Genetic requirement of dact1/2 to regulate noncanonical Wnt signaling and calpain 8 during embryonic convergent extension and craniofacial morphogenesis
Figures
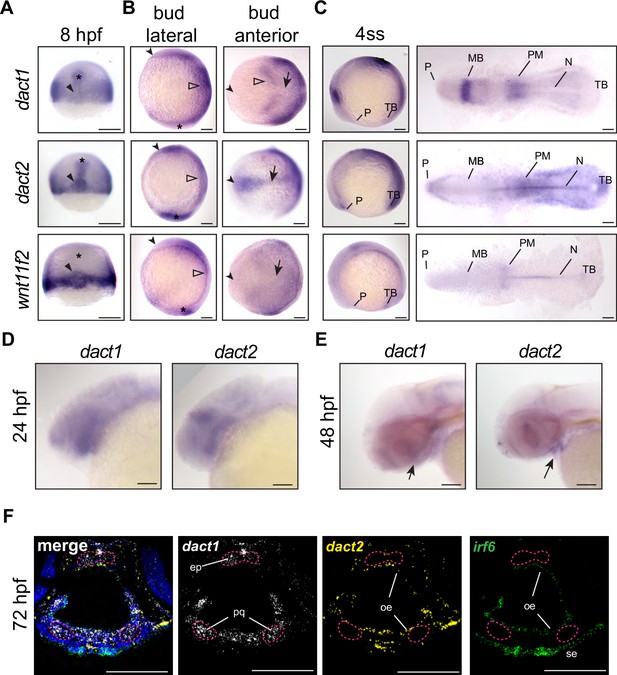
Unique and shared dact1 and dact2 gene expression domains during zebrafish development.
(A–C) Representative images of wholemount in situ hybridization showing dact1, dact2, and wnt11f2 gene expression patterns. (A) At 8 hpf, dact2 and wnt11f2 are highly expressed in the dorsal margin and presumptive Nieuwkoop center of the gastrulating embryo, with dact1 being weakly detected (arrowhead). In contrast to wnt11f2, dact1, and dact2 are expressed in the presumptive dorsal mesoderm (asterisk). (B) Lateral (anterior to the left of page) and anterior (dorsal side toward top of page) views of bud-stage embryos. dact2 and wnt11f2 transcripts are both detected in the tailbud (asterisk) while dact2 is additionally expressed in the axial mesoderm (arrow). dact1 gene expression is concentrated to the paraxial mesoderm and the neuroectoderm (open arrowheads). (C) Lateral and flat-mount views of 4 ss embryos. dact2 is expressed in the anterior neural plate and polster (P), notochord (N), paraxial and presomitic mesoderm (PM) and tailbud (TB). In contrast, dact1 is expressed in the midbrain (MB) and the paraxial and presomitic mesoderm. (D, E) Representative lateral (anterior to left of page) images of wholemount in situ hybridization showing dact1 and dact2 expression patterns. (D) At 24 hpf expression is detected in the developing head. (E) At 48 hpf expression is detected in the developing craniofacial structures (arrow). (F) Representative images of RNAscope in situ hybridization analysis of dact1 (white) and dact2 (yellow) and irf6 (green) expression in transverse section of 72 hpf embryos. dact1 is expressed in the ethmoid plate (ep) and palatoquadrate (pq) orofacial cartilage, while dact2 is expressed in the oral epithelium (oe). The epithelial marker irf6 is expressed in the oe and surface epithelium (se). Dapi (blue). Scale bar: 100 μm.
