LRMP inhibits cAMP potentiation of HCN4 channels by disrupting intramolecular signal transduction
Figures
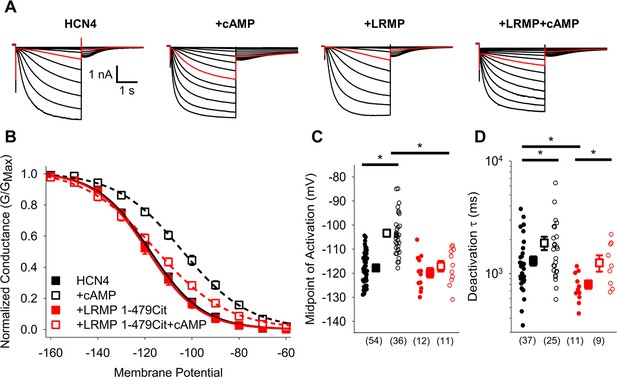
The cytosolic region of LRMP regulates HCN4 but does not antagonize cAMP binding.
(A) Exemplar current recordings from HCN4 in the absence or presence of 1 mM cAMP and/or LRMP 1-479Cit. Currents recorded with a –110 mV activating pulse are shown in red. (B) Voltage dependence of activation for HCN4 alone (black) or co-transfected with LRMP 1-479Cit (red) in the presence or absence of 1 mM intracellular cAMP (open symbols). (C) Average (± standard error of the mean) midpoints of activation for HCN4 in the absence or presence of LRMP 1-479Cit and/or 1 mM cAMP using the same color scheme as (B). (D) Average (± standard error of the mean) time constants of deactivation for HCN4 in the absence or presence of LRMP 1-479Cit and/or 1 mM cAMP using the same color scheme as (B). Small circles in (C) and (D) represent individual cells and values in parentheses are the number of independent recordings for each condition. * indicates a significant (p<0.05) difference. All means, standard errors, and exact p-values are in Table 1.

The pre-coiled-coil region of the LRMP N-terminus is necessary and sufficient to regulate HCN4.
(A) Schematic of LRMP showing the coiled-coil domain (CCD) and ER-transmembrane and luminal domains (ER) as predicted by Alphafold (Q60664). The locations of cut sites in the LRMP coiled-coil and N-terminal domains are indicated (red dotted lines).( B–E) Voltage-dependence of activation for HCN4 in the absence (black) or presence (red) of LRMP 1–227 (B), LRMP 228–539 (C), LRMP 1-108Cit (D), or LRMP 110-230Cit (E), and/or 1 mM intracellular cAMP (open symbols). The midpoints of activation for HCN4 with (dotted line) or without (solid line) 1 mM cAMP in the absence of LRMP are shown. (F) Average (± standard error of the mean) midpoints of activation for HCN4 in the absence or presence of LRMP constructs and/or 1 mM cAMP using the same color scheme as (B–E). Small circles represent individual recordings and values in parentheses are the number of independent recordings for each condition. * indicates a significant (p<0.05) difference. All means, standard errors, and exact p-values are in Table 1.

Mouse LRMP sequence.
Sequence for the mouse LRMP construct used in this study showing the predicted start of the coiled-coil, ER transmembrane, and ER lumenal domains as well as cut sites used for constructs in this study.

The distal HCN4 N-terminus is required for functional regulation by LRMP.
(A) Schematic representation of HCN4 showing truncation sites (red dotted lines) in the non-conserved distal N-terminus (TMD: Transmembrane domain). (B–E) Voltage-dependence of activation for HCN4 Δ1–25 (B), HCN4 Δ1–62 (C), HCN4 Δ1–130 (D), and HCN4 Δ1–185 (E) in the absence (black) or presence of LRMP (red) and/or 1 mM intracellular cAMP (open symbols). (B-E) Insets: Exemplar current recordings for HCN4 Δ1–25 (B), HCN4 Δ1–62 (C), HCN4 Δ1–130 (D), and HCN4 Δ1–185 (E) in the absence of LRMP and cAMP. Currents recorded with a –110 mV activating pulse are shown in red. (F) Average (± standard error of the mean) midpoints of activation for HCN4 Δ1–25, HCN4 Δ1–62, HCN4 Δ1–130, and HCN4 Δ1–185 in the absence or presence of LRMP and/or 1 mM cAMP using the same color scheme as (B–E). Small circles represent individual recordings and values in parentheses are the number of independent recordings for each condition. * indicates a significant (p<0.05) difference. All means, standard errors, and exact p-values are in Table 2.
