Tissue-specific O-GlcNAcylation profiling identifies substrates in translational machinery in Drosophila mushroom body contributing to olfactory learning
Peer review process
This article was accepted for publication as part of eLife's original publishing model.
History
- Version of Record updated
- Version of Record published
- Accepted
- Preprint posted
- Received
Decision letter
-
Mani RamaswamiReviewing Editor; Trinity College Dublin, Ireland
-
K VijayRaghavanSenior Editor; National Centre for Biological Sciences, Tata Institute of Fundamental Research, India
Our editorial process produces two outputs: i) public reviews designed to be posted alongside the preprint for the benefit of readers; ii) feedback on the manuscript for the authors, including requests for revisions, shown below. We also include an acceptance summary that explains what the editors found interesting or important about the work.
Decision letter after peer review:
Thank you for submitting your article "Olfactory learning in Drosophila requires O-GlcNAcylation of mushroom body ribosomal subunits" for consideration by eLife. Your article has been reviewed by 2 peer reviewers, and the evaluation has been overseen by a Reviewing Editor and K VijayRaghavan as the Senior Editor.
The reviewers have discussed their reviews with one another, and the Reviewing Editor has drafted this to help you prepare a revised submission.
Essential revisions (for the authors):
1) Potential reasons for the very low overlap with other methods to identify GlycNac-modified proteins requires more discussion o, as well as potential problems with the authors own approach
2) The current data do not rule out the possibility that altering GLcNAC affects LTM through a mechanism independent of ribosome modification and/or activity dependent translational control. It should be clearly acknowledged in the abstract and discussion that the data fall short of causally connecting ribosomal protein modification with translational control required for LTM. Phrases like "consistent with" may be useful.
3) Please respond carefully and as you consider appropriate all of the other comments of the reviewers.
Reviewer #1 (Recommendations for the authors):
1. This study relies almost exclusively on the over-expression of foreign proteins and interpreting their effects based solely on their known affinity and activity for O-GlcNac modifications. The effects of high levels of over-expression are often hard to identify or know and while there are appropriate controls in their studies, using catalytically inactive mutant forms for comparison, there is no discussion of this issue. The authors do describe that over-expression in optic and antennal lobes for example, provided reason for exclusion by virtue of differences in susceptibility (that term is not explained), indicating there are behavioral effects that can interfere with their assays. Some direct discussion of this potential problem is warranted.
2. Their comparison of several methods of identifying O-GlcNAc-modified proteins was tested in HEK293 cells, providing a measure of their method of proximity-labelling using their Clostridium OGA construct. The set of proteins identified by all three tested methods revealed 48, a modest number compared to the total sets for each method (2119, 273 and 336) for chemoenzymatic, direct capture and TurboID-CpOGA. This level of concordance suggests that different methods might well identify different sets of proteins, a concern for interpreting the functional significance of these changes. Do the authors argue that these results only affect sensitivity or could it possibly bias the classes of O-GlcNAc modified proteins identified?
3. The rescue of associative learning phenotypes mediated by reductions in O-GlcNAc modification by over-expression of dMyc is a very important experiment that supports their conclusion that translational deficits are the basis of the learning defects. The authors should explain further in the text the rationale and basis for this experiment, namely, how dMyc affects the protein synthetic machinery and why these changes could rescue loss of O-GlcNAc modifications.
4. The paper is challenging to read, and I suggest the authors provide it for reading and editing by someone not in their immediate field in order to make changes that make it more accessible to a general reader.
5. Their graphics are terrific and provide great assistance in clarifying the figures. Well done.
Reviewer #2 (Recommendations for the authors):
As it stands the manuscript does not support the idea that O-GlcNAcylation of ribosomal proteins in the mushroom body (MB) is required for protein synthesis and olfactory learning. Experiments targeting specific ribosomal components could be a compelling demonstration. For example, Ser/Thr residues that get O-GlcNAcylated in RPL24 or RPS3 could be mutated. If these mutants showed learning defects it would be an important result. Same type of mutants in ex vivo OPP assays could be performed to determine their effect on protein synthesis. These types of experiments would definitely show specific requirement of ribosomal protein modification for ribosomal activity and associative learning.
https://doi.org/10.7554/eLife.91269.sa1Author response
Essential revisions (for the authors):
1) Potential reasons for the very low overlap with other methods to identify GlycNac-modified proteins requires more discussion o, as well as potential problems with the authors own approach
Thank you for raising this important issue. A lot of effort has been made to identify protein O-GlcNAc modifications and many profiling methods have been established in the past 30 years1. However, as you pointed out, the O-GlcNAcome identified using different methods are quite diverse. There are at least two possible reasons behind the observed low overlap.