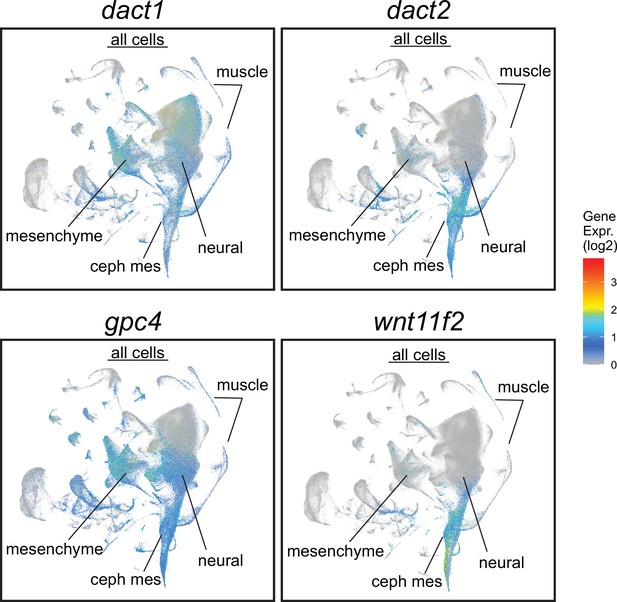
Daniocell single-cell RNAseq analysis with a display of dact1, dact2, gpc4, and wnt11f2 in all cell clusters from 3 to 120 hpf of development (https://daniocell.nichd.nih.gov; Farrell et al., 2018).
Cephalic mesoderm (ceph mes), mesenchyme, neural ectoderm, and muscle clusters are noted.
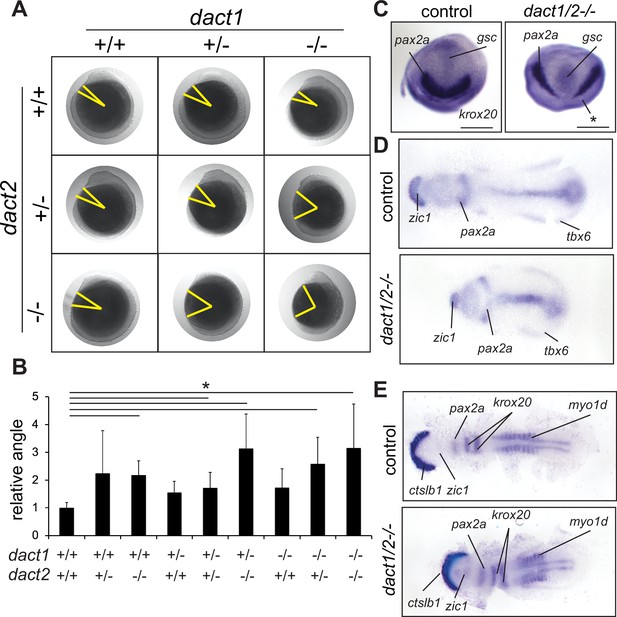
Impaired convergent extension in dact1/2 compound mutants.
(A) Inter-cross of compound heterozygotes yield embryos with different degrees of axis extension that correspond to the dact1 and dact2 genotypes. Representative lateral images of embryos at 12 hpf. The yellow line indicates body axis angle measured from the anterior point of the head, the center of the yolk, to the end of the tail. (B) Quantification of body axis angle. Numbers represent the difference in angle relative to the average wildtype embryo. Asterisk indicates genotypes with angles significantly different from wildtype. ANOVA p < 0.5 n = 3–21 embryos. Error bars: ± SEM. (C) Representative bud stage wildtype and dact1/2−/− mutant embryos stained for gsc (prechordal plate), pax2a (midbrain/hindbrain boundary), and krox20 (rhombomere 3). Asterisk indicates lack of krox20 expression in dact1/2−/− mutant. Scale bar = 200 μm (D) Representative flat mounts of 1–2 ss wildtype and dact1/2 mutant embryos stained for zic1 (telencephalon), pax2a and tbx6 (ventrolateral mesoderm). (E) Representative flat mounts of 10 ss wildtype and dact1/2−/− mutant embryos stained for ctsl1b (hatching gland), zic1, pax2a, krox20, and myo1d (somites).
