circHIPK3 nucleates IGF2BP2 and functions as a competing endogenous RNA
Figures
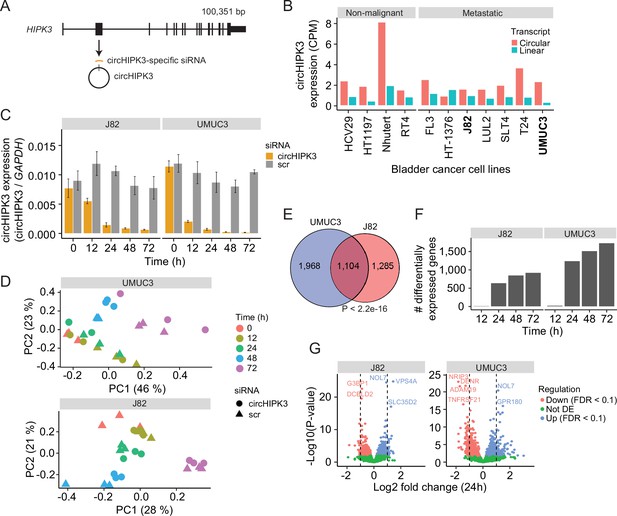
Thousands of genes are deregulated upon circHIPK3 knockdown (KD).
(A) circHIPK3 is produced from the second exon of the HIPK3 gene. We designed a circHIPK3-specific siRNA targeting the backsplice junction of circHIPK3 to specifically KD the expression of circHIPK3. (B) Expression of circHIPK3 and the corresponding linear transcript in 11 bladder cancer cell lines. UMUC3 and J82 were chosen for KD experiments based on circHIPK3 expression levels and cell line stability. CPM = counts per million. (C) KD efficiency of circHIPK3 in UMUC3 and J82 at different time points post circHIPK3 (yellow) or scramble (gray) siRNA transfection. Expression is normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. (D) Principal component analysis (PCA) plot of gene expression in UMUC3 and J82. PCA plots are based on the genes with the 50% most variance across all cell line samples. Gene expression is log transformed (natural logarithm) and added a pseudocount of 1. (E) Overlap of differentially expressed genes across all time points and/or conditions between UMUC3 (n = 3072) and J82 (n = 2389). p-value obtained by Fisher’s exact test. (F) Number of differentially expressed genes at each time point (Scr vs circHIPK3) (Wald test, Benjamini–Hochberg correction with false discovery rate [FDR] <0.1). Only genes with perturbed expression profiles across time and/or conditions in UMUC3 (n = 3072) or J82 (n = 2389) are considered. (G) Differential expression analysis between circHIPK3 KD and scr siRNA samples 24 hr post-transfection (Wald test). The log2 fold changes (circHIPK3 KD vs scr) are plotted against the negative log10(p-values). Colors indicate if genes are significantly down- (red) or upregulated (blue) or not differentially expressed (Not DE, green) after Benjamini–Hochberg correction, FDR <0.1. Vertical lines indicate a log2FC >1 or <−1.
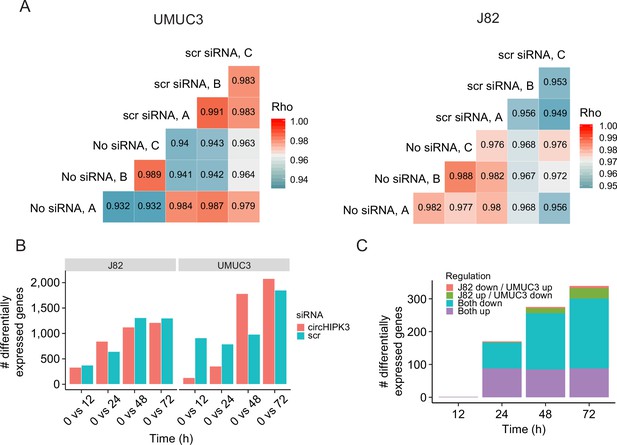
Thousands of genes are deregulated upon circHIPK3 knockdown (KD).
(A) Gene expression correlation upon scr siRNA transfection (scr siRNA, after 1h of incubation) and no transfection (No siRNA) in UMUC3 (left) and J82 (right). Letters (A, B, C) indicate triplicates. Gene expression is highly correlated between scr siRNA samples and No siRNA samples indicating no immediate effect of transfection (Rho > 0.93 for all comparisons, Spearman's rank-order correlation). (B) Number of differentially expressed genes between time points and baseline upon circHIPK3 KD or scr siRNA transfection (Wald test, Benjamini–Hochberg correction with false discovery rate [FDR] < 0.1). Only genes with perturbed expression profiles across time and/or conditions in UMUC3 (n = 3072) or J82 (n = 2389) are considered here. (C) Regulation of shared differentially expressed genes (n = 1104) between UMUC3 and J82 at each time point. Colours indicate regulation upon circHIPK3 KD vs scr siRNA transfection.
