Ice nucleation proteins self-assemble into large fibres to trigger freezing at near 0 °C
Figures
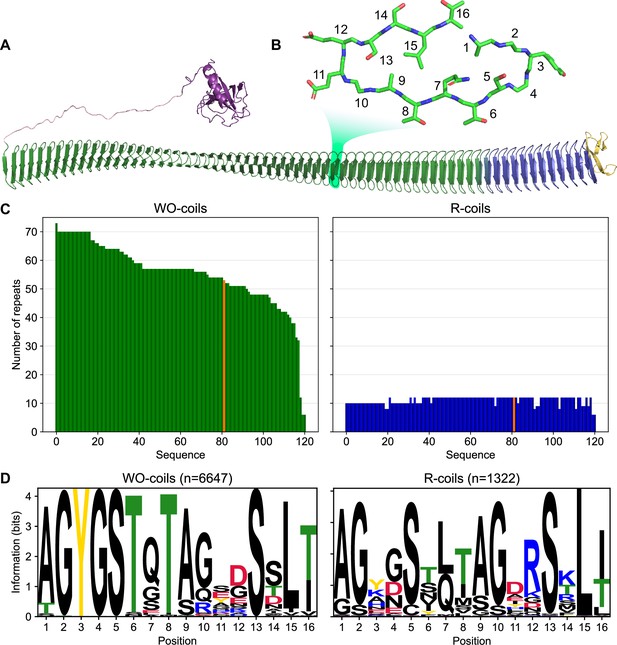
Classification of unique ice nucleation protein (INP) tandem arrays into either WO- or R-coil subdomains.
(A) AlphaFold2 model of PbINP coloured by domain. Purple: N-terminal domain, pink: flexible linker, green: water-organizing (WO) coils, blue: arginine-rich (R) coils, yellow: C-terminal cap. The inset shows a cross-section through the solenoid coil. (B) 16-residue tandem repeat forming one coil from the β-solenoid with positions numbered from N- to C-terminus. (C) The number of repeats in the WO- and R-coils for each unique sequence. PbINP is included and indicated in orange. n = 121. (D) Sequence logos constructed from each 16-residue repeat present in the dataset.
-
Figure 1—source data 1
Raw data table summarizing INP genes sequenced with long-read technology.
- https://cdn.elifesciences.org/articles/91976/elife-91976-fig1-data1-v1.csv
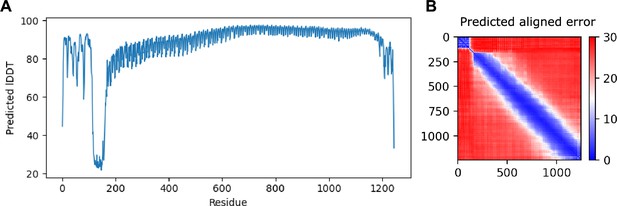
AlphaFold shows high confidence in overall fold of the model.
(A) Predicted location dependent difference test (pLDDT) shows the residue-by-residue confidence of the model generated by AlphaFold. Low values may indicate low confidence or intrinsic disorder. (B) Predicted aligned error (pAE) plots indicate the confidence in the relative orientation of the models. The x- and y-axes indicate residue position of the model, with low (blue) values indicating high confidence and high (red) indicating low. Rigid domains often appear as squares along the diagonal axis.
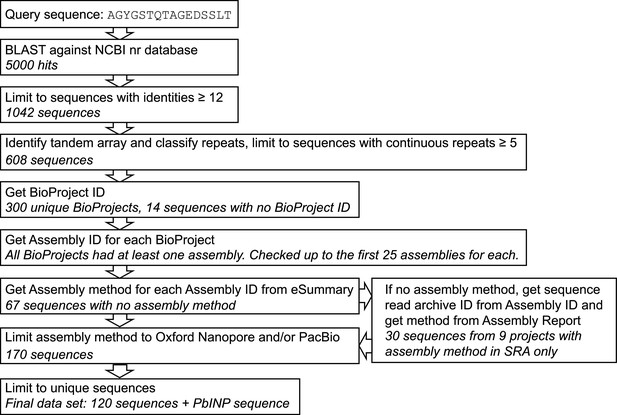
Flowchart and quality control steps in sequence selection for bioinformatic analysis.