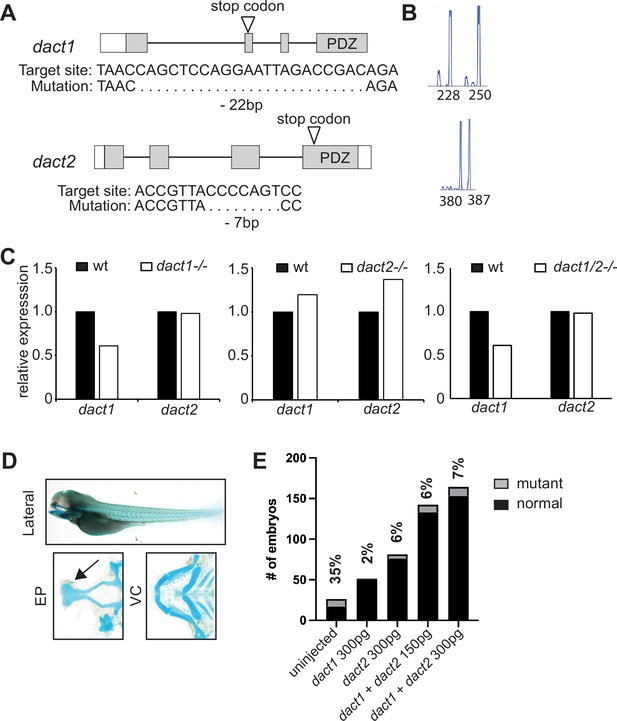
Characterization of CRISPR/Cas9 generated dact1−/− and dact2−/− mutants.
(A) Schematic representations of dact1 and dact2 exons, positions of guide RNA target site, introduced premature stop codon (arrow), and sequences of mutations. (B) DNA fragment analysis of dact1+/- and dact2+/- animals showing wildtype (250 and 387 bp, respectively) and mutant (228 and 380 bp, respectively). (C) Expression levels of dact1 and dact2 mRNA by RT-qPCR in 12 hpf dact1−/− mutants, dact2−/− mutants, and dact1/2−/− compound mutants. Eight embryos were pooled for mRNA isolation per sample. (D) Injection of dact1 mRNA, dact2 mRNA, or a combination of dact1 and dact2 mRNA rescues the rod-shaped ethmoid plate phenotype in dact1/2−/− compound mutants. Representative images of Alcian blue stained dact1/2−/− double mutant treated with 300 pg dact1 mRNA and 300 pg dact2 mRNA. Arrow highlights normal ethmoid plate (EP). Visceral cartilage (VC) also appeared normal. (E) Quantification of the mutant craniofacial phenotype observed in a dact1−/−,dact2+/- breeding in-cross. Without mRNA injection, the mutant phenotype was observed at approximately (35%) the expected Mendelian ratio of 25%. Injection with dact1 mRNA, dact2 mRNA, or a combination of dact1 and dact2 mRNA decreased the frequency that the mutant craniofacial phenotype was observed.
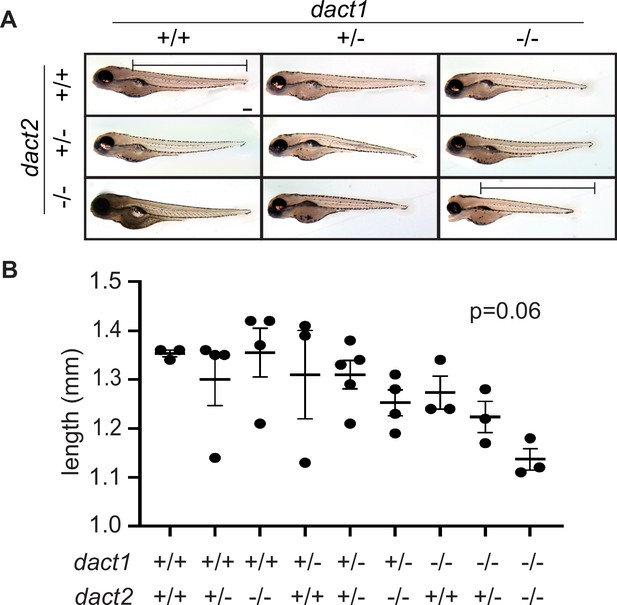
Loss of dact1 and dact2 tends to decrease total body length.
(A) Representative brightfield images of 4 dpf larvae of compound dact1 and dact2 mutant genotypes. Bar represents dact1/2+/+ body length measurement. Scale bar: 100 μm. (B) Scatter plot of body length measurement of compound dact1 and dact2 mutant genotypes at 4 dpf. ANOVA p = 0.06. n = 3–4. Error bars: ± SEM.