First, O-GlcNAcylation is highly responsive to a wide range of intrinsic and extrinsic stimuli, acting as an important cellular metabolic and stress sensing mechanism2. Meanwhile, as a monosaccharide modification, the addition and removal of O-GlcNAc moiety can be very rapid, with cycling rates as short as several minutes3,4. This dynamic nature of O-GlcNAc cycling contributes to the observed diversity among different profiling attempts.
Second, the high lability of the GlcNAc moiety and low abundance of the O-GlcNAcylated proteins pose significant challenges in direct utilization of mass spectrometry for O-GlcNAcylation profiling1,5,6. Different enrichment methods are needed to protect and stabilize the GlcNAc moiety. This unavoidably introduces bias in substrates preference intrinsic to the methods, which also contributes to the limited overlap of the identified O-GlcNAc substrates.
As for our approach reported here, there are several potential caveats: (1) Similar to the direct capture methods, the sensitivity is limited compared to the chemoenzymatic labeling methods. Additionally, our method uses the differential enrichment in the TurboID-CpOGACD experimental group relative to the TurboID-CpOGADM control group to identify putative O-GlcNAc substrates. Therefore, the sensitivity is also influenced by the cutoff chosen in the analysis. (2) Our method relies on ectopic expression of bacterial CpOGA mutants fused with TurboID to label the O-GlcNAcome. The introduction of these foreign proteins could impose stress on the targeted tissue and interfere with its normal functions. Therefore, we included CpOGADM as a control in all the experiments to minimize possible artifacts. (3) The TurboID used in our method can biotinylate all the proximal proteins within ~ 10 nm radius. As a result, the identified proteins can be in complex with other O-GlcNAc substrates but itself is not directly O-GlcNAcylated. To be accurate, we used “putative” or “candidate” substrates and O-GlcNAc “interactome” in our description.
We have updated the Introduction and Discussion to include these aforementioned points.
2) The current data do not rule out the possibility that altering GlcNAc affects LTM through a mechanism independent of ribosome modification and/or activity dependent translational control. It should be clearly acknowledged in the abstract and discussion that the data fall short of causally connecting ribosomal protein modification with translational control required for LTM. Phrases like "consistent with" may be useful.
We appreciate that you bring up this crucial point. Our study demonstrated that: (1) hypo-O-GlcNAcylation resulted in learning defect and reduced protein synthesis in mushroom body; (2) knockdown of several ribosomal subunits to reduce protein synthesis in mushroom body was sufficient to drive learning defect; (3) upregulation of protein synthesis by overexpression of dMyc in mushroom body ameliorated the cognitive defect caused by hypo-O-GlcNAcylation. These observations consolidated a link between hypo-O-GlcNAcylation and cognitive impairment in Drosophila, and suggested that insufficient O-GlcNAcylation in the mushroom body of Drosophila brain reduced local translational activity that contributed to the observed olfactory learning deficits. However, our data indeed fall short for a causal connection between O-GlcNAcylation of ribosomes and translational activity. In fact, our profiling also identified many translational initiation factors whose O-GlcNAcylation could impact translation as well.
Therefore, your point has been well taken, and we have made the following changes accordingly: (1) we changed the Title of our manuscript to put more emphasis on the method itself; (2) we replaced “ribosomal activity” with “translational activity” in all the texts to describe the findings more accurately; (3) we used words like “suggest” and “consistent with” to emphasize it is one of the explanations whenever possible; (4) we specifically pointed out in the Discussion that the impact of O-GlcNAcylation on ribosomal activity needs future structural and biochemical characterizations.
We think these changes have greatly improved the accuracy of our description, and we want to express our gratitude again for this suggestion.
3) Please respond carefully and as you consider appropriate all of the other comments of the reviewers.
We have prepared a point-by-point response to all the comments, and we express our sincere appreciation to you for the meticulous evaluation of our manuscript.
Reviewer #1 (Recommendations for the authors):
1. This study relies almost exclusively on the over-expression of foreign proteins and interpreting their effects based solely on their known affinity and activity for O-GlcNAc modifications. The effects of high levels of over-expression are often hard to identify or know and while there are appropriate controls in their studies, using catalytically inactive mutant forms for comparison, there is no discussion of this issue. The authors do describe that over-expression in optic and antennal lobes for example, provided reason for exclusion by virtue of differences in susceptibility (that term is not explained), indicating there are behavioral effects that can interfere with their assays. Some direct discussion of this potential problem is warranted.
