A single-cell transcriptomic atlas reveals resident dendritic-like cells in the zebrafish brain parenchyma
Figures

Leukocyte heterogeneity in the adult zebrafish brain using blood lineage-specific transgenic lines.
(A) Schematic overview of the experiment. First, cd45:DsRed + cells were sorted, cytospined, and stained with May-Grunwald Giemsa (MGG). In parallel, lines carrying the cd45:DsRed transgene in combination with blood lineage-specific GFP reporters were analyzed by flow cytometry. (B) Morphology of brain-sorted cd45:DsRed + cells stained with MGG. Microglia and/or macrophages, monocytes, dendritic cells, neutrophils, and lymphocytes were identified. The scale bar represents 5 μm. C. Flow cytometry analysis on brain cell suspensions from adult Tg(mpeg1:GFP; cd45:DsRed) identifying mpeg1:GFP+; cd45:DsRed + mononuclear phagocytes (green gate). (D) Proportion of brain immune cell types, as determined by flow cytometry analysis on cell suspensions from fish carrying cd45:DsRed + and a lineage-specific GFP reporter (n=4 fish). The percentage relative to total cd45:DsRed+ leukocytes is shown, with the exception of Tg(ighm:GFP; cd45:DsRed) which are not normalized as the cd45:DsRed transgene is not expressed in the B cell lineage. (E) Flow cytometry analysis of brain cell suspensions from an adult Tg(p2ry12:p2ry12-GFP; cd45:DsRed) fish, identifying p2ry12:p2ry12-GFP+; cd45:DsRed+ microglial cells (light green gate). n refers to the number of biological replicates. Data in (D) are mean ± SEM.

Flow cytometry analyses of brain leukocytes using blood lineage-specific GFP reporter lines.
(A) Proportion of neutrophils (purple gate), identified using the Tg(mpx:GFP; cd45:DsRed) double transgenic line. (B) Proportion of T/NK cells (blue gate), as determined using Tg(lck:GFP; cd45:DsRed) fish. (C) B lymphocytes (red gate), analyzed using Tg(ighm:GFP; cd45:DsRed) animals. Note that, as previously shown, the cd45:DsRed transgene is not expressed in ighm:GFP + B cells. Percentages of each population refer to a single individual and are relative to the total cd45:DsRed+ population (mean ± SEM of 4 fish: see text).

Diversity of brain leukocytes as shown by single-cell transcriptomics.
(A) Schematic overview of the experimental approach. Single-cell profiling of total brain cd45:DsRed+ leukocytes (pool from three individual fish) was performed using the 10 X Genomics platform. (B) Split Uniform Manifold Approximation Projection (UMAP) of brain cd45:DsRed+ cells with annotated cell populations. Clusters in gray shade are not indicative of a specific cell type and were not annotated. (C) UMAP plots depicting the expression pattern of ptprc, also known as cd45 (leukocytes), mpeg1.1 (mononuclear phagocytes), mpx (neutrophils), and lck (T and NK lymphocytes). Gene expression levels from low to high are indicated by a color gradient from yellow to purple (normalized counts in log1p). (D) Heat map of the top differentially up-regulated genes in each cluster (row = gene, column = cell type). Color scale (gradual from purple to yellow) indicates the expression level (average log2 fold change).

Isolation of a pure population of brain leukocytes.
FACS plots illustrating the gating strategy to isolate total brain leukocytes from Tg(cd45:DsRed) transgenic fish. Cells are first selected based on side (SSC-A) and forward scattering (FSC-A) characteristics. Cell doublets are then filtered out based on the width of the side scatter and forward signal. Next, calcein+ live cells are selected and cd45:DsRed+ cells are sort out of the live population. As shown in the last plot, this strategy captures leukocytes from both the lymphoid and myeloid lineages, with the exception of B cells, as they don’t express the cd45:DsRed transgene. In these experiments, brain cd45:DsRed+ cells isolated from three individual fish were pooled for scRNA sequencing.