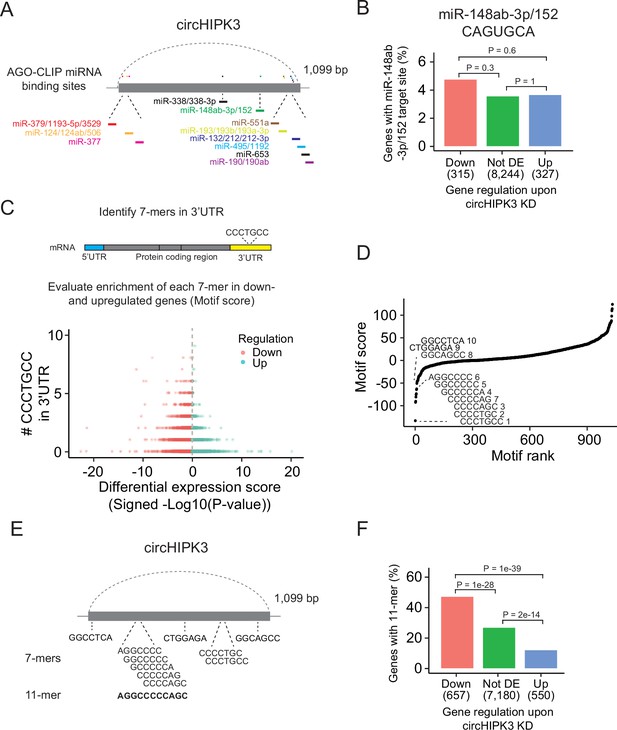
A long motif in circHIPK3 is enriched in downregulated genes upon circHIPK3 knockdown (KD).
(A) Illustration of conserved miRNA-binding sites in circHIPK3 based on AGO-CLIP data. (B) Percentage of genes in each group with miR-148ab-3p/152 target sites (CAGUGCA) in their 3′UTRs. Gene regulation is based on circHIPK3 KD vs scr siRNA transfection 24 hr post-transfection in J82. p-values obtained by Chi-square test. (C) Procedure for motif enrichment analysis using Regmex. First, we extract all 7-mers in the sequence of circHIPK3. For each 7-mer, for example, CCCTGCC, we identify their presence in the 3′UTR of genes. We order all genes according to a differential expression score calculated as the −log10(p-value) multiplied by the fold change direction, for example, 1 for upregulated genes and −1 for downregulated genes. Then we calculate a motif score for each 7-mer based on their occurrences in either down- or upregulated genes. If a 7-mer has a positive motif score it means that it is enriched in the 3′UTR of genes that are upregulated upon circHIPK3 KD. Conversely, 7-mers with a negative motif score are primarily found in genes that are downregulated upon circHIPK3 KD. The 7-mer, CCCTGCC, is used for illustration purposes. (D) Regmex motif scores for circHIPK3 7-mers (UMUC3, 24 hr). Alignment of the ten 7-mers with the most negative motif scores are shown. 7-mers are ranked from most negative to most positive motif scores. Numbers correspond to rank. (E) Illustration of circHIPK3 and position of the ten motifs with the lowest motif scores. The 7-mers with the most negative motif scores found in circHIPK3 comprise a larger 11-mer, AGGCCCCCAGC, present in the sequence of circHIPK3. (F) Percentage of genes in each group containing the 11-mer motif upon circHIPK3 KD in UMUC3 cells (24 hr). p-values obtained by Chi-square test.
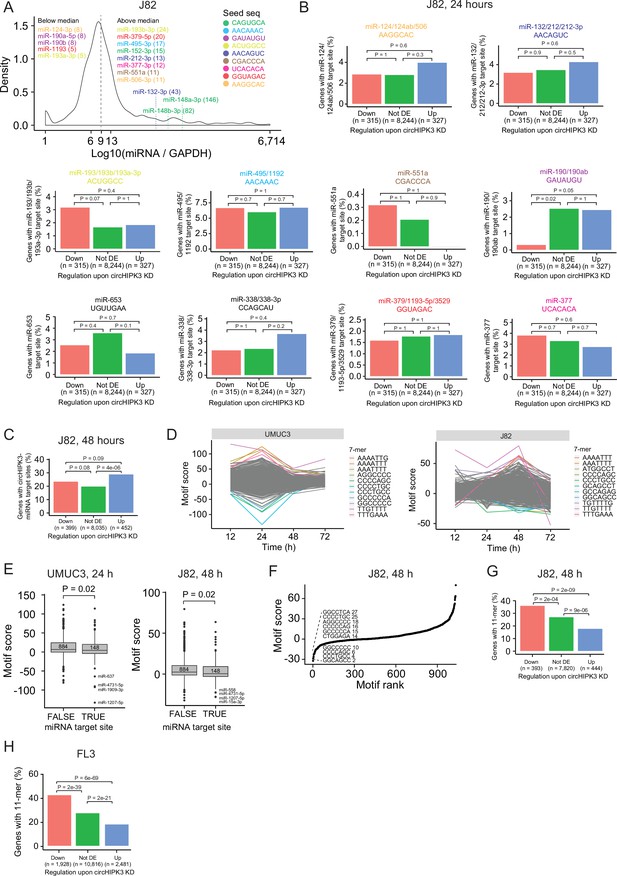
A long motif in circHIPK3 is enriched in downregulated genes upon circHIPK3 knockdown (KD).
