Peptidoglycan-tethered and free forms of the Braun lipoprotein are in dynamic equilibrium in Escherichia coli
Figures
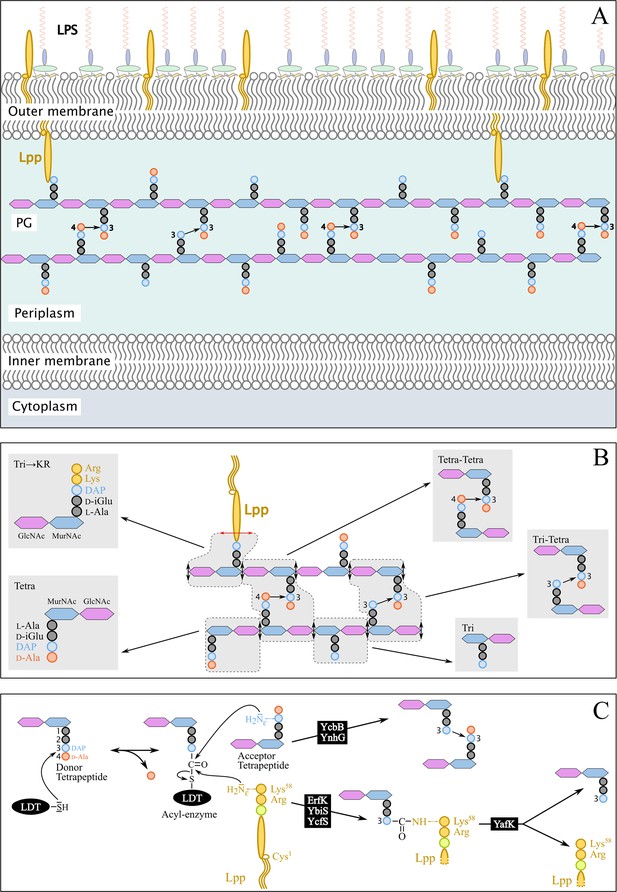
Structure and synthesis of the cell envelope of Escherichia coli.
(A) Structure of the main polymers of the envelope. The envelope of gram-negative bacteria contains two membranes. The inner membrane (IM) is made of phospholipids. The outer membrane (OM) is asymmetric, with phospholipids in the inner leaflet and lipopolysaccharides (LPS) in the outer leaflet. The peptidoglycan (PG) is located in the periplasmic space delimited by the IM and the OM. The PG consists of glycan chains linked to each other by short peptides. The glycan chains consist of alternating β1→4-linked N-acetyl-glucosamine (GlcNAc) and N-acetyl muramic acid (MurNAc) residues. The sequence of the stem peptide linked to the D-lactoyl group of MurNAc, as assembled in the cytoplasm, is L-Ala1-D-iGlu2-DAP3-D-Ala4-D-Ala5, in which DAP is a diaminopimelyl residue. Polymerization of this disaccharide-pentapeptide subunit by glycosyltransferases results in linear glycan strands that are inserted into the expanding PG network by the combined actions of space-making hydrolases and transpeptidases. The majority of the cross-links connecting D-Ala4 of a donor stem peptide to DAP3 of an acceptor stem peptide (4→3 cross-links) are made by D,D-transpeptidases belonging to the penicillin-binding protein (PBP) family that are the essential targets of β-lactam antibiotics. L,D-transpeptidases (LDTs) catalyze the formation of 3→3 cross-links connecting two DAP residues (see panel C). The Braun lipoprotein (Lpp) provides a link between the OM and the PG since its C-terminus is covalently bound to DAP residues (panel C), whereas its N-terminal Cys residue carries three alkyl chains, one of which is carried by the amino group of the C-terminal Cys residue and the other two by a glycerol moiety linked to the sulfhydryl of that residue by a thioether bond. These alkyl chains are inserted into the inner leaflet of the OM (PG-tethered form of Lpp) or into the outer leaflet of that membrane (free form of Lpp facing the bacterial cell surface). (B) Determination of peptidoglycan structure. The sacculi are extracted by the hot SDS procedure, treated with pronase and trypsin (red double arrows), and digested by muramidases that cleave the MurNAc-GlcNAc bonds (black double arrows). The structure of the resulting fragments (enclosed in rounded polygons) is determined by mass spectrometry following the reduction of N‑acetyl muramic acid (MurNAc) residues and separation by rpHPLC. Please note that we used the standard nomenclature for transpeptidation products in which the acyl donor and the acyl acceptor appear left and right, respectively, separated by an arrow to indicate the CO-to-NH polarity of the amide bond. (C) Reactions catalyzed by L,D-transpeptidases. In E. coli, the LDT protein family includes six paralogues that catalyze (i) the formation of 3→3 cross-links (YcbB and YnhG), (ii) the covalent anchoring of Lpp to PG (Ybis, YcfS, and ErfK), and (iii) the hydrolysis of the resulting PG→Lpp bond (YafK). Formation of the 3→3 and PG→Lpp bonds involves a common catalytic intermediate resulting from the nucleophilic attack of the DAP3-D-Ala4 bond of an acyl-donor tetrapeptide stem by the active Cys residue of the LDTs, the release of D-Ala4, and the formation of a D-Ala4-Cys thioester bond. For the formation of a 3→3 cross-link, the side-chain amino group of DAP at the third position of an acyl acceptor stem reacts with the acyl-enzyme. For the formation of a PG→Lpp bond, the side-chain of the C-terminal Lys residue of Lpp (at position 58) reacts with the acyl-enzyme.