Single-cell RNA sequencing identifies several lymphocyte subpopulations in the adult brain.
(A–C) Uniform Manifold Approximation Projection (UMAP) visualization of the expression of selected genes in the annotated T cell clusters Tcells1 and Tcells2 (zap70, cd4-1, and cd8a), NK cluster (ccl38.6, il2rb, gzm3.3), and ILC-like cluster (il4, il13, and gata3). Color scale (gradual from yellow to purple) indicates the expression level for each gene (normalized counts in log1p). (D) Violin plots representing the expression levels of known lymphocyte markers (normalized counts in log1p) within the different clusters. (E) Comparison of the relative expression of lck, zap70, gata3, il13, and il4 transcripts between brain cd45:DsRed + cells isolated by FACS from T cell-deficient rag2-/- mutants (red bars) and their wild-type siblings (gray bars). Each data point represents an individual fish (n=6) and error bars indicate SEM. ***p<0.001, **** Pp<0.0001 (Two-tailed unpaired t-test).

Heterogeneous subsets of mononuclear phagocytes exist in the zebrafish brain.
(A–C) Uniform Manifold Approximation Projection (UMAP) visualization of the expression of selected genes in the microglia (apoeb and lgals3bpb) (A), non-microglia macrophage (marco and f13a1b), (B) and DC-like (xcr1a.1 and siglec15l), (C) cell clusters. Color scale (gradual from yellow to purple) indicates the expression level for each gene (normalized counts in log1p). (D) Violin plot analysis comparing the expression levels of selected genes (y-axis, normalized counts in log1p) between the different mononuclear phagocyte cell clusters. (E) Volcano plot showing the differentially expressed (DE) genes between microglia (MG) and non-microglia macrophages (MF). Lines indicate significantly DE genes (log2 fold-change >|0.5|, -log10 Padj <0.001). Red dots represent up-regulated genes and blue dots down-regulated genes. Labels show representative DE genes identified in the analysis. (F) Volcano plot showing the DE genes between MG and DC-like cells. Lines indicate significantly DE genes (log2 fold-change >|0.5|, -log10 Padj <0.001).

Canonical microglial genes conserved between zebrafish and mammals.
Uniform Manifold Approximation Projection (UMAP) plots depicting the expression pattern of zebrafish orthologs of well-known mammalian microglia signature genes (with mammalian orthologs indicated in parenthesis). From top to bottom: p2ry12 (P2RY12) (Butovsky et al., 2014; Gerrits et al., 2020; Jurga et al., 2020; Van Hove et al., 2019), hexb (HEXB) (Butovsky and Weiner, 2018; Gerrits et al., 2020; Jurga et al., 2020; Van Hove et al., 2019), mertka (MERTK) (Butovsky and Weiner, 2018; Gerrits et al., 2020; Jurga et al., 2020; Van Hove et al., 2019), slco2b1 (SLCO2B1) (Gerrits et al., 2020; Van Hove et al., 2019), c1qa, c1qb, c1qc (C1Q A-C) (Gerrits et al., 2020; Butovsky and Weiner, 2018; Van Hove et al., 2019), lgmn (LGMN) (Gerrits et al., 2020; Butovsky et al., 2014; Van Hove et al., 2019). Color scale (gradual from yellow to purple) indicates the expression level for each gene (normalized counts in log1p).

Functional analysis using the corresponding mammalian orthologs.
(A–B) Pathway analysis of the significantly up-regulated markers (Padj <0.05, log2fc = 0.25) for microglia (A) and DC-like (B) clusters. The histogram bars represent the percentage of the total number of associated genes per term found and the line graph represents the level of statistical significance (Padj -log10) of the enriched Gene Ontology (GO) terms and Reactome pathways. (C–D) Cell type enrichment analysis using PanglaoDB from the Enrichr engine and uploading the same gene list as in A-B.