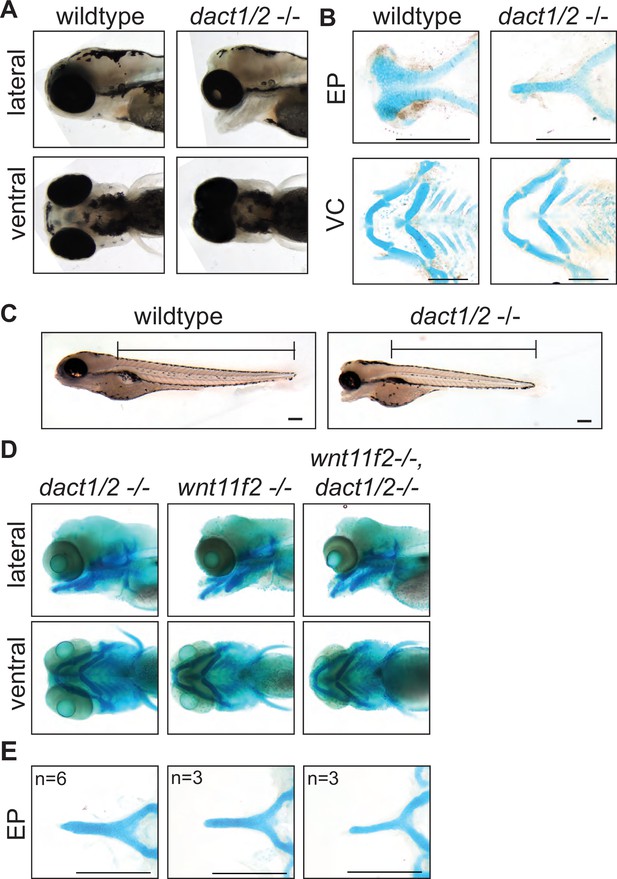
Midface development requires dact1 and dact2.
(A) Representative brightfield images of wildtype and dact1/2−/− compound mutants at 4 dpf. 100 individuals were analyzed from a dact1/2+/- double het cross. Lateral and ventral views show dact1/2−/− compound mutants have a hypoplastic midface, medially displaced eyes, and a displaced lower jaw. (B) Representative flat-mount images of Alcian blue stained ethmoid plate (EP) and visceral cartilage (VC) elements from 4 dpf wildtype and dact1/2−/− compound mutants. dact1/2−/− mutants have a rod-shaped EP with no distinct lateral and medial elements. No obvious differences were found in dact1/2 mutant VC. (C) Representative brightfield image of 4 dpf wildtype and dact1/2−/− mutant. Bar indicates vertebral spine length. Scale bar: 100 μm. (D) Representative images of Alcian blue stained dact1/2−/−, wnt11f2−/−, and wnt11f2−/−,dact1/2−/− compound mutants. Embryos resulted from a dact1+/-,dact2+/-,wnt11f2+/-in-cross. Lateral and ventral views show similar craniofacial phenotypes in each mutant. (E) Representative flat-mount images of Alcian blue stained EP show a similar phenotype between dact1/2−/−, wnt11f2−/−, and wnt11f2−/−,dact1/2−/− compound mutants. Scale bar: 200 μm.
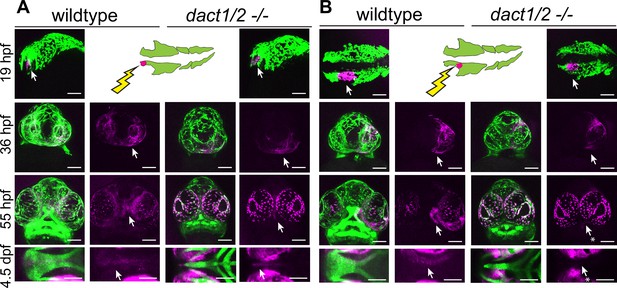
Anterior neural crest cells of the dact1/2−/− mutant migrate to the midline and populate the dysmorphic ethmoid plate.
Lineage tracing of wildtype and dact1/2−/− double mutant zebrafish embryos using Tg(sox10:kaede) line. sox10:kaede fluorescence is shown in green and photo-converted kaede is shown in magenta and highlighted with an arrow. Asterisks indicate that the cell population is absent. (A, B) 19 hpf embryo sagittal views showing photoconversion of anterior-most neural crest population. At 36 hpf frontal images show the migration of photoconverted neural crest cells to the frontal prominence in wildtype and dact1/2−/− double mutants. At 55 hpf, frontal images show photoconverted neural crest cells populating the region of the developing anterior neurocranium (ANC) in wildtype and dact1/2−/− mutants. At 4.5 dpf ventral images show photoconverted neural crest cells populating the medial ethmoid plate in wildtype. Similarly, neural crest cells in dact1/2−/− mutants populate the rod-shaped ethmoid plate. Scale bar: 100 μm. Representative images of three individual experiments.