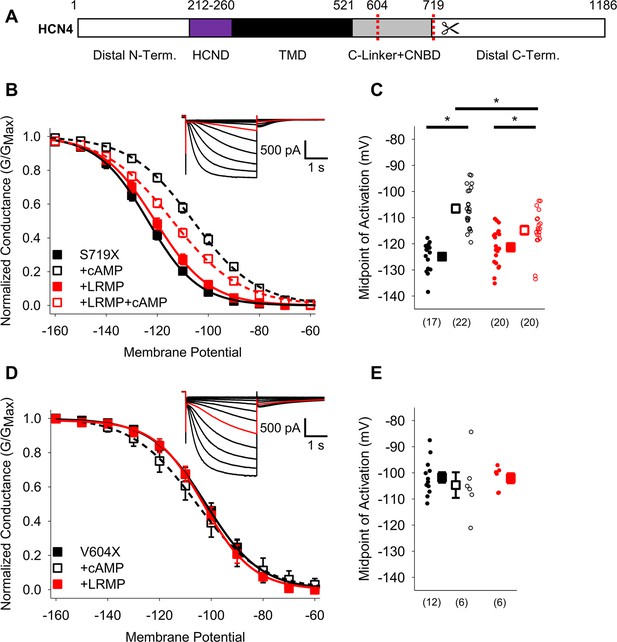
The HCN4 C-terminus is not the primary site for functional regulation by LRMP.
(A) Schematic representation of HCN4 showing truncation sites (red dotted lines) of the distal C-terminus and CNBD (TMD: Transmembrane domain). (B) Voltage-dependence of activation for HCN4 S719X in the absence (black) or presence of LRMP (red) and/or 1 mM intracellular cAMP (open symbols). (C) Average (± standard error of the mean) midpoints of activation for HCN4 S719X in the absence or presence of LRMP and/or 1 mM cAMP using the same color scheme as (B). (D) Voltage-dependence of activation for HCN4 V604X in the absence or presence of LRMP or 1 mM intracellular cAMP using the same color scheme as (B). (E) Average (± standard error of the mean) midpoints of activation for HCN4 V604X in the absence or presence of LRMP or 1 mM cAMP using the same color scheme as (B). Small circles represent individual recordings in (C) and (E) and values in parentheses are the number of independent recordings for each condition. (B and D) insets: Exemplar current recordings for HCN4 S719X (B) and HCN4 V604X (D) in the absence of LRMP and cAMP. Currents recorded with a –110 mV activating pulse are shown in red. * indicates a significant (p<0.05) difference. All means, standard errors, and exact p-values are in Table 2.
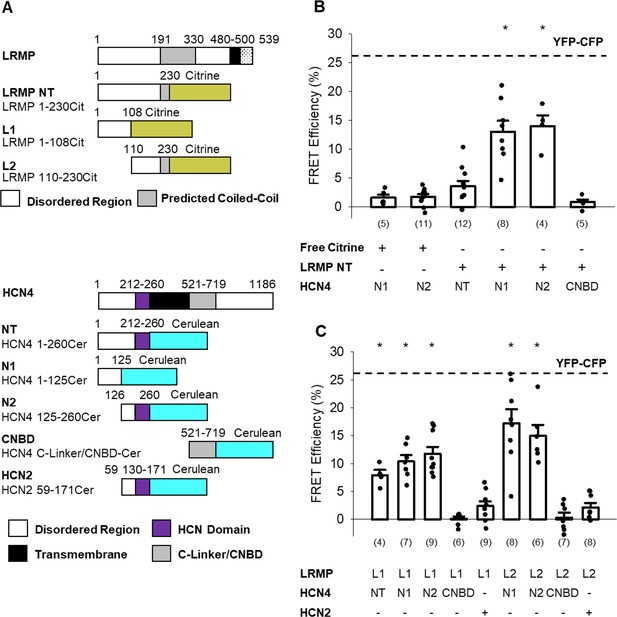
The N-terminus of LRMP FRETs with the N-terminus of HCN4.