Dendritic cell (DC)-like cells localize together with microglia within the brain parenchyma.
(A–F) Immunofluorescence on transversal brain sections (14 µm) from Tg(mpeg1:GFP) (A–C) or Tg(p2ry12:p2ry12- GFP) (D–F) transgenic adult fish co-immunostained with anti-GFP (green) and anti-Lcp1 (magenta) antibodies. (A–C) All mpeg1:GFP+ mononuclear phagocytes in the brain parenchyma display Lcp1 immunostaining, as expected. (D–F) Similarly, all microglial cells, identified by GFP expression in the brain parenchyma of Tg(pr2y12:p2ry12-GFP) fish, are Lcp1+, as expected. (G–J) In sections of adult Tg(p2ry12:p2ry12-GFP; mpeg1:mCherry) double transgenic animals, GFP labeling is not observed in all mCherry+ cells. GFP (green), mCherry (gray), Lcp1 (magenta), and merge of the three channels. All images were taken using a 20 X objective and correspond to orthogonal projections. White arrowheads point to microglial cells (GFP+; Lcp1+or GFP+; mCherry+; Lcp1+) and yellow arrowheads to DC-like cells (GFP-; Lcp1+ or GFP-; mCherry+; Lcp1+). Scale bars: 50 µm. (K–N) Confocal imaging of a midbrain vibratome section (100 µm) from an adult Tg(mhc2dab:GFP; cd45:DsRed) brain. GFP (green), DsRed (magenta) and merge of the two channels are shown. Images correspond to orthogonal projections, white arrowheads point to GFP+; DsRed+ cells, and yellow arrowheads to GFP-; DsRedhigh. Scale bar in (K): 500 µm, scale bar in (L–N): 50 µm. Images are representative of brain tissue sections from 2 to 3 fish.

Distribution of dendritic cell (DC)-like cells and immunofluorescence staining for neutrophils and lymphoid cells in the adult brain.
(A–E) Vibratome sections (100 µm) from an adult Tg(mhc2dab:GFP; cd45:DsRed) brain. (A–B) Anterior telencephalon sections contain few cd45high; mhc2+ cells (yellow arrowhead) and these are more abundant in posterior telencephalic sections (dashed area). Scale bars: 200 µm. (C–D) Posterior midbrain sections and hindbrain (E). Scale bars: 500 µm. DiV, diencephalic ventricle; TeV, telencephalic ventricle; Vv, ventral nucleus of ventral telencephalic area; PPa, parvocellular preoptic nucleus anterior part; TL, torus longitudinallis; OTe, tectum opticum; DIL, lobus caudali cerebelli; CCe corpus cerebelli. (F–N) Blood lineage-specific reporter lines labeling neutrophils (Tg(mpx:GFP)) (F–H), T and NK cells (Tg(lck:GFP)) (I–K) and B lymphocytes (Tg(ighm:GFP)) (L–N) were used in these experiments. Sections were co-stained for GFP (green, left panels) and the pan-leukocytic marker L-plastin (Lcp1) (magenta, middle panels) to validate the hematopoietic identity of GFP-expressing cells. Merged images are represented in the right panels. Representative midbrain sections are shown. (F–H) In line with our flow cytometry analyses, neutrophils are scarce in the zebrafish brain. A rare neutrophil is shown lining the borders of the diencephalic ventricle (DiV). (I–K) T/NK cells are mainly located in the periphery or lining the ventricles (dashed line) and occasionally within the brain parenchyma. (L–N) B cells are rarely found in the brain or within the brain parenchyma. Scale bars: 50 µm.

Transcriptomic analysis of microglia (p2ry12+; cd45+or mhc2dab+; cd45low) and dendritic cell (DC)-like cells (mhc2dab+; cd45high).
(A) Schematic overview of the experiments. Microglia were isolated using Tg(p2ry12:p2ry12-GFP; cd45:DsRed) or Tg(mhc2dab:GFP; cd45:DsRed) transgenic fish, and DC-like cells using the Tg(mhc2dab:GFP; cd45:DsRed) reporter line. (B) Representative flow cytometry plot identifying microglial cells in brain cell suspensions from Tg(p2ry12:p2ry12-GFP; cd45:DsRed) fish. (C) Representative flow cytometry plot identifying mhc2dab:GFP+; cd45:DsRedlow microglia from mhc2dab:GFP+; cd45:DsRedhigh DC-like cells in brain cell suspensions from Tg(mhc2dab:GFP; cd45:DsRed) fish. (D) Volcano plot showing the differentially expressed (DE) genes between mhc2dab+; cd45high DC-like cells and p2ry12+; cd45+microglia. Red dots represent up-regulated genes and blue dots represent down-regulated genes. Lines indicate significantly DE genes (log2 fold-change >|2|, -log10 Padj <0.01). Labels show marker genes for DC-like cells and microglia identified in the scRNA-sequencing analysis. (E) Volcano plot showing the DE genes between mhc2dab+; cd45high DC-like cells (blue) and mhc2dab +cd45 low microglia (red). Lines indicate significantly DE genes (log2 fold-change >|2|, -log10 Padj <0.01).