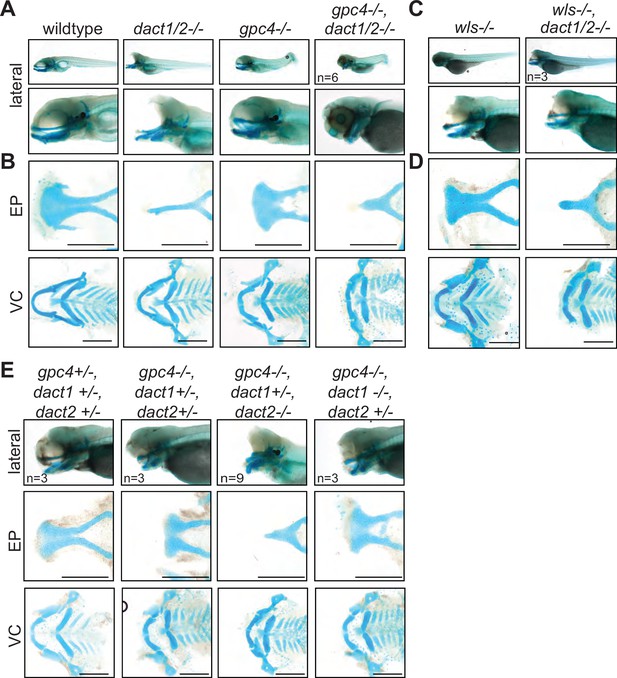
A nonoverlapping functional role for dact1, dact2, and gpc4 and wls.
(A) Representative Alcian blue stained wholemount images of wildtype, dact1/2−/− double mutant, gpc4−/− mutant, and gpc4/dact1/2−/− triple mutants at 4 dpf. Low magnification lateral images of embryos showing tail truncation in dact1/2−/− mutants, shortened and kinked tail in gpc4−/− mutants, and a combinatorial effect in gpc4/dact1/2−/− triple mutants. Higher magnification lateral images show a shortened midface and displaced lower jaw in dact1/2−/− mutants, a shortened midface in gpc4−/− mutant, and a combinatorial effect in gpc4/dact1/2−/− triple mutants. (B) Representative flat-mount images of dissected Alcian blue-stained cartilage elements. dact1/2−/− mutants have a narrow rod-shaped ethmoid plate (EP) while gpc4−/− mutants have a broad and shortened EP. dact1/2/gpc4 triple mutants have a combinatorial effect with a short, broad rod-shaped EP. In ventral cartilages (VC), dact1/2−/− mutants have a relatively normal morphology while Meckel’s cartilage in gpc4−/− mutants and gpc4/dact1/2−/− triple mutants is truncated. (C, D) Same as above except wls−/− mutant and wls/dact1/2−/− triple mutant, with similar findings. (E) Combinatorial genotypes of dact1, dact2, and gpc4. dact2−/− contributed the dact/gpc4 compound phenotype while dact1−/− did not. Scale bar: 200 μm.