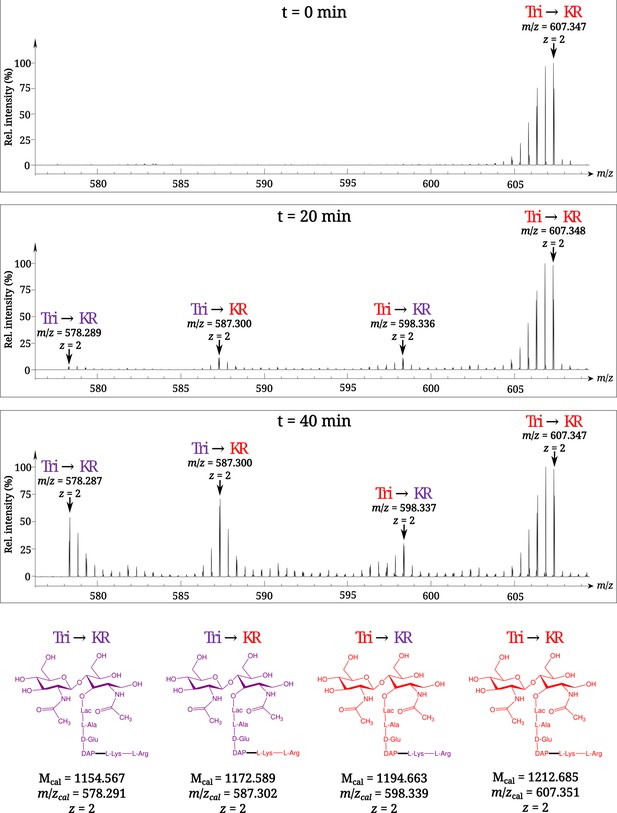
Time-resolved appearance of differentially labeled Tri→KR muropeptide isotopologues containing new (light) and old (heavy) lipoprotein (Lpp) and peptidoglycan (PG) moieties.
Light and heavy residues are indicated in purple and in red, respectively. Mcal, calculated monoisotopic mass (see Atze et al., 2022) for the detailed description of the method used to determine Mcal values.
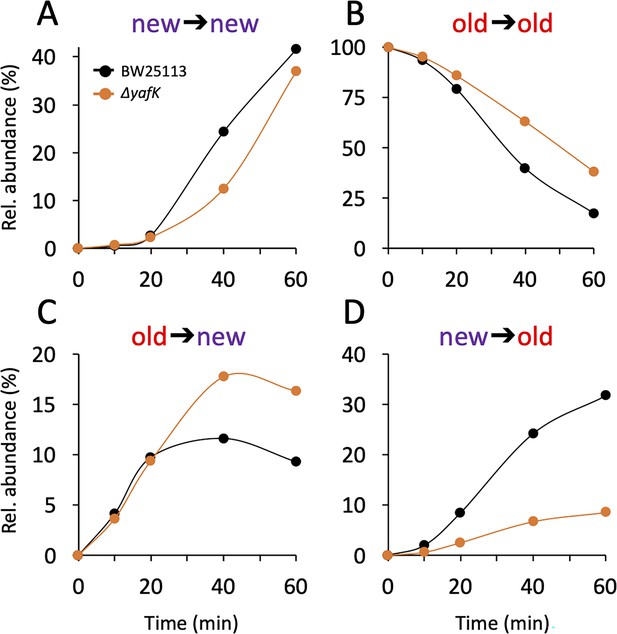
Kinetics of variations in the relative abundance of Tri→KR isotopologues.
(A) Unlabeled isotopologue containing newly synthesized muropeptide and lipoprotein (Lpp) moieties (new→new). (B) Fully labeled isotopologue containing existing muropeptide and Lpp moieties (old→old). (C) Hybrid isotopologue containing old and new muropeptide and Lpp moieties, respectively (old→new). (D) Hybrid isotopologue containing new and old muropeptide and Lpp moieties, respectively (new→old). Light and heavy moieties are indicated in purple and in red, respectively. Muropeptide analysis was performed for the BW25113 strain (black) and the ΔyafK mutant (orange). Data are the average of three to five biological replicates. The full data set appears in Supplementary file 2. Rel. abundance, relative abundance, is defined as the ratio (%) between the intensity of the indicated isotopologue and the sum of the intensities of all four isotopologues. This implies that an isotopologue that is present at the medium switch and is neither synthesized nor degraded during the following incubation of 60 min (one generation) will display a 50% reduction in its relative abundance because the total number of stem peptides doubles in one generation.