Ten known ice nucleation proteins (INPs) from literature were used to generate a consensus sequence for WO-coils which was then used as a query in a BLAST against NCBI’s non-redundant protein database to identify INPs. NCBI E-Utils were used to generate a dataset using only genes from long-read DNA sequencing data.
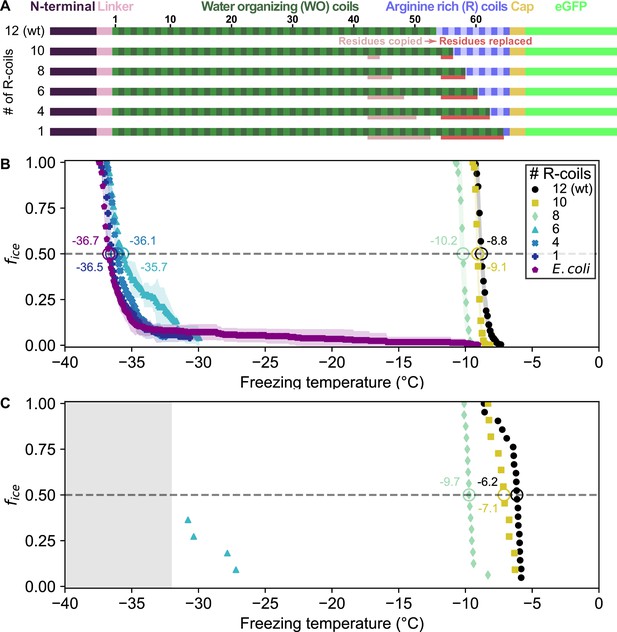
Ice nucleation activity of mutant ice nucleation proteins (INPs) in which R-coils are incrementally replaced with WO-coils.
(A) Diagram indicating the domain map of PbINP and from it, the design of the INP mutants. Sections of the WO-coils used in the replacement of the R-coils are indicated. wt: wild type. (B) Ice nucleation temperatures measured by WISDOM for E. coli cells expressing PbINP and mutants with different numbers of R-coils. The temperature at which 50% of the droplets froze (T50) is indicated with a hollow circle, and its corresponding value written nearby. The shaded region indicates standard deviation. (C) The same constructs assayed for ice nucleation using a nanoliter osmometer. The grey box indicates temperatures beyond the lower limit of the NLO apparatus for detecting ice nucleation in this experiment.
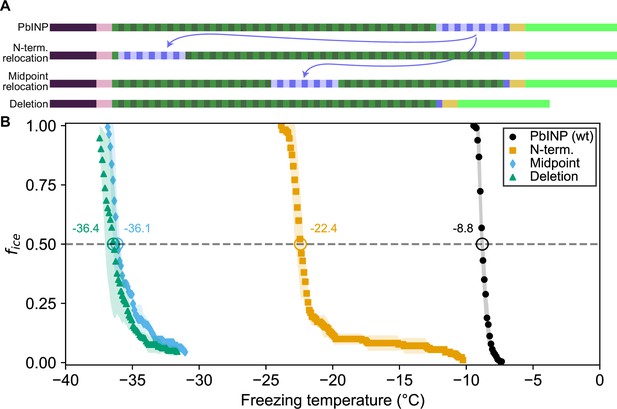
Ice nucleation activity of mutant ice nucleation proteins (INPs) with entirely relocated or deleted R-coil subdomain.
(A) Diagram indicating the design of the constructs. Eleven of the twelve R-coil repeats were either moved within the construct or deleted. (B) Freezing curves with T50 and number of unfrozen droplets indicated where applicable.
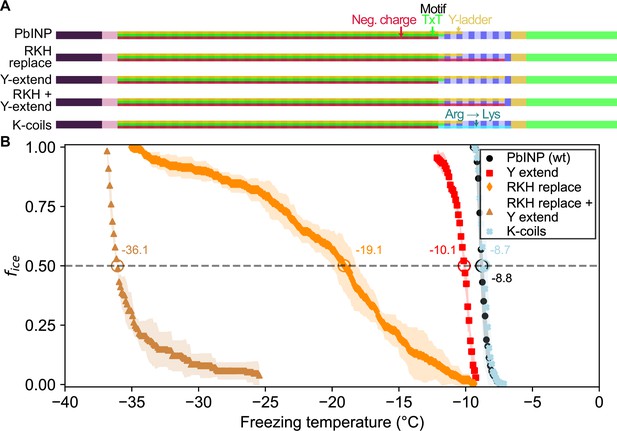
Site-specific mutagenesis of noteworthy motifs in the R-coil subdomain.