Differential expression analysis reveals that brain ccl34b.1:GFP-; mpeg1.1:mCherry +cells have a dendritic cell (DC)-like transcriptome.
(A) Volcano plot showing the differentially expressed (DE) genes between two parenchymal populations previously described in Wu et al., 2020 as ccl34b.1+; mpeg1.1+ phagocytic microglia and ccl34b.1-; mpeg1.1+ regulatory microglia. Red dots represent up-regulated genes and blue dots represent down-regulated genes. Lines indicate significantly DE genes (log2 fold-change >|2|, -log10 Padj <0.01). Labels show that these cells differentially express marker genes identified for DC-like and microglia in our scRNA-sequencing analysis. (B) Venn diagram showing 130 overlapping genes amongst the significantly up-regulated DE genes (log2 fold-change >|1|, -log10 Padj <0.01) between DC-like and microglia from our dataset and ccl34b.1-; mpeg1.1+ and ccl34b.1+; mpeg1.1+ cells from Wu et al., 2020. The majority of DC-like marker genes are shared, suggesting that DC-like and ccl34b.1-mpeg1.1+ cells have a similar expression profile. (C) Venn diagram showing 134 overlapping genes amongst the significantly down-regulated DE genes between DC-like cells and microglia from our dataset and ccl34b.1-; mpeg1.1+ and ccl34b.1+; mpeg1.1+ cells from Wu et al., 2020. The majority of microglia marker genes are shared, suggesting that microglia and ccl34b.1+; mpeg1.1+ cells have a similar expression profile. (D) Hierarchical clustering of the normalized expression of the top DE-expressed genes in each sample (a total of 1252 genes; log2fc 1.2 Padj <0.01). DC-like cells (mhc2dab+; cd45high) cluster together with ccl34b.1-; mpeg1.1+ cells, while microglia (either as p2ry12+; cd45+or mhc2dab+; cd45low) cluster together with ccl34b.1+; mpeg1.1+ cells. Color scale indicates normalized expression for each gene. (E) Correlation matrix (Spearman) performed using the list of the top DE-expressed genes (a total of 1252 genes; log2fc 1.2 Padj <0.01) shows that DC-like and ccl34b.1-; mpeg1.1+ cells are positively correlated, suggesting their similar identity, while they show a decreased relationship with microglia (p2ry12+; cd45+or mhc2dab+; cd45low) and ccl34b.1+; mpeg1.1+ cells.