(A) Schematic representations of the Citrine-tagged LRMP fragments and Cerulean-tagged HCN4 and HCN2 fragments used in FRET experiments. (B) Average (± standard error of the mean) acceptor photobleaching FRET efficiency between free Citrine or the Citrine-tagged N-terminal region of the LRMP (LRMP NT) and the Cerulean-tagged HCN4 N-terminus (NT), halves of the HCN4 N-terminus (N1 and N2), or the HCN4 C-Linker/CNBD. The dotted line is the average FRET in YFP-CFP concatemers from a prior study (Wang et al., 2020b). (C) Average (± standard error of the mean) acceptor photobleaching FRET efficiency between Citrine-tagged fragments of the LRMP N-terminus (L1 and L2) and Cerulean-tagged fragments of HCN4 or HCN2. Small circles in (B and C) represent individual recordings and values in parentheses are the number of independent recordings for each condition. * indicates a significant (p<0.05) difference compared to control FRET in cells co-transfected with free Citrine and with Cerulean-tagged HCN4 N-terminal fragments. All means, standard errors, and exact p-values are in Table 3.
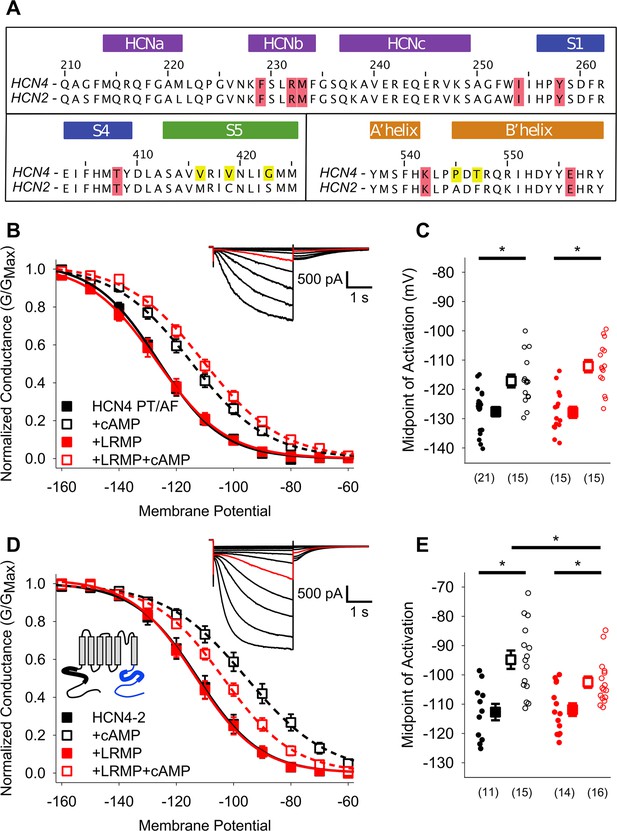
Mutants in the HCN4 C-linker disrupt LRMP’s functional effects.