(A) miRNA expression profiling in J82 by NanoString. miRNA expression is normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression levels. Dotted line indicates median miRNA expression. Conserved miRNAs with target sites in circHIPK3 (circHIPK3-miRNAs) are shown. miRNAs are coloured according to the sequence of their seed site (seed seq). Numbers in parentheses indicate miRNA expression levels. Colors refer to Figure 2A. (B) Percentage of genes in each group with target sites in their 3’UTRs for each circHIPK3-miRNA individually. Gene regulation is based on circHIPK3 KD vs scr siRNA transfection 24 hr post-transfection in J82. Headers indicate the name of the miRNA and the seed site. Colors refer to Figure 2A and panel A. The expression of miR-338-3p and miR-653 is not evaluated/detected in J82. p-values obtained by Chi-square test. (C) Percentage of genes in each group with circHIPK3-miRNA target sites in their 3’UTRs. Gene regulation is based on circHIPK3 KD vs scr siRNA transfection 48 hr post-transfection in J82. p-values obtained by Chi-square test. (D) Regmex motif scores upon circHIPK3 KD at each time point in UMUC3 (left) and J82 (right). (E) Regmex motif scores for 7-mers corresponding to miRNA target sites (TRUE) and not (FALSE). Numbers correspond to observations in each group. p-values obtained by Wilcoxon Rank Sum Test. (F) Regmex motif scores for circHIPK3-7-mers in J82 (48 hr). The ten 7-mers with the most negative motif scores in UMUC3 are shown. 7-mers are ranked from most negative to most positive motif scores. Numbers correspond to rank. (G) Percentage of genes in each group containing the 11-mer motif in J82 (48 hr) and FL3 (24 hr) cells. p-values obtained by Chi-square test.
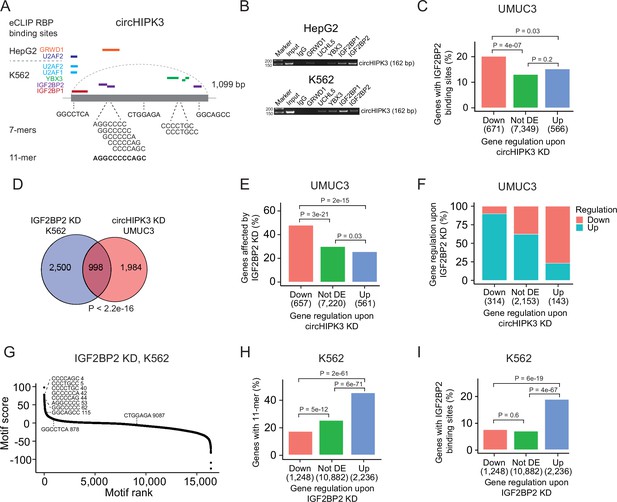
The 11-mer motif in circHIPK3 constitutes a binding site for IGF2BP2.
(A) Illustration of RNA-binding protein (RBP)-binding sites in circHIPK3 based on eCLIP data in the ENCODE cell lines HepG2 and K562. (B) RNA immunoprecipitation of circHIPK3–RBPs and others in HepG2 and K562 cells. Bands semi-quantitatively confirm that circHIPK3 interacts with IGF2BP1 and IGF2BP2. Marker indicates 50 bp. (C) Percentage of genes in each group containing IGF2BP2-binding sites (K562) in UMUC3 (24 hr). p-values obtained by Chi-square test. (D) Overlap between genes that are affected by circHIPK3 knockdown (KD) in UMUC3 and IGF2BP2 KD in K562. p-value obtained by Fisher’s exact test. (E) Percentage of genes in each group affected by IGF2BP2 KD in K562. p-values obtained by Chi-square test. (F) Regulation of genes affected by both circHIPK3 KD (UMUC3, 24 hr) and IGF2BP2 KD (K562). The x-axis indicates gene regulation upon circHIPK3 KD in UMUC3. Percentage on y-axis and colors show how these genes are regulated upon IGF2BP2 KD. Downregulated genes upon circHIPK3 KD are mainly upregulated upon IGF2BP2 KD and vice versa. (G) Regmex motif enrichment analysis upon IGF2BP2 KD in K562. 7-mers are ranked from most positive to most negative motif scores. The ten 7-mers with the most negative motif scores in UMUC3 are shown. Numbers indicate motif rank. All possible 7-mers are evaluated (n = 16,384). Percentage of genes in each group containing the 11-mer motif (H) and IGF2BP2-binding sites (I) upon IGF2BP2 KD in K562. p-values obtained by Chi-square test.
-
Figure 3—source data 1
Original file of the agarose gel shown in Figure 3B.
Bands included in the main figure are marked.