Thank you for this valuable advice. The reason we chose a bacterial OGA to manipulate O-GlcNAcylation levels in flies are following.
First, the function of OGA is highly conserved during evolution (Author response image 1). The N-terminal O-GlcNAc hydrolase catalytic domain of CpOGA has sequence homology to Drosophila OGA7,8. Previous studies have shown that CpOGA exhibits remarkable catalytic activity on Drosophila O-GlcNAcylated proteins9. We have codon-optimized CpOGA sequence to Drosophila, and constructed the transgenic flies.
Second, the first insight into the structure of OGA catalytic domain came from the crystal structure of CpOGA10. Until now the structural information of Drosophila OGA is not available. The crystal structure of CpOGA clearly reveals that the Asp298 residue is crucial for its catalytic activity, and Asp401 residue for O-GlcNAc peptide binding. This structural and mechanistic information can guide the rational design of active or control OGA, as well as the TurboID-fused OGA mutants for profiling, during the generation of transgenic lines.
Third, in OGT mutated patient-derived fibroblasts, the expression of OGA is often compensatorily downregulated via mechanisms that are currently not fully understood. We decided to use a bacterial OGA to bypass the potential feedback between OGT and OGA. Indeed, we found that the expression of CpOGAWT in Drosophila reduced total O-GlcNAcylation level without altering the mRNA abundance of endogenous oga or sxc11.
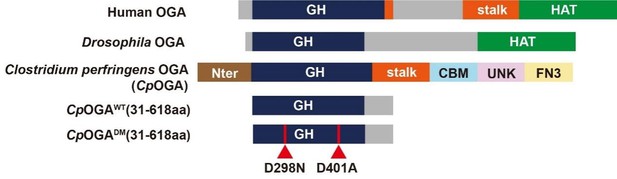
Schematic of the domain structures of human OGA, Drosophila OGA, and CpOGA.
The O-GlcNAcase activity resides in the GH domain (blue). The C-terminal of Drosophila OGA harbors a putative histone acetyltransferase domain (HAT-like). Residues 31–618 of CpOGA (corresponding to the GH domain plus an additional C-terminal) are used in this study.
As for the experiments in the optic and antennal lobes, we found ectopic expression of CpOGAWT in these brain regions altered odor acuity toward MCH or OCT when compared with CpOGADM control. This difference in olfactory susceptibility invalidated the learning tests. Therefore, we excluded these flies from subsequent olfactory learning test. The reason why expression of CpOGAWT in optic and antennal lobes impacted olfactory perception is not clear. We think the multisensory nature of Drosophila behavior and perception might be involved. Perhaps there are functional interactions between olfactory processing neuronal circuit and visual as well as sensory neuronal circuits.
We have updated the Results and Discussion accordingly to accommodate the key points.
2. Their comparison of several methods of identifying O-GlcNAc-modified proteins was tested in HEK293 cells, providing a measure of their method of proximity-labelling using their Clostridium OGA construct. The set of proteins identified by all three tested methods revealed 48, a modest number compared to the total sets for each method (2119, 273 and 336) for chemoenzymatic, direct capture and TurboID-CpOGA. This level of concordance suggests that different methods might well identify different sets of proteins, a concern for interpreting the functional significance of these changes. Do the authors argue that these results only affect sensitivity or could it possibly bias the classes of O-GlcNAc modified proteins identified?
Thank you for raising this important issue. The possible reasons for the low overlap among datasets generated by different profiling methods are elaborated in Essential revisions 1. We have also updated the Introduction and Discussion accordingly.
As for the question of whether it only affects sensitivity or introduces bias to different classes of substrates, we performed gene ontology (GO) analysis on these three datasets and found that they were enriched in similar biological processes (Figure S2E). Therefore, despite different enrichment strategies unavoidably introducing bias in substrate preference, we think the main classes of O-GlcNAcylated substrates from different profiling attempts remain comparable.
3. The rescue of associative learning phenotypes mediated by reductions in O-GlcNAc modification by over-expression of dMyc is a very important experiment that supports their conclusion that translational deficits are the basis of the learning defects. The authors should explain further in the text the rationale and basis for this experiment, namely, how dMyc affects the protein synthetic machinery and why these changes could rescue loss of O-GlcNAc modifications.