(A) Sequence alignments of the HCN channel HCND (purple), voltage-sensor (blue), pore (green), and C-linker regions (orange) known to regulate cAMP-transduction. Non-conserved HCN4 residues in the S5 and C-linker regions are highlighted in yellow, and some of the residues believed to participate in cAMP-transduction are highlighted in red. (B) Voltage-dependence of activation for HCN4 P545A/T547F (PT/AF) in the absence (black) or presence of LRMP (red) and/or 1 mM intracellular cAMP (open symbols). (C) Average (± standard error of the mean) midpoints of activation for HCN4 PT/AF in the absence or presence of LRMP and/or 1 mM cAMP using the same color scheme as (B). (D) Voltage-dependence of activation for HCN4-2 (HCN4 1–518+HCN2 442-863) in the absence or presence of LRMP and/or 1 mM intracellular cAMP using the same color scheme as (B). Schematic Inset: Schematic of the chimeric HCN4-2 channel with HCN4 sequence shown in black and HCN2 in blue. The HCN and cyclic-nucleotide binding domains are indicated as thicker line segments. (E) Average (± standard error of the mean) midpoints of activation for HCN4-2 in the absence or presence of LRMP and/or 1 mM cAMP using the same color scheme as (B). (B and D) insets: Exemplar current recordings for HCN PT/AF (B) and HCN4-2 (D) in the absence of LRMP and cAMP. Currents recorded with a –110 mV activating pulse are shown in red. Small circles represent individual recordings in (C) and (E) and values in parentheses are the number of independent recordings for each condition. * indicates a significant (p<0.05) difference. All means, standard errors, and exact p-values are in Table 2.
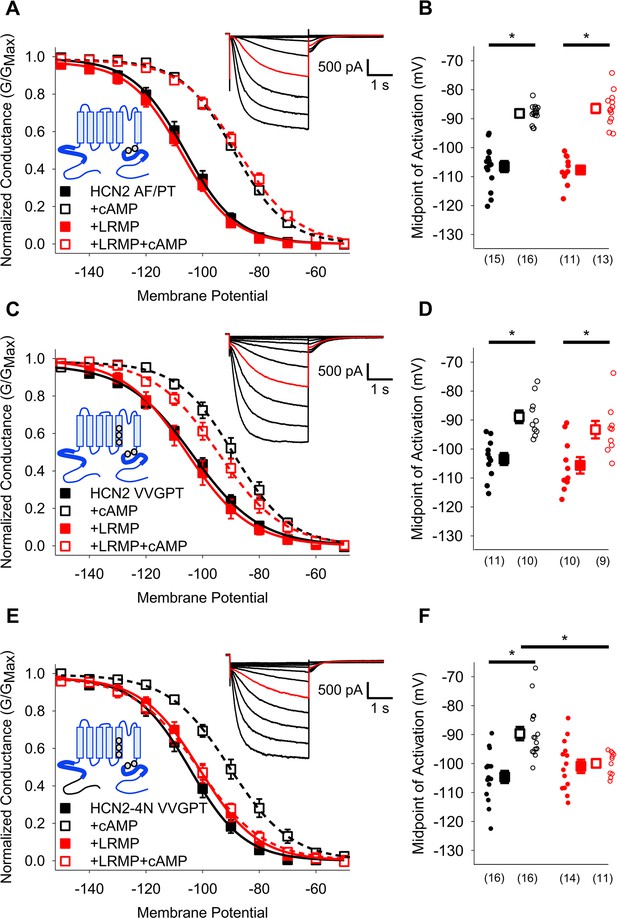
HCN4-specific residues and the HCN4 N-terminus confer LRMP regulation on HCN2.
(A) Voltage-dependence of activation for HCN2 A467P/F469T (AF/PT) in the absence (black) or presence of LRMP (red) and/or 1 mM intracellular cAMP (open symbols). (B) Average (± standard error of the mean) midpoints of activation for HCN2 AF/PT in the absence or presence of LRMP and/or 1 mM cAMP using the same color scheme as (A). (C) Voltage-dependence of activation for HCN2 VVGPT (M338V/C341V/S345G/A467P/F469T) in the absence or presence of LRMP and/or 1 mM intracellular cAMP using the same color scheme as (A). (D) Average (± standard error of the mean) midpoints of activation for HCN2 VVGPT in the absence or presence of LRMP and/or 1 mM cAMP using the same color scheme as (A). (E) Voltage-dependence of activation for HCN2-4N VVGPT (HCN4 1–212+HCN2 135-863 M338V/C341V/S345G/A467P/F469T) in the absence or presence of LRMP and/or 1 mM intracellular cAMP using the same color scheme as (A). (F) Average (± standard error of the mean) midpoints of activation for HCN2-4N VVGPT in the absence or presence of LRMP and/or 1 mM cAMP using the same color scheme as (A). Sample current insets: Exemplar current recordings for HCN2 AF/PT (A), HCN2 VVGPT (C), and HCN2-4N VVGPT (E) in the absence of LRMP and cAMP. Currents recorded with a –110 mV activating pulse are shown in red. Schematic Insets: Schematics of the chimeric channels with HCN4 sequence shown in black and HCN2 in blue. The HCN and cyclic-nucleotide binding domains are indicated as thicker line segments. Small circles represent individual recordings in (B, D) and (F) and values in parentheses are the number of independent recordings for each condition. * indicates a significant (p<0.05) difference. All means, standard errors, and exact p-values are in Table 2.