- https://cdn.elifesciences.org/articles/91783/elife-91783-fig3-data1-v1.zip
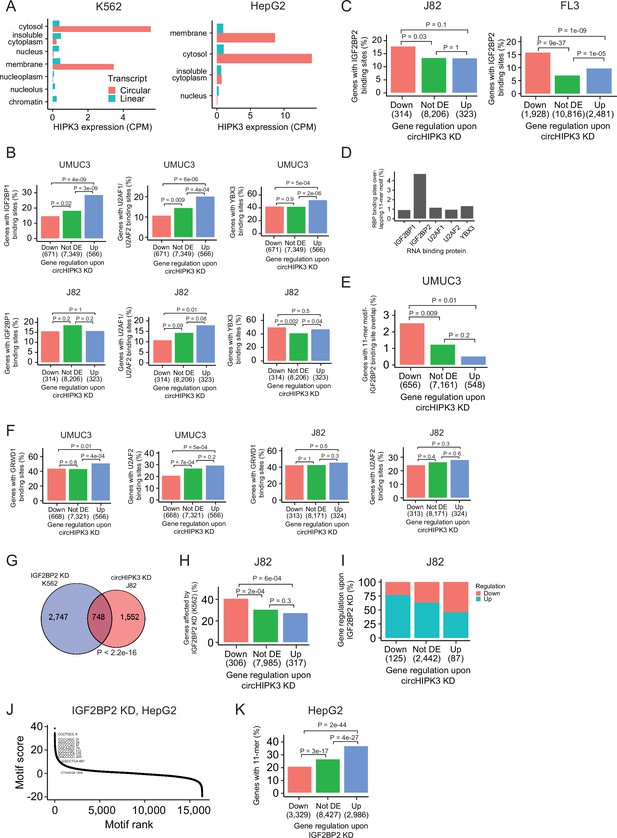
The 11-mer motif in circHIPK3 constitutes a binding site for IGF2BP2.
(A) Expression of circHIPK3 and the corresponding linear transcript in subcellular fractions of K562 (right) and HepG2 (left). CPM = Count per million. (B) Percentage of genes in each group containing RNA-binding protein (RBP) binding sites for individual circHIPK3-RBPs in K562. Gene regulation is based on circHIPK3 knockdown (KD) vs scr siRNA transfection 24 hr post-transfection (24 hr) in UMUC3 (upper panel) and J82 (lower panel). p-values obtained by Chi-square test. (C) Percentage of genes in each group containing IGF2BP2 binding sites (K562) in J82 (24 hr) and FL3 cells. p-values obtained by Chi-square test. (D) Percentage of times circHIPK3-RBP binding sites (K562) overlap the 11-mer motif. (E) Percentage of genes in each group where IGF2BP2 binding sites (K562) overlap the 11-mer motif (UMUC3, 24 hr). p-values obtained by Chi-square test. (F) Percentage of genes in each group containing RBP binding sites for individual circHIPK3-RBP in HepG2 (UMUC3 and J82, 24 hr). p-values obtained by Chi-square test. (G) Overlap between genes that are affected by circHIPK3 KD in J82 and IGF2BP2 KD in K562. p-value obtained by Fisher’s exact test. (H) Percentage of genes in each group (J82, 24 hr) affected by IGF2BP2 KD in K562. p-values obtained by Chi-square test. (I) Regulation of genes affected by both circHIPK3 KD (J82, 24 hr) and IGF2BP2 KD (K562). Downregulated genes upon circHIPK3 KD are mainly upregulated upon IGF2BP2 KD and vice versa. (J) Regmex motif enrichment analysis upon IGF2BP2 KD in HepG2. 7-mers are ranked from most positive to most negative motif scores. The ten 7-mers with the most negative motif scores in UMUC3 are shown. Numbers indicate motif rank. All possible 7-mers are evaluated (n = 16,384). (K) Percentage of genes in each group containing the 11-mer motif upon IGF2BP2 KD in HepG2. p-values obtained by Chi-square test.
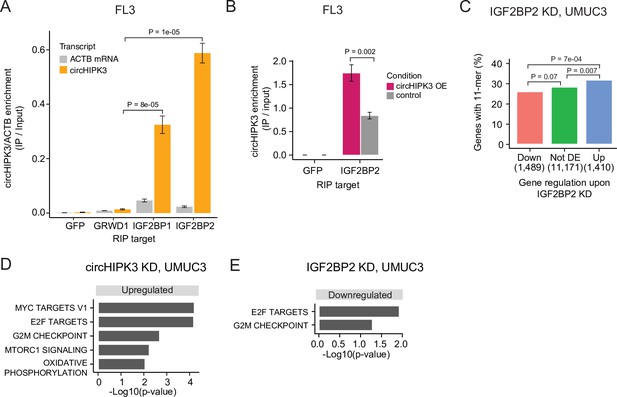
circHIPK3 interacts with IGF2BP2 and affects genes controlling cell cycle progression.