Thank you for highlighting this point. One well-recognized function of MYC is to serve as a direct regulator of ribosome biogenesis, promoting protein synthesis through transcriptional control of RNA and protein components of ribosomes, as well as factors involved in processing and nuclear export of these ribosomal subunits 12-14. MYC binds to sequences surrounding rDNA and directly promotes Pol I transcription throughout this region15-17. It also contributes to driving 5S transcription by Pol III18. These activities rely on the ability of MYC to both interact with the factors that promote Pol I recruitment to rDNA promoters and induce changes in local chromatin organization within rDNA repeats. Moreover, MYC directly enhances the transcription of key factors involved in rRNA transcription, such as UBF and components of the SL1 complex16. Furthermore, MYC boosts Pol II-mediated transcription of genes that encode ribosomal proteins18-20. The Drosophila dMyc, like its vertebrate homologs, activates numerous target genes associated with ribosome biogenesis and translation12,21-24.
We think dMyc boosts ribosome biogenesis at transcription level, which compensates the reduced translational activity caused by the insufficient post-translational O-GlcNAc modification. We have updated the RESULTS to better explain the rationale for this experimental design.
4. The paper is challenging to read, and I suggest the authors provide it for reading and editing by someone not in their immediate field in order to make changes that make it more accessible to a general reader.
Thank you for the suggestion. We have thoroughly edited the manuscript to make it more accessible to general readers.
5. Their graphics are terrific and provide great assistance in clarifying the figures. Well done.
Thank you very much for the encouragement.
Reviewer #2 (Recommendations for the authors):
As it stands the manuscript does not support the idea that O-GlcNAcylation of ribosomal proteins in the mushroom body (MB) is required for protein synthesis and olfactory learning. Experiments targeting specific ribosomal components could be a compelling demonstration. For example, Ser/Thr residues that get O-GlcNAcylated in RPL24 or RPS3 could be mutated. If these mutants showed learning defects it would be an important result. Same type of mutants in ex vivo OPP assays could be performed to determine their effect on protein synthesis. These types of experiments would definitely show specific requirement of ribosomal protein modification for ribosomal activity and associative learning.
We agree that the current data fall short for a direct connection between O-GlcNAcylation of ribosomes and translational activity. Therefore, we have modified the TITLE and thoroughly edited the descriptions throughout the manuscript to assure accuracy (see Essential revisions 2).
Because the whole translational machinery is heavily O-GlcNAcylated in the mushroom body, and our profiling method can not identify individual O-GlcNAcylation sites on the substrates, it is very difficult to design the mutational experiments as you suggested. Future biochemical and structural characterizations of ribosomes might reveal the key O-GlcNAcylation sites and verify the connection between O-GlcNAc modifications of ribosomes and their activity.
Reference
1. Dupas, T., Betus, C., Blangy-Letheule, A., Pele, T., Persello, A., Denis, M., and Lauzier, B. (2022). An overview of tools to decipher O-GlcNAcylation from historical approaches to new insights. Int J Biochem Cell Biol 151, 106289. 10.1016/j.biocel.2022.106289.
2. Zachara, N.E., O'Donnell, N., Cheung, W.D., Mercer, J.J., Marth, J.D., and Hart, G.W. (2004). Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J Biol Chem 279, 30133-30142. 10.1074/jbc.M403773200.
3. Miller, M.W., Caracciolo, M.R., Berlin, W.K., and Hanover, J.A. (1999). Phosphorylation and glycosylation of nucleoporins. Arch Biochem Biophys 367, 51-60. 10.1006/abbi.1999.1237.
4. Roquemore, E.P., Chevrier, M.R., Cotter, R.J., and Hart, G.W. (1996). Dynamic O-GlcNAcylation of the small heat shock protein α B-crystallin. Biochemistry 35, 3578-3586. 10.1021/bi951918j.
5. Yin, R., Wang, X., Li, C., Gou, Y., Ma, X., Liu, Y., Peng, J., Wang, C., and Zhang, Y. (2021). Mass Spectrometry for O-GlcNAcylation. Front Chem 9, 737093. 10.3389/fchem.2021.737093.
6. Ma, J., and Hart, G.W. (2014). O-GlcNAc profiling: from proteins to proteomes. Clin Proteomics 11, 8. 10.1186/1559-0275-11-8.
7. Alonso, J., Schimpl, M., and van Aalten, D.M. (2014). O-GlcNAcase: promiscuous hexosaminidase or key regulator of O-GlcNAc signaling? J Biol Chem 289, 34433-34439. 10.1074/jbc.R114.609198.
8. Muha, V., Fenckova, M., Ferenbach, A.T., Catinozzi, M., Eidhof, I., Storkebaum, E., Schenck, A., and van Aalten, D.M.F. (2020). O-GlcNAcase contributes to cognitive function in Drosophila. J Biol Chem 295, 8636-8646. 10.1074/jbc.RA119.010312.