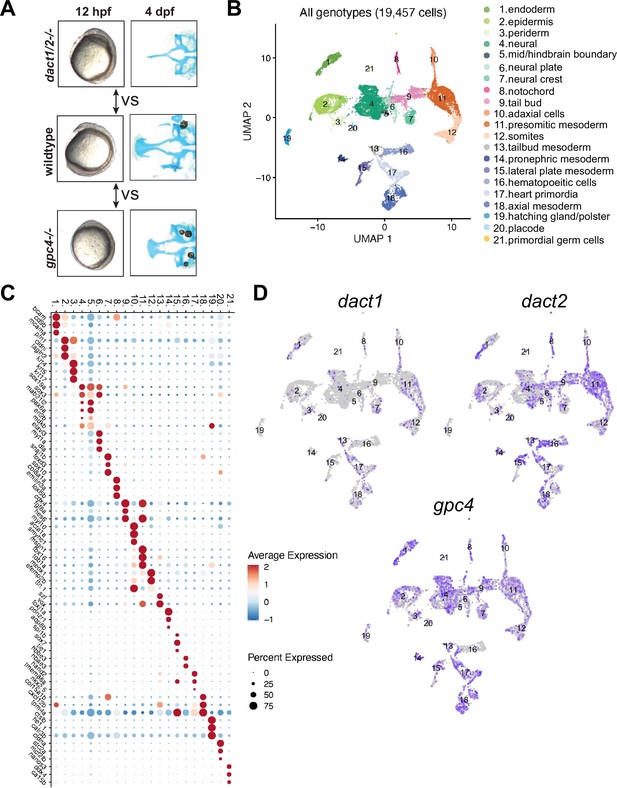
Single-cell RNAseq of 4 ss wildtype, dact1/2−/− mutant, and gpc4−/− mutants.
(A) Summary schematic showing similar phenotypes in dact1/2−/− and gpc4−/− mutants at 12 hpf and divergent phenotypes at 4 dpf. Single-cell RNAseq was performed during axis extension to compare and contrast dact1/2−/− and gpc4−/− transcriptional programs. Uniform manifold approximation and projection (UMAP) showing cluster identification. (B) UMAP of cell clusters identified by single-cell RNAseq. (C) Dot plot showing the most differentially expressed genes between clusters. (D) UMAP showing dact1, dact2, and gpc4 expression in wildtype embryos.
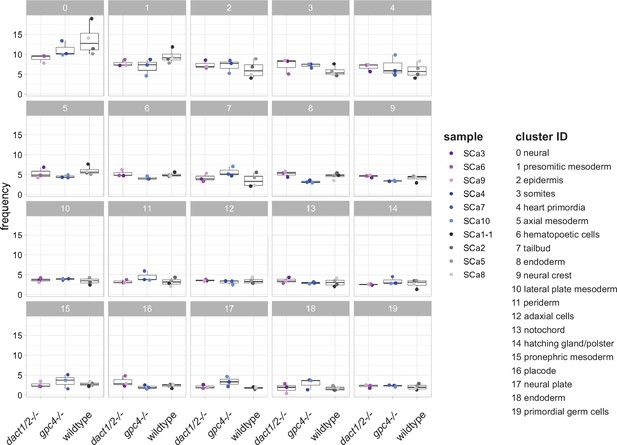
Cluster abundance across genotype groups.
Scatter box plots showing the frequency of each identified cell cluster between dact1/2−/−, gpc4−/−, and wildtype samples.
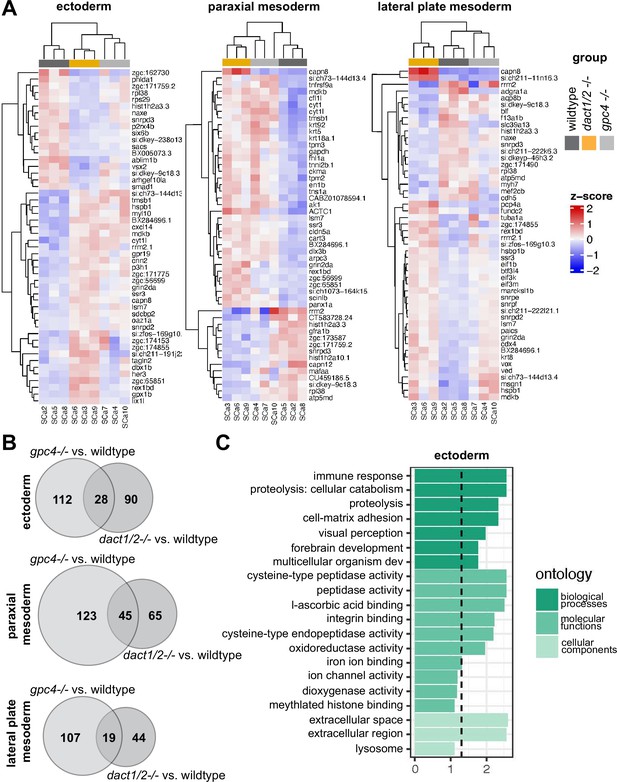
Pseudobulk differential expression analysis of single-cell RNAseq data.