(A) Relative enrichment of circHIPK3 levels or ACTB mRNA between IP and input for the three RNA-binding proteins (RBPs) GRWD1, IGF2BP1, and IGF2BP2 in the bladder cancer cell line FL3. Green fluorescent protein (GFP) was used as a negative control. The statistical difference in circHIPK3 enrichment for GRWD1 and IGF2BP1 or IGF2BP2 is indicated by the p-value (T-test). (B) Relative enrichment of circHIPK3 levels between IP and input for IGF2BP2 upon circHIPK3 overexpression (OE – 24 hr). GFP was used as a negative control. Error bars reflect standard deviation of biological triplicates. (C) Percentage of genes in each group containing the 11-mer motif upon IGF2BP2 knockdown (KD) in UMUC3. p-values obtained by Chi-square test. (D) Gene set enrichment analysis of 50 hallmarks of cancer upon circHIPK3 KD (24 hr) in UMUC3 cells (false discovery rate [FDR] <0.1 for all shown pathways). (E) Downregulated hallmarks of cancer upon IGF2BP2 KD in UMUC3 (p-value <0.05 for all shown pathways).
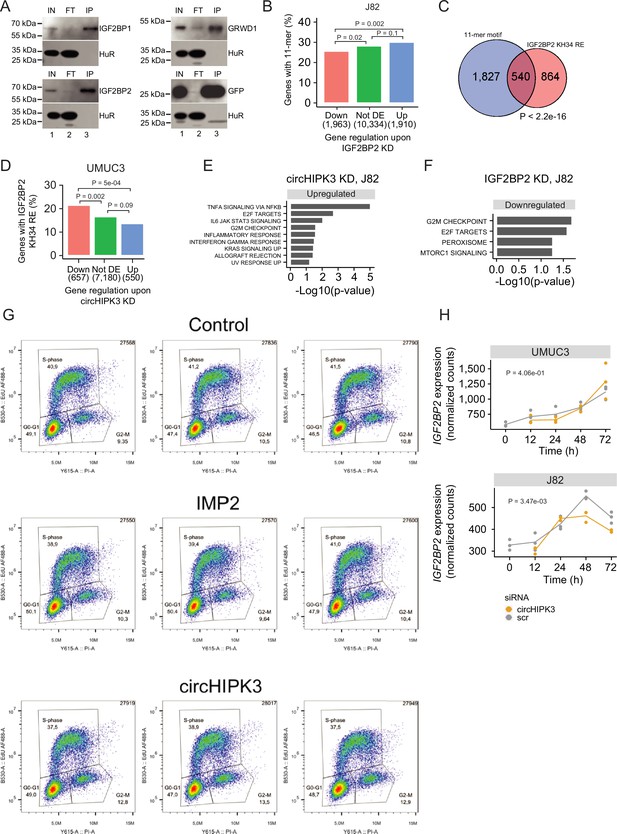
circHIPK3 interacts with IGF2BP2 and affects genes controlling cell cycle progression.
(A) Western blot analyzing protein samples from RNA IP (Figure 4A). Indicated twin-streptagged proteins (”IGF2BP1”, ”IGF2BP2”, ”GRWD1” or ”GFP”) were detected using and anti-streptag antibody. Anti-HuR (”HuR) was used as a loading control. ”IN” = Input, ”FT” = flowthrough, and ”IP” = Immunoprecipitate. (B) Percentage of genes in each group containing the 11-mer motif upon IGF2BP2 knockdown (KD) in J82. p-values obtained by Chi-square test. (C) Overlap between genes that contain the 11-mer motif and the IGF2BP2 KH34 RE in their 3’UTRs. p-value obtained by Fisher’s exact test. (D) Percentage of genes in each group containing the IGF2BP2 KH34 RE upon circHIPK3 KD (UMUC3, 24 hr). p-values obtained by Chi-square test. (E) Upregulated hallmarks of cancer upon circHIPK3 KD (48 hr) in J82 cells (p-value < 0.05 for all shown pathways). (F) Downregulated hallmarks of cancer upon IGF2BP2 KD in J82 (p-value < 0.05 for all shown pathways). (G) Distribution of FL3 cells in the cell cycle. Flow cytometry of cells transiently incubated with EdU followed by AlexaFluor488 labeling (Click-It chemistry). Intensities of propidium iodide (PI) stained DNA (X-axis) and AlexaFluor488 labeled newly synthesized DNA (Y-axis) is plotted. Boxes mark the gates used to estimate the fraction of cells in the indicated phase of the cell cycle. Upper panel: biological triplicates of control siRNA-treated cells, middle panel: IGF2BP2 (IMP2) KD, and lower panel: circHIPK3 KD. (H) IGF2BP2 mRNA expression in time-course experiments in UMUC3 (left) and J82 (right). Expression represents DESeq2 normalized counts. p-values obtained by likelihood ratio test.
-
Figure 4—figure supplement 1—source data 1
Original file for the western blot analysis shown in Figure 4—figure supplement 1 (anti-HuR).
- https://cdn.elifesciences.org/articles/91783/elife-91783-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Original file for the western blot analysis shown in Figure 4—figure supplement 1 (anti-streptag).