9. Mariappa, D., Selvan, N., Borodkin, V., Alonso, J., Ferenbach, A.T., Shepherd, C., Navratilova, I.H., and vanAalten, D.M.F. (2015). A mutant O-GlcNAcase as a probe to reveal global dynamics of protein O-GlcNAcylation during Drosophila embryonic development. Biochem J 470, 255-262. 10.1042/BJ20150610.
10. Rao, F.V., Dorfmueller, H.C., Villa, F., Allwood, M., Eggleston, I.M., and van Aalten, D.M. (2006). Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J 25, 1569-1578. 10.1038/sj.emboj.7601026.
11. Zhang, Y., Yu, H., Wang, D., Lei, X., Meng, Y., Zhang, N., Chen, F., Lv, L., Pan, Q., Qin, H., et al. (2023). Protein O-GlcNAcylation homeostasis regulates facultative heterochromatin to fine-tune sog-Dpp signaling during Drosophila early embryogenesis. J Genet Genomics. 10.1016/j.jgg.2023.05.014.
12. Gallant, P. (2013). Myc Function in Drosophila. Cold Spring Harbor Perspectives in Medicine 3, a014324-a014324. 10.1101/cshperspect.a014324.
13. Jiao, L., Liu, Y., Yu, X.Y., Pan, X., Zhang, Y., Tu, J., Song, Y.H., and Li, Y. (2023). Ribosome biogenesis in disease: new players and therapeutic targets. Signal Transduct Target Ther 8, 15. 10.1038/s41392-022-01285-4.
14. van Riggelen, J., Yetil, A., and Felsher, D.W. (2010). MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10, 301-309. 10.1038/nrc2819.
15. Arabi, A., Wu, S., Ridderstrale, K., Bierhoff, H., Shiue, C., Fatyol, K., Fahlen, S., Hydbring, P., Soderberg, O., Grummt, I., et al. (2005). c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol 7, 303-310. 10.1038/ncb1225.
16. Grandori, C., Gomez-Roman, N., Felton-Edkins, Z.A., Ngouenet, C., Galloway, D.A., Eisenman, R.N., and White, R.J. (2005). c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 7, 311-318. 10.1038/ncb1224.
17. Shiue, C.N., Berkson, R.G., and Wright, A.P. (2009). c-Myc induces changes in higher order rDNA structure on stimulation of quiescent cells. Oncogene 28, 1833-1842. 10.1038/onc.2009.21.
18. Kawai, H., Kanegae, T., Christensen, S., Kiyosue, T., Sato, Y., Imaizumi, T., Kadota, A., and Wada, M. (2003). Responses of ferns to red light are mediated by an unconventional photoreceptor. Nature 421, 287-290. 10.1038/nature01310.
19. Boon, K., Caron, H.N., van Asperen, R., Valentijn, L., Hermus, M.C., van Sluis, P., Roobeek, I., Weis, I., Voute, P.A., Schwab, M., and Versteeg, R. (2001). N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J 20, 1383-1393. 10.1093/emboj/20.6.1383.
20. Popay, T.M., Wang, J., Adams, C.M., Howard, G.C., Codreanu, S.G., Sherrod, S.D., McLean, J.A., Thomas, L.R., Lorey, S.L., Machida, Y.J., et al. (2021). MYC regulates ribosome biogenesis and mitochondrial gene expression programs through its interaction with host cell factor-1. ELife 10. 10.7554/eLife.60191.
21. Orian, A., van Steensel, B., Delrow, J., Bussemaker, H.J., Li, L., Sawado, T., Williams, E., Loo, L.W., Cowley, S.M., Yost, C., et al. (2003). Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev 17, 1101-1114. 10.1101/gad.1066903.
22. Grewal, S.S., Li, L., Orian, A., Eisenman, R.N., and Edgar, B.A. (2005). Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nature Cell Biology 7, 295-302. 10.1038/ncb1223.
23. Hulf, T., Bellosta, P., Furrer, M., Steiger, D., Svensson, D., Barbour, A., and Gallant, P. (2005). Whole-genome analysis reveals a strong positional bias of conserved dMyc-dependent E-boxes. Mol Cell Biol 25, 3401-3410. 10.1128/MCB.25.9.3401-3410.2005.
24. Marshall, L., Rideout, E.J., and Grewal, S.S. (2012). Nutrient/TOR-dependent regulation of RNA polymerase III controls tissue and organismal growth in Drosophila. EMBO J 31, 1916-1930. 10.1038/emboj.2012.33.
https://doi.org/10.7554/eLife.91269.sa2