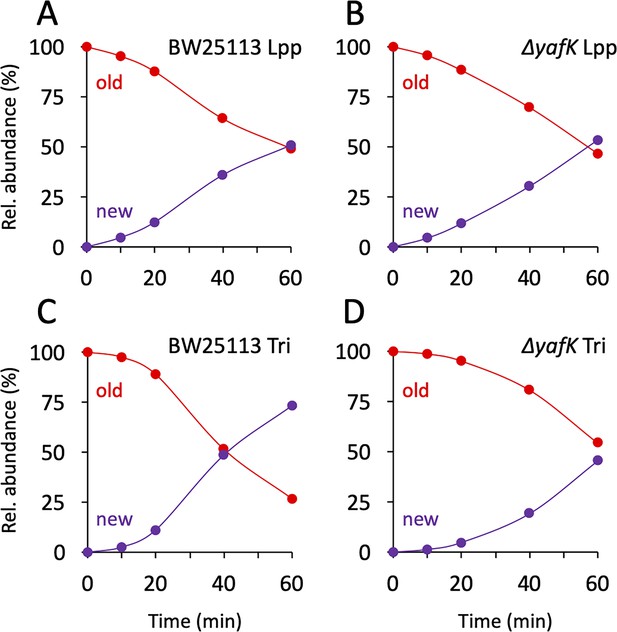
Kinetics of variations in the proportions of old and new moieties in the Tri→KR muropeptide.
(A, B) The relative abundance of old (red) and new (purple) lipoprotein (Lpp) moieties in the isotopologues from the BW25113 strain and the ΔyafK mutant, respectively. The relative abundances were calculated from data appearing in Figure 3 by adding the relative abundances of the new→old and old→old isotopologues (red curves; old Lpp) or of the new→new and old→new isotopologues (purple curves; new Lpp). (C, D) The relative abundance of old (red) and new (purple) tripeptide moieties in the isotopologues from the BW25113 strain and the ΔyafK mutant, respectively. The relative abundances were calculated from data appearing in Figure 3 by adding the relative abundances of the old→old and old→new isotopologues (red curves; old tripeptide) or of the new→new and new→old isotopologues (purple curves; new tripeptide). Data are the average of three to five biological replicates. The full data set appears in Supplementary file 3.
Tables
MS–MS analysis of the four isotopologues of the Tri⟶KR muropeptide.
| Deduced structure of fragments | Mass of ions generated by fragmentation of indicated isotopologues* | |||||||
| Tri⟶KR | Tri⟶KR | Tri⟶KR | Tri⟶KR | |||||
| Mcal | Mobs | Mcal | Mobs | Mcal | Mobs | Mcal | Mobs | |
| GlcNAc-MurNAcR-ʟ-Ala-D-iGlu-DAP⟶ʟ-Lys-ʟ-Arg† | 1,154.57 | 1,154.56 | 1,172.59 | 1,172.58 | 1,194.66 | 1,194.65 | 1,212.69 | 1,212.68 |
| MurNAcR-ʟ-Ala-D-iGlu-DAP⟶ʟ-Lys-ʟ-Arg | 951.49 | 951.49 | 969.51 | 969.52 | 982.56 | 982.56 | 1,000.58 | 1,000.58 |
| ʟ-Ala-D-iGlu-DAP⟶ʟ-Lys-ʟ-Arg | 674.37 | 674.38 | 692.39 | 692.39 | 693.41 | 693.41 | 711.43 | 711.43 |
| D-iGlu-DAP⟶ʟ-Lys-ʟ-Arg | 603.33 | 603.34 | 621.36 | 621.37 | 618.37 | 618.36 | 636.39 | 636.39 |
| DAP⟶ʟ-Lys-ʟ-Arg | 474.29 | 474.29 | 492.31 | 492.31 | 483.31 | 483.31 | 501.33 | 501.33 |
| ʟ-Lys-ʟ-Arg | 302.21 | 302.21 | 320.23 | 320.23 | 302.21 | 302.21 | 320.23 | 320.23 |
| ʟ-Arg | 174.11 | 174.11 | 184.12 | 184.12 | 174.11 | 174.11 | 184.12 | 184.12 |
-
*
Newly synthesized and existing moieties are indicated in purple and red, respectively. Mcal and Mobs, calculated and observed monoisotopic masses (Da), respectively.
-
†
Parental ion.
Additional files
-
Supplementary file 1
Time-resolved appearance of differentially labeled Tri➔KR muropeptide isotopologues containing new (light) and old (heavy) lipoprotein (Lpp) and PG moieties.
This file contains mass spectral data obtained for the kinetics analyses for the BW25113 strain and the ΔyafK mutant.
- https://cdn.elifesciences.org/articles/91598/elife-91598-supp1-v1.pdf
-
Supplementary file 2
Kinetic analysis of the relative abundance of Tri➔KR isotopologues in wild-type and ΔyafK.
- https://cdn.elifesciences.org/articles/91598/elife-91598-supp2-v1.docx
-
Supplementary file 3
Kinetic analysis of the content of Tri➔KR isotopologues in new and old Tri and KR moieties.
- https://cdn.elifesciences.org/articles/91598/elife-91598-supp3-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91598/elife-91598-mdarchecklist1-v1.docx