- https://cdn.elifesciences.org/articles/91783/elife-91783-fig4-figsupp1-data2-v1.zip
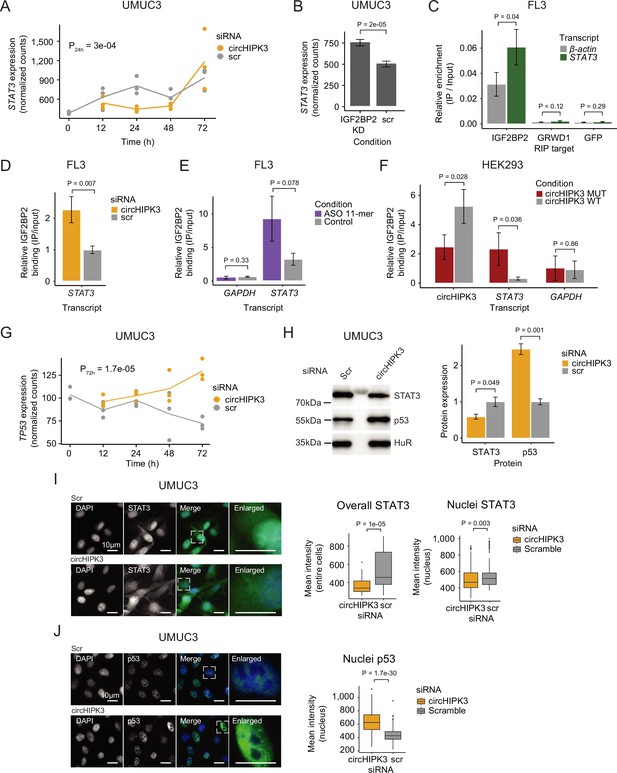
circHIPK3 functions as a competing endogenous RNA for IGF2BP2.
(A) STAT3 mRNA expression upon circHIPK3 knockdown (KD) in the time-course perturbation experiment in UMUC3 cells. Expression represents DESeq2 normalized counts. p-value (24 hr) obtained by Wald test. (B) Expression of STAT3 upon IGF2BP2 KD in UMUC3 cells. p-value obtained by Wald test. (C) Relative enrichment of STAT3 and β-actin levels between IGF2BP2 IP and input in FL3 cells. Green fluorescent protein (GFP) was used as a negative control. p-value obtained by T-test. (D) Relative enrichment of STAT3 levels between IGF2BP2 IP and input upon circHIPK3 KD in FL3 cells. p-value obtained by T-test. Control sample (scr) has been normalized to 1. (E) Relative enrichment of STAT3 and GAPDH levels between IGF2BP2 IP and input after 11-mer antisense oligonucleotide (ASO) transfection in FL3 cells. p-values obtained by T-test. (F) Relative enrichment of circHIPK3, STAT3, and GAPDH levels between IGF2BP2 IP and input after transfection of wildtype and mutated circHIPK3 in HEK293 cells. p-values obtained by T-test. (G) TP53 mRNA expression upon circHIPK3 KD in the time-course perturbation experiment in UMUC3 cells. Expression represents DESeq2 normalized counts. p-value (72 hr) obtained by Wald test. (H) Western blot of STAT3 and p53 protein upon circHIPK3 KD in UMUC cells. Quantifications on the left are internally normalized to HuR and then scr is normalized to 1. p-values obtained by T-test. Immunofluorescence staining for STAT3 (I) or p53 (J) in fixed UMUC3 cells (as indicated – green) subjected to control (scr) or circHIPK3 KD. Nuclei were counterstained using 4',6-diamidino-2-phenylindole (DAPI) (as indicated – blue). Mean signal intensities are quantified within nuclei or entire cells to the right. p-values obtained by Wilcoxon Rank Sum Test. (B–F, H) Error bars reflect standard deviation of biological triplicates.
-
Figure 5—source data 1
Original file for the western blot analysis shown in Figure 5H (anti-STAT3, anti-p53 and anti HuR).
- https://cdn.elifesciences.org/articles/91783/elife-91783-fig5-data1-v1.zip
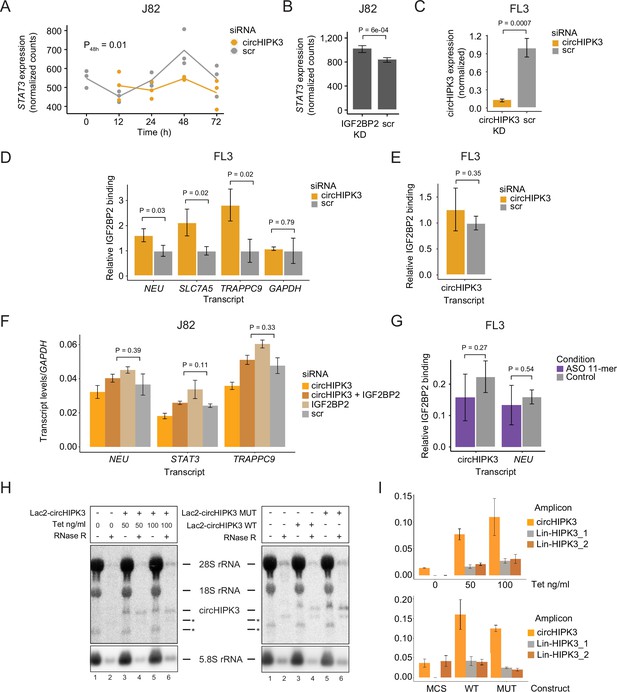
circHIPK3 functions as a competing endogenous RNA for IGF2BP2.