(A) Heatmaps showing the 50 most differentially expressed genes (DEGs) in 3 major cell types; ectoderm (clusters 4, 5, 6, 7), paraxial mesoderm (clusters 10, 11, 12), and lateral plate mesoderm (clusters 15, 16, 17,18) between dact1/2−/− mutants and wildtype and gpc4−/− mutants and wildtype. (B) Venn diagrams showing unique and overlapping DEGs in dact1/2−/− and gpc4−/− mutants. (C) Gene Ontology (GO) analysis of dact1/2−/− mutant-specific DEGs in ectoderm showing enrichment for proteolytic processes.
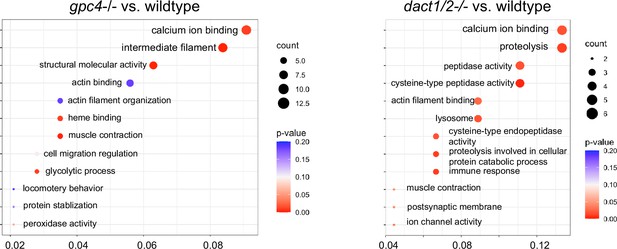
Loss of gpc4 and loss of dact1/2 lead to distinct changes in gene expression profiles but with some overlapping functions.
Gene Ontology (GO) analysis of differentially expressed genes (DEGs) identified between gpc4−/− and wildtype embryos and dact1/2−/− and wildtype embryos found changes in calcium ion binding and actin interaction in both mutants.
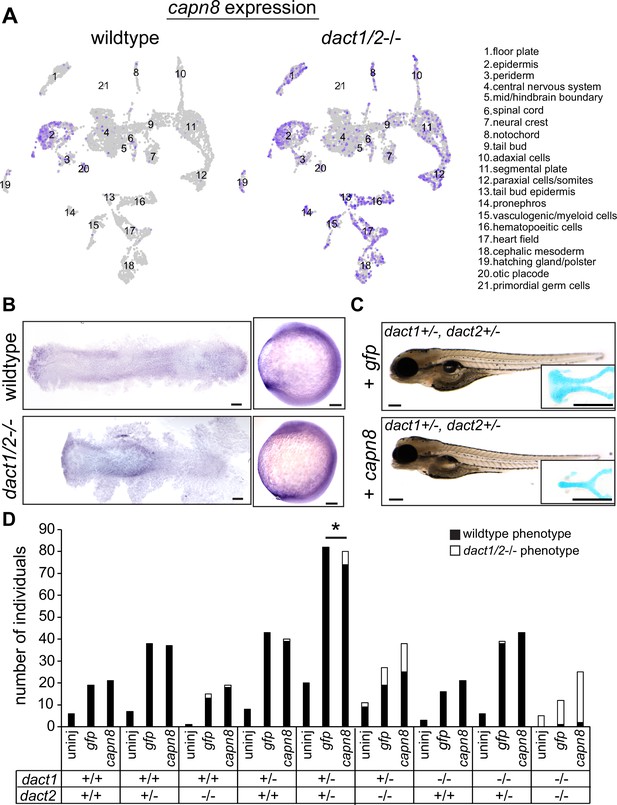
Expression of capn8 is significantly dysregulated in dact1/2−/− mutants.