(A) Diagram indicating the design of the constructs. Translucent bars indicate continuity of three conserved motifs along the length of the central repetitive domain. Neg. charge: negative residues present in positions 11, 12, and 14 of the repeat. TxT: Thr–Xaa–Thr motif in positions 6–8 of the repeat. Y-ladder: an entirely conserved Tyr in position 3 of the motif. Three mutants were created in which these motifs were extended into the R-coils. K-coils: all arginines in the R-coils were replaced with lysines. (B) Freezing curves with T50 and number of unfrozen droplets indicated where applicable.
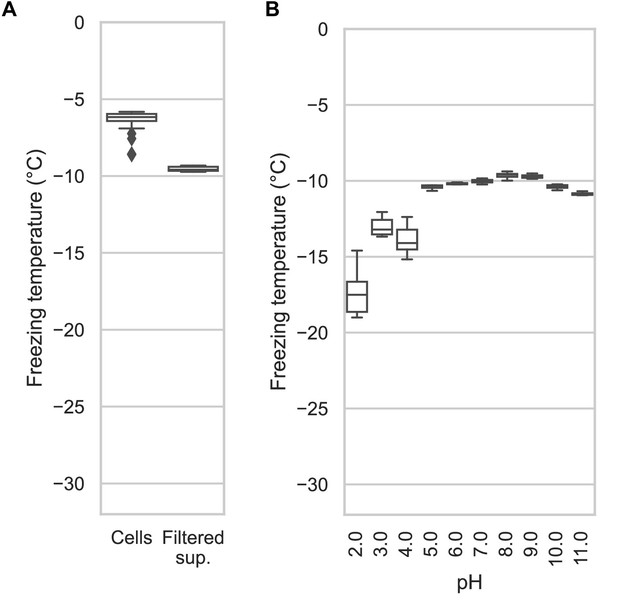
Assaying the activity of INP-containing supernatants across the pH scale.
(A) A comparison of the nucleation temperatures of PbINP when assayed using intact E. coli cells and when assayed with filtered supernatant. (B) A box and whisker plot showing the nucleation temperatures of filtered supernatant containing PbINP under different pH conditions. Boxes and bars indicate quartiles, with medians indicated by a centre line. Outliers are indicated by diamonds.
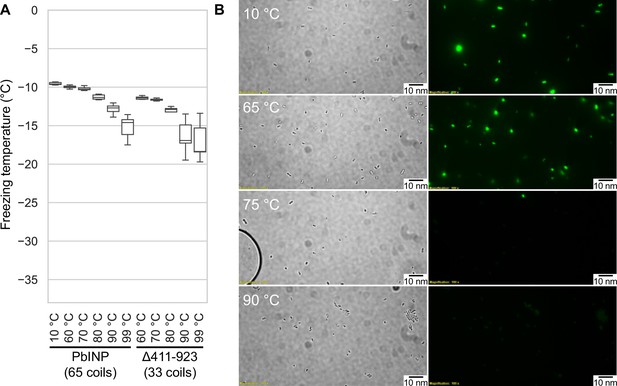
Assaying the heat stability of INPs.
(A) Measured freezing temperatures of heat-treated droplets containing either PbINP or PbINP with the first 32 repeats (counting from the N-terminal end) deleted. Wild-type PbINP freezing without heat treatment (kept at roughly 10 °C) is indicated on the left. (B) Fluorescent microscopy images of recombinant E. coli cells expressing PbINP tagged with GFP viewed under bright-field (BF) or fluorescent excitatory (GFP) light. Representative images are shown (n = 3). Note: cells that retain their fluorescence after 75 °C treatment are rarely observed.