(A) STAT3 mRNA expression upon circHIPK3 knockdown (KD) in the time-course perturbation experiments in J82 cells. Expression represents DESeq2 normalized counts. p-value (48 hr) obtained by Wald test. (B) Expression of STAT3 upon IGF2BP2 KD in J82. p-value obtained by Wald test. (C) Expression of circHIPK3 upon circHIPK3 KD in FL3 cells. p-values obtained by T-test. (D, E) Relative enrichment of NEU, SLC7A5, and TRAPPC9 levels (D) and circHIPK3 (E) between IGF2BP2 IP and input upon circHIPK3 KD. p-values obtained by T-test. Control samples (scr) have been normalized to 1. GAPDH was used as a negative control. (F) Rescue experiment showing normalization of target gene expression (NEU, STAT3 and TRAPPC9) upon KD of both circHIPK3 and IGF2BP2. p-values obtained by T-test. (G) Relative enrichment of circHIPK3 and NEU levels between IGF2BP2 IP and input after 11-mer antisense oligonucleotide (ASO) transfection in FL3 cells. (H) Northern blot using RNA from Lac2-circHIPK3 expressing HEK293 FlpIn T-Rex cells or HEK293 cells transfected transiently with Lac2-circHIPK3 expression vectors. * denotes non-specific bands. (I) qRT-PCR using RNA from H circHIPK3 vs linear transcript. Primer efficiencies for amplicons were calculated from standard curves (not shown). (B-G and I) Error bars reflect standard deviation of biological triplicates.
-
Figure 5—figure supplement 1—source data 1
Original file for the northen blot analysis shown in Figure 5—figure supplement 1 (backsplicing junction probe).
- https://cdn.elifesciences.org/articles/91783/elife-91783-fig5-figsupp1-data1-v1.zip
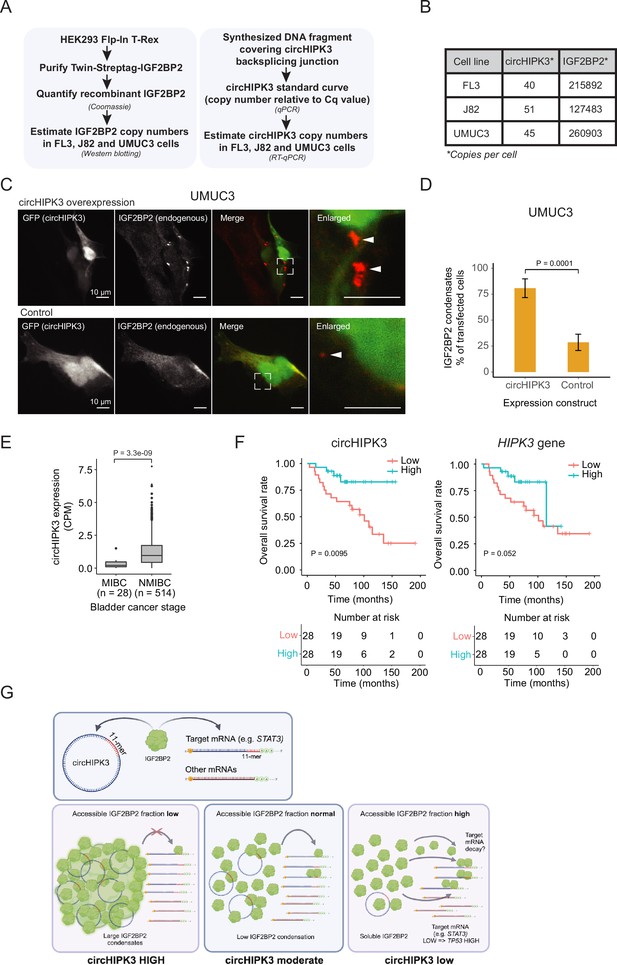
Absolute quantifications suggest low levels of circHIPK3 compared to IGF2BP2, where high circHIPK3 levels correlate with increased survival of bladder cancer (BC) patients.