Tables
Midpoints of activation in HCN4 in the presence of LRMP fragments.
| Control(mV) | cAMP (1 mM)(mV) | ΔV½ in cAMP | p-Value Control vs. cAMP | |
|---|---|---|---|---|
| HCN4 Control | –117.8±0.9 (54) | –103.4±1.5 (36) | 14.4 mV | p<0.0001 |
| HCN4 LRMP 1-479Cit | –119.8±2.0 (12) p=0.3847 | –117.1±2.2 (11) p<0.0001 | 2.7 mV | p0.3710 |
| HCN4 LRMP 1–227 | –117.9±1.9 (13) p=0.9399 | –118.1±1.4 (11) p<0.0001 | –0.2 mV | p=0.9642 |
| HCN4 LRMP 228–539 | –116.1±2.6 (9) p=0.5265 | –106.3±2.0 (8) p=0.3100 | 9.8 mV | p=0.0069 |
| HCN4 LRMP 1-108Cit | –123.1±1.8 (7) p=0.0747 | –103.0±2.5 (8) p=0.8890 | 20.1 mV | p<0.0001 |
| HCN4 LRMP 110-230Cit | –118.0±4.0 (9) p=0.9423 | –106.4±1.3 (12) p=0.2118 | 11.6 mV | p=0.0005 |
-
Average midpoint of activation (mV) ± standard error of the mean (Number of independent cells). ΔV½ values reflect the difference in population midpoints for whole-cell experiments in the presence vs. absence of cAMP.
Midpoints of Activation for HCN Channel Constructs.
| Control(mV) | +LRMP(mV) | LRMP vs. Control | Control ΔV½ in cAMP (mV) | LRMP ΔV½ in cAMP (mV) | |
|---|---|---|---|---|---|
| HCN4 +cAMP | –117.8±0.9 (54)* –103.4±1.5 (36) p<0.0001 | –120.1±2.2 (14)* –114.7±2.6 (16) p=0.0724 | p=0.3530 p<0.0001 | 14.4 mV | 5.4 mV |
| HCN2 +cAMP | –109.3±1.5 (8) –90.3±3.2 (8) p<0.0001 | –114.4±1.9 (8) –87.9±1.6 (8) p<0.0001 | p=0.1101 p=0.4293 | 19.0 mV | 26.5 mV |
| HCN4 Δ1–25 +cAMP | –118.1±2.2 (19) –101.1±2.6 (13) p<0.0001 | –121.0±2.7 (10) –116.5±1.5 (12) p=0.2236 | p=0.3859 p=0.0001 | 17.0 mV | 4.5 mV |
| HCN4 Δ1–62 +cAMP | –116.5±1.7 (8) –99.1±2.2 (10) p<0.0001 | –118.8±1.9 (10) –107.9±1.4 (8) p=0.0003 | p=0.4089 p=0.0027 | 17.4 mV | 10.9 mV |
| HCN4 Δ1–130 +cAMP | –115.2±2.2 (11) –101.3±2.3 (8) p=0.0003 | –117.4±1.3 (6) –106.9±3.7 (7) p=0.0152 | p=0.5651 p=0.1481 | 13.9 mV | 10.5 mV |
| HCN4 Δ1–185 +cAMP | –117.1±2.1 (12) –103.1±3.3 (13) p<0.0001 | –125.5±2.3 (8) –103.6±3.1 (8) p<0.0001 | p=0.0500 p=0.8913 | 14.0 mV | 21.9 mV |
| HCN4 V604X +cAMP | –101.7±2.0 (12) –104.7±4.9 (6) p=0.4698 | –102.0±1.9 (6) — | p=0.9407 — | –3.0 mV | — |
