG, light chain specific | Jackson ImmunoResearch | Code: 115-035-174 Lot #: 12150 RRID:AB_2338512 | WB (1:10,000) |
| Cell line | Flp-In-CHO (Chinese hamster ovary) cell line | Invitrogen/Thermo Fisher Scientific | Catalog #: R75807 Lot #: 1819218 (Oct 2017) | Used in automated patch experiments |
| Cell line | CHO (Chinese hamster ovary) cell line | PMID: 15862463 | Used in manual patch expts | |
| Transfected construct | Human KCNQ2 cDNA in pcDNA3 | PMID: 9872318; PMID: 16525039 | NP_742106 | Manual patch expts; gift of Thomas Jentsch |
| Transfected construct | Human KCNQ3 cDNA in pcDNA3 | PMID: 9872318; PMID: 16525039 | NM_004519.4 | Manual patch expts; gift of Thomas Jentsch |
| Transfected construct | Human KCNQ2 cDNA in pIRES2-EGFP | PMID: 35104249 | NM_172108 | Automated patch expts |
| Transfected construct | Human KCNQ3 cDNA in pcDNA5/FRT | PMID: 35104249 | NM_004519.4 | Automated patch expts; from Thomas Jentsch |
| Strain, strain background (Mus musculus) | Kcnq2G256W (C57BL/6J-Kcnq2em4(G256W)Lutzy/J) | This paper; Jackson Laboratory Mutant Mouse Resource and Research Center | JR 29407 | Heterozygous G256W mice |
| Strain, strain background (Mus musculus) | Kcnq2E254fs*16 (C57BL/6J-Kcnq2em5(7del)Lutzy/J) | This paper; Jackson Laboratory Mutant Mouse Resource and Research Center | JR 29408 | Heterozygous E254fs mice |
| Recombinant DNA reagent | pIRES2-EGFP mammalian expression vector | BD Biosciences-Clontech | ||
| Recombinant DNA reagent | pcDNA3 mammalian expression vector | ThermoFisher Scientific | ||
| Recombinant DNA reagent | pcDNA5/FRT mammalian expression vector | ThermoFisher Scientific | ||
| Chemical compound, drug | ProLong Gold Antifade Mountant | ThermoFisher Scientific | Cat. No. P36931 | |
| Commercial assay, kit | RNeasy Lipid Tissue Mini Kit | Qiagen | Cat. No. 74804 | |
| Commercial assay, kit | SuperScript III First-Strand Synthesis SuperMix | Invitrogen | Ref 18080–400 | |
| Commercial assay, kit | TaqMan Fast Advanced Master Mix for qPCR | Applied Biosystems | Cat. No. 4444557 | |
| Commercial assay, kit | Kcnq2 Taqman gene expression assay | Thermo Fisher Scientific | Assay ID: Mm00440084_mH | Allele non-specific assay |
| Commercial assay, kit | Kcnq2 Taqman gene expression assay | Thermo Fisher Scientific | Assay ID: AP2XHY6 | WT specific assay |
| Commercial assay, kit | Kcnq3 Taqman gene expression assay | Thermo Fisher Scientific | Assay ID: Mm00548884_m1 | |
| Commercial assay, kit | Gapdh Taqman gene expression assay | Thermo Fisher Scientific | Assay ID: Mm99999915_g1 | |
| Commercial assay, kit | QuantStudio 3 Real-Time PCR System, 96-well, 0.2 mL, laptop | Thermo Fisher Scientific | A28567 | |
| Sequence-based reagent | Kcnq2 exon 4–7 region forward primer | This paper | PCR Primers | 5’-CGG TAG TCT ACG CTC ACA GC-3’ |
| Sequence-based reagent | Kcnq2 exon 4–7 region reverse primer | This paper | PCR Primers | 5’-TCT TGG ACT TTC AGG GCA AA–3’ |
| Sequence-based reagent | Kcnq2 G256W mutagenic forward primer | This paper | PCR Primers | 5’-gcagagaaaTggg agaacgaccactttgacacctac–3’ |
| Sequence-based reagent | Kcnq2 e G256W mutagenic reverse primer | This paper | PCR Primers | 5’-ttctcccAtttctctgcc aagtacaccaggaacgag-3’ |
| Commercial assay, kit | DNA Clean & Concentrator –25 Kit | Zymo Research | D4033 | |
| Commercial assay, kit | ApexHot Start 2 X Master Mix Blue Apex Buffer 1 | Genesee Scientific | 42–143 | |
| Chemical compound, drug | 4 X Protein Sample Loading Buffer for Western Blots | Li-Cor | Selected P/N: 928–40004 | |