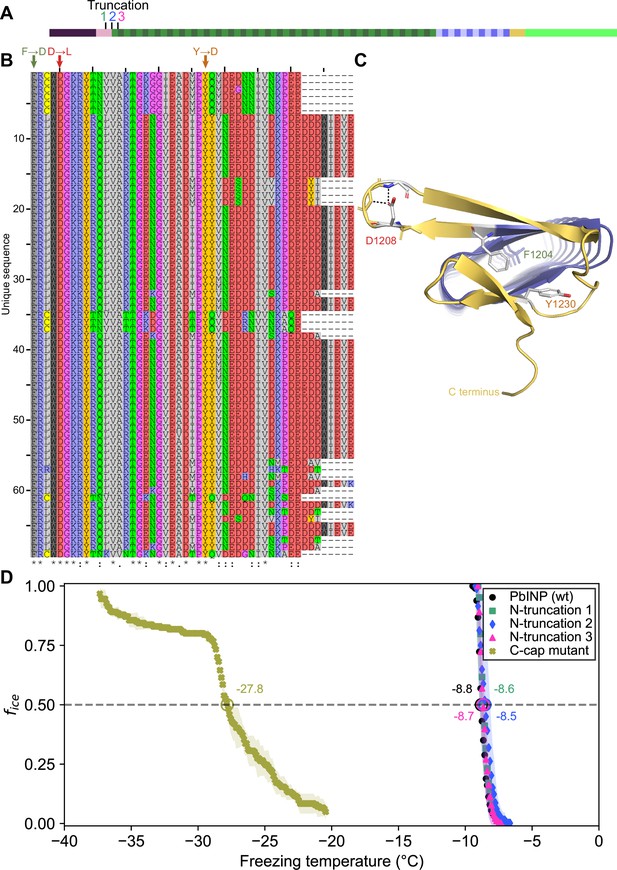
Investigating putative INP cap structures through a series of mutant proteins.
(A) Sites of N-terminal truncations to PbINP, indicating the location of the starting residue in the shortened construct. (B) Alignment of representative ice nucleation protein (INP) C-terminal domains from the genus Pseudomonas. Mutated residues and their one-letter codes are indicated above. Symbols at the bottom indicate consensus (* for fully conserved, : for conservation of strongly similar chemical properties, . for conservation of weakly similar chemical properties). (C) Predicted location of mutated residues in the PbINP C-terminal cap with side chains shown and predicted H-bonds for D1208 shown as dashed lines. (D) The ice nucleation curves for the N- and C-terminal cap mutants.
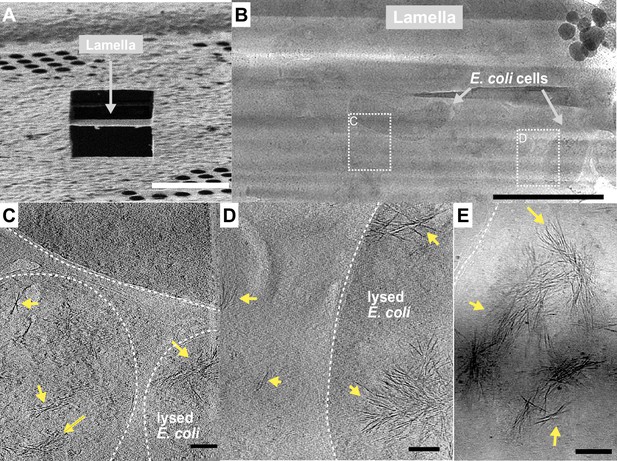
Fibrous bundles observed by cryo-focused-ion-beam (cryo-FIB) and cryo-electron tomography (cryo-ET) in in E. coli cells expressing ice nucleation protein (INP).
(A) Ion-beam image of a thin lamella containing E. coli cells expressing INP obtained from cryo-FIB milling. (B) Zoomed-in view of a cryo-transmission electron microscopy (TEM) image of the lamella in (A). Boxes with dashed lines indicate areas where tilt series were collected. (C, D) Snapshots from 3D cryo-tomograms reconstructed from tilt series collected in the boxed regions in (B) showing striking fibrous bundles (yellow arrowheads). The E. coli cell envelopes are indicated with thick dash lines. (E) Further examples of the fibrous bundles produced by INP-expressing E. coli. Size markers in (A) is 10 µm, in (B) is 2 µm, and in (C), (D), and (E) are 100 nm, respectively.