(A) Single-cell RNAseq gene expression analysis of capn8 in wildtype and dact1/2−/− mutants. In wildtype embryos, capn8 expression is restricted predominantly to the epidermis whereas capn8 is widely expressed throughout the embryo in dact1/2−/− mutants, especially in the mesoderm. (B) Wholemount in situ hybridization of capn8 expression in wildtype and dact1/2−/− mutant embryos at 2 ss. Staining corroborates the single-cell RNAseq data, with expanded ectopic expression of capn8 throughout the embryo. Flat mounts are oriented anterior to the left. Scale bar: 100 μm. (C) Brightfield images and Alcian blue staining of the ethmoid plate show ectopic expression of capn8 mRNA (200 pg) at the 1 cell stage in dact1+/-,dact2+/- embryos recapitulates the dact1/2−/− compound mutant craniofacial phenotype. The mutant craniofacial phenotype did not manifest in gfp mRNA (200 pg) injected 1 cell-stage dact1+/-,dact2+/- embryos. Scale bar: 100 μm (D) Quantification of mutant and normal craniofacial phenotype in 4 dpf larvae after mRNA injection at the 1 cell stage. Larvae were derived from dact1/2+/- interbreeding. Larvae were uninjected or injected with 200 pg gfp control or capn8 mRNA. A Fisher exact test showed a significant effect of capn8 mRNA injecting in the dact1/2 double heterozygotes. Asterisk indicates a significant difference between conditions (p = 0.013).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (Danio rerio) | WT (AB) RRID:ZIRC_ZL1 | Zebrafish International Resource Center | ZDB-GENO-960809-7 | |
| Strain (Danio rerio) | WT (Tubingen) RRID:NCBITaxon_7955 | Zebrafish International Resource Center | ZDB-GENO-990623-3 | |
| Strain (Danio rerio) | wnt11f2 RRID:ZFIN_ZDB-GENO-200617-11 | Zebrafish International Resource Center | wnt11f2tx226/+ | |
| Strain (Danio rerio) | gpc4 RRID:ZFIN_ZDB-GENO-070209-132 | Zebrafish International Resource Center | gpc4hi1688Tg/+ | |
| Strain (Danio rerio) | wls | Gift. Rochard et al., 2016 PMID:27287801 | ||
| Strain (Danio rerio) | sox10:kaede | Dougherty et al., 2012 PMID:22948622 | ||
| Strain (Danio rerio) | dact1 | This paper | Methods: Animals and CRISPR/Cas9 targeted mutagenesis | |
| Strain (Danio rerio) | dact2 | This paper | Methods: Animals and CRISPR/Cas9 targeted mutagenesis | |
| Commercial kit | RNeasy Plus Mini Kit | QIAGEN | ID_source:identifier 74134 | |
| Commercial kit | High Capacity cDNA Reverse Transcription Kit | Thermo Fisher | ID_source:identifier 4368814 | |
| Commercial assay or kit | dact1 gene expression assay | Thermo Fisher | Dr03152516_m1 | |
| Commercial assay or kit | dact2 gene expression assay | Thermo Fisher | Dr03426298_s1 | |
| Commercial assay or kit | 18S rRNA gene expression assay | Thermo Fisher | Hs03003631_g1 | |
| Recombinant DNA reagent | pCS2+8 destination plasmid | Addgene Gökirmak et al., 2012 PMID:23124201 | #34931 | |
| Commercial assay or kit | ImMessage mMachine | Invitrogen | ID_source:identifier AM1344 | |
| Commercial assay or kit | RNAscope probe dact1 | ACDbio | ID_source:identifier 857191-C2 | |
| Commercial assay or kit | RNAscope probe dact2 | ACDbio | ID_source:identifier 857201-C3 | |
| Commercial assay or kit | RNAscope probe irf6 | ACDbio | ID_source:identifier 555101 | |
| Commercial assay or kit | Chromium Single Cell 3′ kit (version 3) | 10X Genomics | ID_source:identifier 1000268 | |
| Software, algorithm | Cellranger (version 6.1.0) 10x Genomics Cellranger DNA (RRID:SCR_023221) | Zheng et al., 2017 PMID:28091601 | ||
| Software, algorithm | Seurat (version 4.1.0) SEURAT (RRID:SCR_007322) | Hao et al., 2021 PMID:34062119 | ||
| Software, algorithm | Harmony (version 0.1.0) Harmony (RRID:SCR_022206) | Korsunsky et al., 2019 PMID:31740819 | ||
| Software, algorithm | DESeq2 (v1.34.0) DESeq2 (RRID:SCR_015687) | Love et al., 2014 PMID:25516281 | ||
| Software, algorithm | clusterProfiler (version 4.2.2) clusterProfiler (RRID:SCR_016884) | Wu et al., 2021 | ||
| Software, algorithm | ZiFiT Targeter v4.2 | Sander et al., 2007 PMID:17526515 | ||
| Software, algorithm | ChopChop CHOPCHOP (RRID:SCR_015723) | Montague et al., 2014 PMID:24861617 |