Brain dendritic cell (DC)-like cells are lost in batf3-/- mutant fish.
(A–J) Immunofluorescence on transverse brain sections (14 µm) from adult wild-type (A–E) and batf3-/- mutant (F–J) fish carrying the Tg(p2ry12:p2ry12-GFP; mpeg1:mCherry) double transgene and immunostained for GFP (green), mCherry (gray), and Lcp1 (magenta). Illustrative case of the merge of the three channels (A, F) allowing to identify GFP+; mCherry+; Lcp1+ microglia (white arrowheads) versus GFP-; mCherry+; Lcp1+ DC-like cells (yellow arrowheads). While DC-like cells are found in high numbers within the ventral part of control parenchyma (A), these are dramatically decreased following genetic loss of batf3 (F). Scale bars: 100 µm. (B–E, G–J). Single channels high magnification of the insets in A (B–E) and F (G–J). Scale bars: 50 µm. Images were taken using a 20 X objective and correspond to orthogonal projections. (K–M) Quantification of cell density for GFP+; mCherry+; Lcp1+ microglia and GFP-; mCherry+; Lcp1+ DC-like cells in the dorsal midbrain area or optic tectum (K), ventral midbrain area (L), and the entire section (M) of control (gray bars) and batf3-/- (green bars) fish. Each dot represents a single fish and data are mean ± SEM. *p<0.05 (Mann-Whitney test), ***p<0.0001 (Two-tailed unpaired t-test). (N) Flow cytometry analysis of brain cell suspensions from wild-type and batf3-/- adult fish carrying the Tg(p2ry12:p2ry12-GFP; mpeg1:mCherry) reporter. The GFP+; mCherry+ fraction identifies microglia (green circle), whereas the GFP-; mCherry+ fraction contains mainly DC-like cells (blue frame). (O) Percentage of microglia and DC-like cells in brain cell suspensions for each genotype, relative to the whole living brain population, as shown in (N) (wild-type, n=6; batf3-/-, n=10). ***p<0.001 (Two-tailed unpaired t-test). n refers to number of biological replicates.
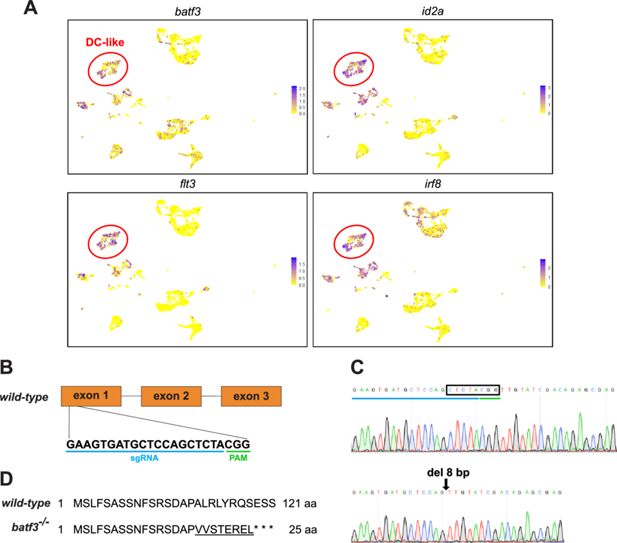
Generation of batf3-/- CRISPR mutants.
(A) Uniform Manifold Approximation Projection (UMAP) visualization of the expression pattern of selected zebrafish dendritic cell (DC)-like cluster genes whose orthologs represent known markers of cDC1 in mammals, including batf3, id2a, flt3, and irf8. Color scale (from yellow to purple) indicates the expression level for each gene (normalized counts in log1p). (B) Schematic view of exons 1–3 of the batf3 gene, with the CRISPR/Cas9 targeting sequence in exon 1 underlined in blue and the PAM sequence underlined in green. (C) Sequence chromatograms of wild-type (upper row) and mutant (lower row) sequences, showing the 8 bp deletion (black box) in batf3ulb31 fish. (D) Predicted Batf3 protein sequence in the wild-type and the mutant. The 8 bp deletion in the batf3 gene induces a frameshift (underlined) that results in the production of a truncated protein due to the presence of three consecutive premature stop codons.

Characterization of brain immune cells in the batf3-/- mutant.
(A–C) Cell density quantification of non-myeloid cells (GFP-; mCherry-; Lcp1+) and total leukocyte (all Lcp1+) in the dorsal midbrain area or optic tectum (A), ventral midbrain area (B) and the whole section (C) of wild-type and batf3-/- fish from Figure 7. Data represent mean ± SEM (n=4 fish). (D, E) Q-PCR expression for dendritic cell (DC)-like markers siglec15l and ccl19a.1 in p2ry12:GFP-; mpeg1:mCherry +DC like cells (D) and for microglia-specific genes p2ry12, apoeb and ccl34b.1 in p2ry12:GFP+; mpeg1:mCherry+ microglia (E) sorted from wild-type (gray bars) and batf3-/- (colored bars) adult brains. *p<0.05, ***p<0.001 (Two-tailed unpaired t-test). Data are represented as mean ± SEM. n refers to the number of biological replicates. (F) Number of cells (y-axis) versus mCherry fluorescence intensity (x-axis) histogram plot of brain cell suspensions from controls (gray) and batf3-/- (blue) fish carrying the Tg(p2ry12:GFP; mpeg1:mCherry) double transgene. It shows that all residual mpeg1:mCherry+ cells in the batf3 mutant display lower fluorescence intensity as compared to their wild-type counterparts. (G) Proportion of mpeg1:mCherry+ cells according to their low or high fluorescence intensity as shown in F. Data represent mean ± SEM (n=4). n refers to the number of biological replicates. ****p<0.0001 (Two-tailed unpaired t-test). (H–K) Illustrative case of vibratome midbrain sections of Tg(cd45:DsRed) transgenic wild-type (H,I) and batf3-/- (J,K) fish (n=3). The endogenous fluorescence of DsRedhigh DC-like cells permits their identification in the ventral part of wild-type brains (n=3). However, these cells are not found in the batf3-/- mutant (n=3). Scale bar: 500 µm. (I, K) High magnification of the insets (white frame) in H (I) and J (K). Scale bar: 100 µm. Note that cells with lower fluorescence intensity (e.g. cd45low microglia) are not detectable using this approach due to signal loss (quenching) following fixation.