| HCN4 S719X +cAMP | –124.9±1.3 (17) –106.5±1.6 (22) p<0.0001 | –121.4±1.6 (20) –114.1±1.6 (18) p=0.0018 | p=0.1217 p=0.0010 | 18.4 mV | 7.3 mV |
| HCN4 PT/AF +cAMP | –127.6±1.6 (21) –117.1±2.2 (15) p=0.0002 | –127.8±1.9 (15) –112.1±2.1 (15) p<0.0001 | p=0.9311 p=0.0785 | 10.5 mV | 15.7 mV |
| HCN2 AF/PT +cAMP | –106.5±1.9 (15) –88.2±0.7 (16) o<0.0001 | –107.7±1.5 (11) –86.4±1.6 (13) p<0.0001 | p=0.5858 p=0.3987 | 18.3 mV | 21.3 mV |
| HCN4-2 +cAMP | –112.7±2.8 (11) –94.8±3.1 (15) p<0.0001 | –111.9±2.2 (14) –102.5±1.9 (16) p=0.0092 | p=0.8290 p=0.0284 | 17.9 mV | 9.4 mV |
| HCN2 VVGPT +cAMP | –103.4±2.1 (11) –88.9±2.2 (10) p=0.0002 | –105.6±2.8 (10) –93.3±3.0 (9) p=0.0018 | p=0.5305 p=0.2278 | 14.5 mV | 12.3 mV |
| HCN2-4N VVGPT +cAMP | –104.6±2.1 (16) –89.6±2.3 (16) p<0.0001 | –100.9±2.3 (14) –99.9±1.2 (11) p=0.7574 | p=0.2183 p=0.0019 | 15.0 mV | 1.0 mV |
-
Average midpoint of activation ± standard error of the mean (Number of independent cells). ΔV½ values reflect the difference in population midpoints for whole-cell experiments in the presence vs. absence of cAMP.
-
*
HCN4 control and cAMP data in the absence of LRMP is the same as in Table 1.
Acceptor photobleaching FRET between LRMP and HCN channel fragments.
| Citrine | Cerulean | FRET efficiency (%) | p-Value vs. control |
|---|---|---|---|
| Free Citrine | HCN4 1–125 or 125–260 HCN4 1–125 HCN4 125–260 | 1.7±0.3 1.6±0.6 (5) 1.8±0.4 (11) | |
| LRMP 1–230 | HCN4 1–260 HCN4 1–125 HCN4 125–260 HCN4 C-Linker/CNBD | 3.6±0.9 (12) 13.1±1.9 (8) 14.0±1.9 (4) 0.8±0.5 (5) | p=0.8471 p<0.0001 p<0.0001 p=1.0000 |
| LRMP 1–108 | HCN4 1–260 HCN4 1–125 HCN4 125–260 HCN4 C-Linker/CNBD HCN2 N-Term | 7.9±1.0 (4) 10.4±1.1 (7) 11.7±1.3 (9) 0.0±0.5 (6) 2.4±0.8 (9) | p=0.0265 p<0.0001 p<0.0001 p=0.9869 p=1.0000 |
| LRMP 110–230 | HCN4 1–125 HCN4 125–260 HCN4 C-Linker/CNBD HCN2 N-Term | 17.2±2.5 (8) 15.0±2.0 (6) 0.3±0.9 (7) 2.1±0.8 (8) | p<0.0001 p<0.0001 p=0.9949 p=1.0000 |
-
Average midpoint of activation ± standard error of the mean (Number of independent cells).
