A kinase to cytokine explorer to identify molecular regulators and potential therapeutic opportunities
Figures
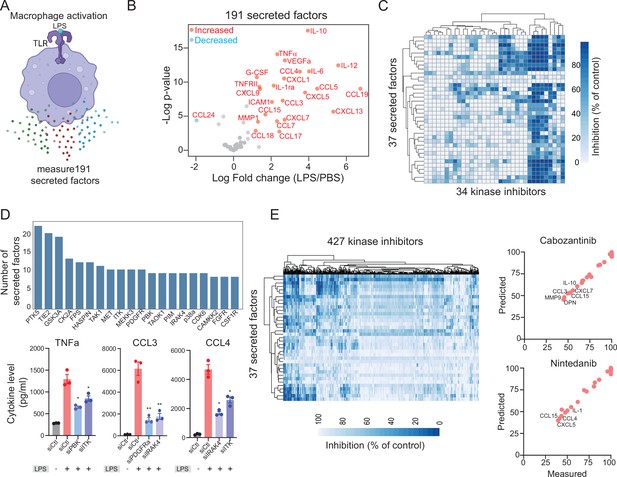
Modeling lipopolysaccharide (LPS)-mediated changes in secreted factors in human macrophages.
(A) A schematic showing LPS-mediated activation of human macrophages. (B) A plot showing changes in the levels of 191 secreted factors in response to LPS stimulation. Significantly increased or decreased factors are also indicated. (C) A heatmap showing changes in LPS-mediated 37 secreted factors in response to 34 kinase inhibitors. (D) Key kinases that regulate cytokines and chemokines. Top, a plot showing the top kinase and the number of secreted factors they regulate. Bottom, experimental validation of change in TNF-α, CCL3, and CCL4 upon knockdown of indicated kinases. (E) A heatmap showing predicted changes in 37 secreted factors in response to 427 kinase inhibitors. Right, experimental validation showing correlation between measured and predicted changes in 37 cytokines in macrophages treated with FDA-approved cabozantinib and nintedanib at 500 nM.
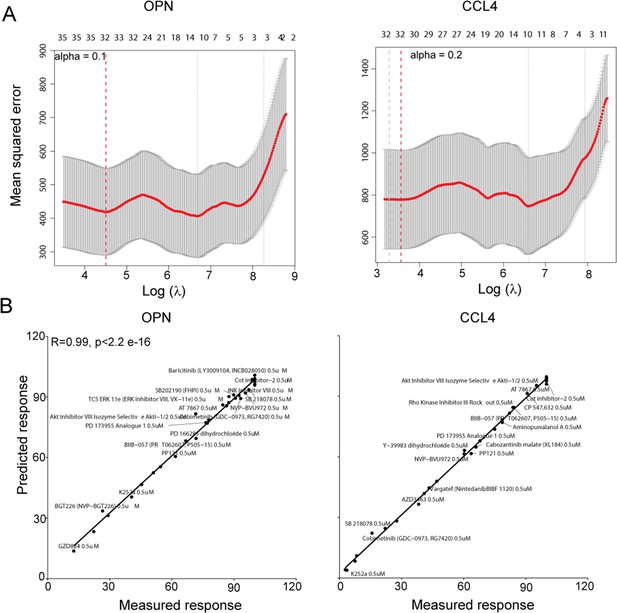
Representative KiR models for OPN and CCL4 release.
(A) Plots showing leave-one-out cross validation (LOOCV) error using elastic net regularization fit for OPN (left) and CCL4 (right). The error bars represent cross-validation error plus 1 SD. The kinases identified at absolute minima (red dashed line) were termed the most informative kinases. (B) Plots showing a correlation between observed and predicted changes in indicated cytokines in response to 32 kinase inhibitors.
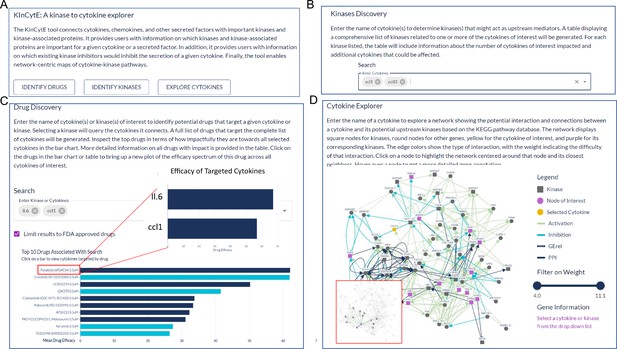
An illustration showing the steps for using KinCytE.
(A) KinCytE offers three sections for users to explore potential cytokine(s) or chemokine(s) inhibiting drugs (Identify Drugs), related kinases (Identify Kinase), and cytokine signaling network (Explore Cytokines). Users can click on any of these sections to access the desired information. (B) Users can obtain information on kinase(s) relevant to the expression of selected cytokines by selecting the desired cytokine(s) from the available options. This will generate a table with the relevant information. (C) Users select cytokine(s) to view a bar plot showing the top 10 most potent drugs in inhibiting the expression of these cytokine(s), with a table recording the effectiveness of all 427 drugs. Users can click on the drug name in either the table or the bar plot to access a more detailed cytokine efficacy profile of that drug. (D) To produce a network showing the connection between a selected cytokine and its related kinases, users need to select the desired cytokine. This will generate a network with edges representing the connections between the cytokine and its related kinases. Users can slide the weight bar to prune edges and click on a node to get the annotation of the gene(s) represented by that node. All tables in KinCytE are fully sortable, filterable, and downloadable for the convenience of users.
Tables
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91472/elife-91472-mdarchecklist1-v1.docx
-
Supplementary file 1
Fold change of average cytokine level in LPS-treated macrophage compared to control.
- https://cdn.elifesciences.org/articles/91472/elife-91472-supp1-v1.xlsx
-
Supplementary file 2
Percentage changes in cytokine level in kinase inhibitor-treated macrophages compared to control.
- https://cdn.elifesciences.org/articles/91472/elife-91472-supp2-v1.xlsx
-
Supplementary file 3
Summary table of predicted kinases involved in cytokine models.
- https://cdn.elifesciences.org/articles/91472/elife-91472-supp3-v1.zip
-
Supplementary file 4
Predicted cytokine levels in macrophares treated with kinase inhibitors.
- https://cdn.elifesciences.org/articles/91472/elife-91472-supp4-v1.zip