Examination of microglia and dendritic cell (DC)-like cells in myeloid–deficient mutant lines.
(A–D) Immunofluorescence on transverse brain sections from Tg(p2ry12:p2ry12-GFP) transgenic adult wild-type (A– D), irf8-/- (E–H), csf1ra-/- (I–L), csf1rb-/- (M–P) and csf1ra-/-; csf1rb-/- (csf1rDM) (Q–T) fish, co-stained with anti-GFP (green) and Lcp1 (magenta) antibodies. (A, E, I, M, Q) For each genotype, illustrative case of the merge of the two channels, allowing to discriminate in the parenchyma GFP+; Lcp1+ microglia (white arrowheads) from GFP-; Lcp1+ DC-like cells (yellow arrowheads). Single channels high magnification of the insets (dashed frame) in A (B–D), E (F–H), I (J–L), M (N–P), and Q (R–T). Outline yellow arrowheads indicate the absence of GFP signal in corresponding yellow arrowhead-pointed cells. Scale bar in (A), (E), (I), (M), and (Q) represents 100 µm and scale bar in other images 50 µm. (U–V) Quantification of the cell density for GFP+ Lcp1+ microglia and GFP-Lcp+ DC-like cells in the dorsal (U), ventral (V), and whole area (W) of the brain for each genotype (n=3). Data in U-W are mean ± SEM. *p<0.05, **p<0.01 (Kruskal-Wallis test with Dunn’s post-hoc).