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo-sapiens) | HEK-293 | ATCC | CRL-1573 | |
| Cell line (Homo-sapiens) | HEK-HCN4 | Dr. Martin Biel; Zong et al., 2012 | ||
| Cell line (Homo-sapiens) | HEK-HCN4 | This paper | ATCC CRL-1573; pcDNA3.1 mHCN4 | HEK-293 stably expressing HCN4 |
| Cell line (Homo-sapiens) | HEK-HCN2 | This paper | ATCC CRL-1573; pcDNA3.1 mHCN2 | HEK-293 stably expressing HCN2 |
| Cell line (Homo-sapiens) | HEK-HCN4 Δ1–62 | This paper | ATCC CRL-1573; pTwist-CMV-WPRE-Neo mHCN4 Δ1–62 | HEK-293 stably expressing HCN4 Δ1–62 |
| Cell line (Homo-sapiens) | HEK-HCN4 Δ1–130 | This paper | ATCC CRL-1573; pTwist-CMV-WPRE-Neo mHCN4 Δ1–130 | HEK-293 stably expressing HCN4 Δ1–130 |
| Cell line (Homo-sapiens) | HEK-HCN4 Δ1–185 | This paper | ATCC CRL-1573; pTwist-CMV-WPRE-Neo mHCN4 Δ1–185 | HEK-293 stably expressing HCN4 Δ1–185 |
| Cell line (Homo-sapiens) | HEK-HCN4 Δ1–200 | This paper | ATCC CRL-1573; pTwist-CMV-Hygro mHCN4 Δ1–200 | HEK-293 stably expressing HCN4 Δ1–200 |
| Cell line (Homo-sapiens) | HEK-HCN4 PT/AF | This paper | ATCC CRL-1573; pcDNA3.1 mHCN4 PT/AF | HEK-293 stably expressing HCN4 P545A/T547F |
| Cell line (Homo-sapiens) | HEK-HCN2 AF/PT | This paper | ATCC CRL-1573; pTwist-CMV-WPRE-Neo mHCN2 AF/PT | HEK-293 stably expressing HCN2 A467P/F469T |
| Recombinant DNA reagent | pcDNA3.1 mHCN1 | Dr. Eric Accili; Proenza et al., 2002 | ||
| Recombinant DNA reagent | pcDNA3.1 mHCN1 | This paper; Liao et al., 2012 | mHCN2 (sequence NP_032252.1) subcloned from pcDNA4 | |
| Recombinant DNA reagent | pcDNA6 mHCN4 Δ1–25 | Dr. Richard Aldrich; Liu and Aldrich, 2011 | ||
| Recombinant DNA reagent | pTwist-CMV-WPRE-Neo mHCN2 A467P/F469T | This paper | Synthesized by Twist Bioscience based on sequence NP_032252.1 | |
| Recombinant DNA reagent | pTwist-CMV-WPRE-Neo mHCN4 | This paper | NP_001074661.1; codon optimized | Synthesized by Twist Bioscience |
| Recombinant DNA reagent | pTwist-CMV-WPRE-Neo mHCN4 Δ1–62 | This paper | Deletions made using site-directed mutagenesis in pTwist-CMV-WPRE-Neo HCN4 | |
| Recombinant DNA reagent | pTwist-CMV-WPRE-Neo mHCN4 Δ1–130 | This paper | Deletions made using site-directed mutagenesis in pTwist-CMV-WPRE-Neo HCN4 | |
| Recombinant DNA reagent | pTwist-CMV-WPRE-Neo mHCN4 Δ1–185 | This paper | Deletions made using site-directed mutagenesis in pTwist-CMV-WPRE-Neo HCN4 | |
| Recombinant DNA reagent | pTwist-CMV-Hygro mHCN4 Δ1–200 | This paper | Synthesized by Twist Bioscience based on sequence NP_001074661.1 | |
| Recombinant DNA reagent | pcDNA3.1 mHCN4 P545T/A547F | This paper | Site-directed mutagenesis of pcDNA3.1 HCN4 by Applied Biological Materials | |
| Recombinant DNA reagent | pcDNA3.1 mHCN4 V604X | This paper | Site-directed mutagenesis of pcDNA3.1 HCN4 by Applied Biological Materials | |
| Recombinant DNA reagent | pcDNA3.1 mHCN4 S719X | Proenza Lab; Liao et al., 2012 | ||