| Chemical compound, drug | Precision Plus Protein Standards All Blue | Biorad | 161–0373 | |
| Commercial assay, kit | Chameleon Duo Pre-stained Protein Ladder | Li-Cor | 928–60000 | |
| Commercial assay, kit | 7.5% Mini-PROTEAN TGX Precast Protein Gels, 12-well, 20 µl | Biorad | #4561025 | |
| Chemical compound, drug | Pierce protease inhibitor mini tablets, EDTA-free | Thermo Fisher Scientific | Ref: A32955 | |
| Commercial assay, kit | ECLPrime Western Blotting Detection Reagents | Cytiva Life Sciences | RPN2232 | |
| Commercial assay, kit | Amersham Hybond 0.2 µm PVDF | Amersham | Cat. No. 10600021 | |
| Commercial assay, kit | BCA Protein Assay Kit | Pierce | Prod # 23227 | |
| Commercial assay, kit | CoverWell Incubation Chamber Gasket | Thermo Fisher Scientific | Cat. No. C18150 | |
| Commercial assay, kit | Digital Sonifier 450 | Branson | Cat. No. 15338553 | |
| Commercial assay, kit | 1/8” Sonifier Doublestep Microtip | Branson | Cat. No. 101063212 | |
| Commercial assay, kit | MaxCyte STx electroporator | MaxCyte | ||
| Commercial assay, kit | Axopatch 200B amplifier | Molecular Devices | ||
| Commercial assay, kit | Multiclamp 700B amplifier | Molecular Devices | ||
| Commercial assay, kit | cFlow perfusion controller | Cell MicroControls | ||
| Commercial assay, kit | mPre8 manifold | Cell MicroControls | ||
| Commercial assay, kit | Leica VT1200S | Leica | ||
| Commercial assay, kit | Single Channel Temperature Controller | Warner Instruments | Cat. No. TC-324C | |
| Software, algorithm | Prism 9.0 | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | NIS-Elements | Nikon | RRID:SCR_002776 | |
| Software, algorithm | Snapgene | Snapgene | RRID:SCR_015052 | |
| Software, algorithm | Image Studio v5.2 | Li-Cor | RRID:SCR_015795 | |
| Software, algorithm | Pclamp/ Clampfit | Molecular Devices | RRID:SCR_011323 | |
| Software, algorithm | PatchController384 V.1.3.0 | Nanion Technologies | ||
| Software, algorithm | Rstudio | Rstudio | RRID:SCR_000432 | |
| Software, algorithm | Chimera-1.16-win64; ChimeraX-1.6.1 | UCSF | RRID:SCR_004097; RRID:SCR_015872 | |
| Software, algorithm | LabChart | AD Instruments | RRID:SCR_017551 | EEG data analysis |
Additional files
-
Source data 1
Single marker greyscale images and selected 2 color merged images of CA1.
In upper panels, the simultaneously acquired AnkG color channel is not included in the merge, for clarity. In middle and lower panels, the yellow boxed regions of upper panels are shown as single channel greyscale and as the indicated merges of 2 channels. In lower merge images, yellow arrows indicate KCNQ2/KCNQ3 overlap, white and green arrows indicate AnkG-only labeling of proximal AIS. Scales: 50 μm, upper; 10 μm, middle and lower.
- https://cdn.elifesciences.org/articles/91204/elife-91204-data1-v2.pdf
-
Source data 2
Single marker grey-scale images and selected merged images of CA1.
In middle and lower panels, the yellow boxed in upper panels is shown as single channel greyscale and as the indicated merges of 2 channels. In lower merged images, yellow arrows indicate portions of AISs showing KCNQ2/KCNQ3 overlap, and white and green arrows indicate AnkG-only labeling of proximal AISs. Scales: 50 μm, upper; 10 μm, middle and lower.
- https://cdn.elifesciences.org/articles/91204/elife-91204-data2-v2.pdf
-
Source data 3
Single marker grey-scale images and selected merged images of CA3.
Scales: 50 μm.
- https://cdn.elifesciences.org/articles/91204/elife-91204-data3-v2.pdf
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91204/elife-91204-mdarchecklist1-v2.docx