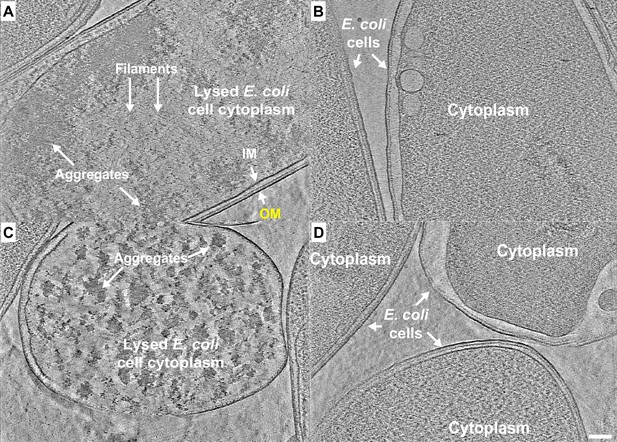
E. coli expressing ice nucleation protein (INP) mutant lacking R-coils show no fibre clusters as observed in those cells overexpressing wild-type INP.
(A–D) Representative snapshots from 3D cryo-tomograms showing cytoplasmic and extracellular features of various E. coli cells overexpressing an INP mutant in which all but the C-terminal R-coil have been replaced by WO-coils. All four images are in the same scale and the scale bar represents 100 nm.
Tomogram of an E. coli cell expressing an ice nucleation protein (INP).
The tomogram shows fibrous bundles in the cytoplasm of a cell recombinantly expressing the INP from Pseudomonas borealis (PbINP). Tilt angle range is ±48°. Scale bar indicates 100 nm.
Tomogram of an E. coli cell expressing an ice nucleation protein (INP).
Another example of fibrous bundles seen upon expression of INP from Pseudomonas borealis (PbINP). Tilt angle range is ±48°. Scale bar indicates 100 nm.
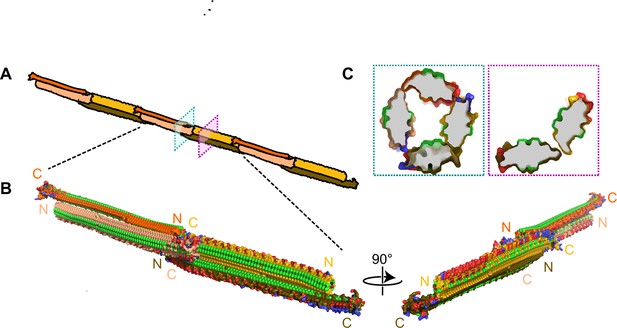
Filamentous multimer model for bacterial ice nucleation proteins (INPs).
(A) A possible assembly of INP solenoids to form long fibres composed of antiparallel INP dimers (indicated by orange and yellow pairs). (B) Dimers are formed along the tyrosine ladder, a previously proposed dimerization interface. They are joined end to end by forming electrostatic interactions between negatively (red) and positively (blue) charged surfaces. All threonines are coloured light green, displaying the arrays of TxT WO motifs. The termini of the INP solenoids are labeled N and C and coloured to match panel A. This illustration uses a manually flattened AlphaFold model of PbINP. (C) Cross-sections of the model at positions indicated in (A). Monomers are rotated approximately 90° to each other and dimerized along their tyrosine ladders (purple). Towards their termini, a pair of dimers can be matched by oppositely charged electrostatic surfaces (teal).
Apparent steric clash when proposed ice nucleation protein (INP) dimers are aligned in a parallel orientation.
Unfavourable steric interactions are observed at the C-terminal end of the model when aligned parallel along the tyrosine ladder, a previously proposed dimerization site. Structures are AlphaFold-predicted models of the INP from Pseudomonas borealis (PbINP) coloured to indicate WO-coils (dark green), R-coils (dark blue), C-terminal cap (yellow), TxT and SxT motifs (light green and cyan, respectively), and negative and positive charged side chains (red and blue, respectively).
Additional files
-
Source data 1
Droplet freezing assay results.
- https://cdn.elifesciences.org/articles/91976/elife-91976-data1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/91976/elife-91976-mdarchecklist1-v1.pdf