(A) Flow chart used to perform absolute quantification of circHIPK3 and IGF2BP2 protein levels in BC cells. (B) Estimated levels of circHIPK3 and IGF2BP2 (copies per cell). (C) Immunofluorescence using IGF2BP2 antibody (red channel) with cells transfected with circHIPK3 expression plasmid (upper panel) or control plasmid (lower panel). Both plasmids encode enhanced green fluorescent protein (EGFP) from a separate expression cassette in order to identify transfected cells (green channel). White arrows indicate IGF2BP2 condensates (D) Quantification of transfected cells containing large IGF2BP2 condensates – % positive cells (28 and 34 cells were counted). Error bars reflect standard error of mean (SE). (E) Expression of circHIPK3 in samples from patients with non-muscle invasive bladder cancer (NMIBC) and muscle invasive bladder cancer (MIBC). CPM = counts per million. p-value obtained by Wilcoxon Rank Sum Test. (F) Kaplan–Meier overall survival plots of circHIPK3 (left) and HIPK3 (right). Median expression of circHIPK3 (0.192 CPM) and HIPK3 (7.96 FPKM) used as cutoff. p-values obtained by Log-Rank Test. (G) Working model that illustrates potential mechanism for circHIPK3. Our studies show that circHIPK3 contains an 11-mer motif (illustrated by red bases) that comprise a binding site for IGF2BP2 (in green). At normal levels, circHIPK3 interacts strongly with IGF2BP2 (middle panel), potentially involving nucleation of many IGF2BP2 proteins on single circHIPK3 molecules – exacerbated at high circHIPK3 levels, which induce condensates (left panel). This allows for only a few accessible IGF2BP2 molecules to bind target mRNAs including STAT3. Upon circHIPK3 knockdown (KD) and low circHIPK3 levels (right panel), IGF2BP2 is more accessible to interact more strongly with IGF2BP2 target genes, which contain the 11-mer motif in their 3′UTRs, incl. STAT3, and which are subsequently downregulated. Conversely, upon IGF2BP2 KD, IGF2BP2 target genes containing 11-mer are upregulated (not illustrated).
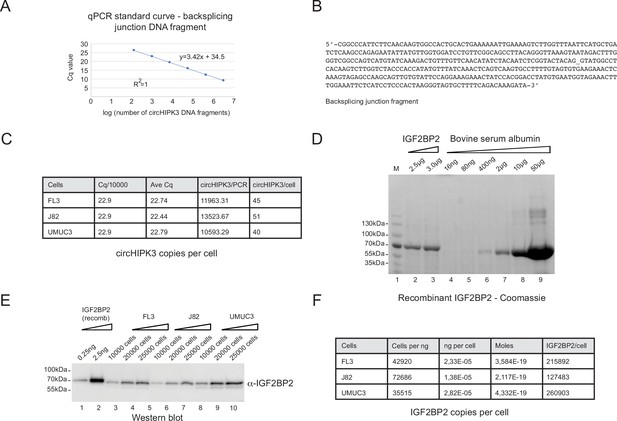
Absolute quantification of IGF2BP2 and circHIPK3 in bladder cancer cells.
(A) qPCR standard curve using a DNA fragment spanning the backsplicing junction of circHIPK3. (B) Sequence of DNA fragment used in the qPCR titration experiment. (C) Table summarizing key numbers used to estimate circHIPK3 copy numbers in FL3, J82 and UMUC3 cells (assuming an RT efficiency of 25% compared to direct DNA amplification). Cq/10000 = Cq value for 10.000 molecules. Ave Cq = average Cq value for 3 independent experiments. (D) Twin-streptagged-IGF2BP2 was expressed and purified from HEK293-Flp-In T-Rex cells and used for SDS page analysis (lanes 2-3). IGF2BP2 protein levels were quantified by comparison to known amounts of bovine serum albumin (BSA) using Gelcode Blue. (E) Known amounts of recombinant Twin-streptagged IGF2BP2 were subsequently used for western blotting to estimate levels in FL3, J82 and UMUC3 cells. (F) Table summarizing numbers used to estimate IGF2BP2 abundance in FL3, J82 and UMUC3 cells. IGF2BP2/Cell = number of copies of IGF2BP2 per cell in the indicated cell type.
-
Figure 6—figure supplement 1—source data 1
Original file for the scanned Coomassie brilliant blue stained gel shown in Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/91783/elife-91783-fig6-figsupp1-data1-v1.zip
-
Figure 6—figure supplement 1—source data 2
Original file for the western blot analysis shown in Figure 6—figure supplement 1 (anti-IGF2BP2).
- https://cdn.elifesciences.org/articles/91783/elife-91783-fig6-figsupp1-data2-v1.zip
Additional files
-
Supplementary file 1
qPCR primers for detecting circHIPK3.
- https://cdn.elifesciences.org/articles/91783/elife-91783-supp1-v1.xlsx
-
Supplementary file 2
Conserved miRNA-binding sites in circHIPK3 based on AgoClip data.
- https://cdn.elifesciences.org/articles/91783/elife-91783-supp2-v1.xlsx
-
Supplementary file 3
miRNA expression in J82.
- https://cdn.elifesciences.org/articles/91783/elife-91783-supp3-v1.xlsx
-
Supplementary file 4
7-mer motifs in circHIPK3 and motif scores from RegMex analysis based on gene regulations 24 hr post-transfection in UMUC3 and 48 hr post-transfection in J82.
- https://cdn.elifesciences.org/articles/91783/elife-91783-supp4-v1.xlsx
-
Supplementary file 5
Subcellular localization of circHIPK3–RBPs in HepG2.
- https://cdn.elifesciences.org/articles/91783/elife-91783-supp5-v1.xlsx
-
Supplementary file 6
DESeq2 output from J82 cells.
- https://cdn.elifesciences.org/articles/91783/elife-91783-supp6-v1.xlsx
-
Supplementary file 7
DESeq2 output from UMUC3 cells.
- https://cdn.elifesciences.org/articles/91783/elife-91783-supp7-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91783/elife-91783-mdarchecklist1-v1.pdf