Location of microglia in the brain ventricles of irf8-deficient fish.
(A–D). Immunofluorescence of brain sections from irf8-/- mutant fish carrying the Tg(p2ry12:p2ry12-GFP) reporter, co-stained with GFP (green) and Lcp1 (magenta) antibodies. (B) Microglial cells in the mutant are mostly found lining the ventricles (A-D, white arrowheads). Nuclear DAPI staining is shown in A to better visualize the position of the GFP+ and Lcp1+ cells along the ventricle. Ly, lymphocyte (outline white arrowhead). Scale bar represents 50 µm. (B) GFP+ cells. (C) Lcp1+ cells. (D) merged channels. Scale bar: 50 µm. TeV, telencephalic ventricle.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Danio rerio) | Tg(mhc2dab:GFPLT)sd67 | Wittamer et al., 2011 | ZFIN: sd67 | |
| Genetic reagent (Danio rerio) | Tg(ptprc:DsRedexpress)sd3 | Wittamer et al., 2011 | ZFIN: sd3 | |
| Genetic reagent (Danio rerio) | Tg(mpeg1.1:eGFP)gl22 | Ellett et al., 2011 | ZFIN: gl22 | |
| Genetic reagent (Danio rerio) | Tg(mpeg1.1:mCherry)gl23 | Ellett et al., 2011 | ZFIN: gl23 | |
| Genetic reagent (Danio rerio) | TgBAC(p2ry12:p2ry12-GFP)hdb3 | Sieger et al., 2012 | ZFIN: hdb3 | |
| Genetic reagent (Danio rerio) | Tg(lck:lck-eGFP)cz1 | Langenau et al., 2004 | ZFIN: cz1 | |
| Genetic reagent (Danio rerio) | TgBAC(cd4-1:mcherry)UMC13 | Dee et al., 2016 | ZFIN: UMC13 | |
| Genetic reagent (Danio rerio) | Tg(Cau.Ighv-ighm:EGFP)sd19 | Page et al., 2013 | ZFIN: sd19 | |
| Genetic reagent (Danio rerio) | Tg(mpx:GFP)i113 | Mathias et al., 2009 | ZFIN: i113 | |
| Genetic reagent (Danio rerio) | pantherj4e1 | Parichy et al., 2000 | ZFIN: i4e1 | |
| Genetic reagent (Danio rerio) | csf1rbsa1503 | Sanger Institute Zebrafish Mutation Project | ZFIN: sa1503 | |
| Genetic reagent (Danio rerio) | irf8std96 | Shiau et al., 2015 | ZFIN: std96 | |
| Genetic reagent (Danio rerio) | rag2E450fs | Tang et al., 2014 | ZFIN: E450fs | |
| Genetic reagent (Danio rerio) | batf3ulb31 | This manuscript | ZFIN: ulb31 | |
| Antibody | Anti-GFP (chicken polyclonal) | Abcam | RRID:AB_300798 | 1:500 |
| Antibody | Anti-Lcp1 (rabbit polyclonal) | In house | 1:1000 | |
| Antibody | Anti-mCherry (mouse monoclonal) | Takara Bio | RRID:AB_2307319 | 1:500 |
| Antibody | Alexa Fluor 488-conjugated anti-chicken IgG (goat polyclonal) | Abcam | RRID:AB_2636803 | 1:500 |
| Antibody | Alexa Fluor 594-conjugated anti-rabbit IgG (donkey polyclonal) | Abcam | RRID:AB_2782993 | 1:500 |
| Antibody | Alexa Fluor 647-conjugated anti-mouse IgG (donkey polyclonal) | Abcam | RRID:AB_2890037 | 1:500 |
| Commercial assay or kit | SP6 RNA Polymerase | New England BioLabs | Cat# M0207 | |
| Commercial assay or kit | High Pure PCR Cleanup Microkit | Roche | Cat# 498395500 | |
| Commercial assay or kit | RNeasy Plus mini kit | Qiagen | Cat# 74134 | |
| Chemical compound, drug | SYTOX Red | Invitrogen | Cat# S34859 | |
| Chemical compound, drug | qScript cDNA SuperMix | Quanta Biosciences | Cat# 95048–100 | |
| Software, algorithm | Flow-Jo LLC | TreeStar | RRID:SCR_008520 | |
| Software, algorithm | Black Zen software | Zeiss, Germany | RRID:SCR_018163 | |
| Software, algorithm | Blue Zen software | Zeiss, Germany | RRID:SCR_013672 | |
| Software, algorithm | R Statistical software v. 4.0.3 | R Project for Statistical Computing | RRID:SCR_001905 | |
| Software, algorithm | GraphPad Prism 8 | GraphPad software, USA | RRID:SCR_002798 |
Additional files
-
Supplementary file 1
List of cluster marker genes for identified cell types.
- https://cdn.elifesciences.org/articles/91427/elife-91427-supp1-v1.csv
-
Supplementary file 2
Top 50 markers for each identified cell type.
- https://cdn.elifesciences.org/articles/91427/elife-91427-supp2-v1.xlsx
-
Supplementary file 3
Differentially expressed genes between mononuclear phagocyte clusters.
- https://cdn.elifesciences.org/articles/91427/elife-91427-supp3-v1.xlsx
-
Supplementary file 4
.Pathway enrichment analysis for mononuclear phagocyte cluster markers.
- https://cdn.elifesciences.org/articles/91427/elife-91427-supp4-v1.xlsx
-
Supplementary file 5
Differentially expressed genes between dendritic cell (DC)-like versus microglia clusters (bulk RNA seq analyses).
- https://cdn.elifesciences.org/articles/91427/elife-91427-supp5-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91427/elife-91427-mdarchecklist1-v1.docx