| Recombinant DNA reagent | pcDNA4 mHCN4-2 | Proenza Lab; Liao et al., 2012 | HCN4 residues 1–518 plus HCN2 residues 442–863 | |
| Recombinant DNA reagent | pTwist-CMV-BG-WPRE-Neo mHCN2-4N VVGPT | This paper | Synthesized by Twist Bioscience based on sequence NP_001074661.1 and NP_032252.1 | |
| Recombinant DNA reagent | pTwist-CMV-BG-WPRE-Neo mHCN2 VVGPT | This paper | Synthesized by Twist Bioscience based on sequence NP_032252.1 | |
| Recombinant DNA reagent | pCMV6 Kan/Neo mLRMP | Origene | CAT#: MC201923 | Untagged mouse LRMP construct |
| Recombinant DNA reagent | pCMV6 Kan/Neo Myc-mLRMP | Proenza Lab; Peters et al., 2020 | N-terminal Myc-tagged LRMP construct | |
| Recombinant DNA reagent | pTwist-CMV mLRMP 1–227 | This paper | Synthesized by Twist Bioscience based on GenBank AAH52909.1 | |
| Recombinant DNA reagent | pTwist-CMV mLRMP 228–539 | This paper | Synthesized by Twist Bioscience based on GenBank AAH52909.1 | |
| Recombinant DNA reagent | pcDNA3.1 mHCN4 125-260Cer | This paper | C-terminal Cerulean; see DNA constructs section of the methods | |
| Recombinant DNA reagent | pcDNA3.1 mHCN4 1-260Cer | This paper | C-terminal Cerulean; see DNA constructs section of the methods | |
| Recombinant DNA reagent | pcDNA3.1 mHCN4 521-719Cer | This paper | C-terminal Cerulean; see DNA constructs section of the methods | |
| Recombinant DNA reagent | pcDNA3.1 mHCN4 1-125Cer | This paper | C-terminal Cerulean; see DNA constructs section of the methods | |
| Recombinant DNA reagent | pcMVBG mLRMP 1-479Cit | This paper | C-terminal Citrine; see DNA constructs section of the methods | |
| Recombinant DNA reagent | pcMVBG mLRMP 1-230Cit | This paper | C-terminal Citrine; see DNA constructs section of the methods | |
| Recombinant DNA reagent | pcMVBG mLRMP 1-108Cit | This paper | C-terminal Citrine; see DNA constructs section of the methods | |
| Recombinant DNA reagent | pcMVBG mLRMP 110-230Cit | This paper | C-terminal Citrine; see DNA constructs section of the methods | |
| Commercial assay or kit | Q5 Site-Directed Mutagenesis Kit | New England Biolabs | CAT#: E0554S | |
| Commercial assay or kit | In-Fusion HD Cloning | Clontech | Clontech:639647 | |
| Chemical compound, drug | FuGENE 6 | Promega | CAT#: E2691 | |
| Chemical compound, drug | Lipofectamine 2000 | Thermo-Fisher Scientific | CAT#: 11668027 | |
| Software, Algorithm | pClamp and clampfit | Molecular Devices | RRID:SCR_011323 | |
| Software, Algorithm | ImageJ | NIH DOI: https://doi.org/10.1038/nmeth.2089 | RRID:SCR_003070 | |
| Software, Algorithm | Sigmaplot 12.0 | Systat Software Inc | RRID:SCR_003210 | |
| Software, Algorithm | JMP14 | SAS Institute | RRID:SCR_014242 |
Additional files
-
Source data 1
Individual data points, averages, and standard errors of the mean for patch-clamp and FRET data.
- https://cdn.elifesciences.org/articles/92411/elife-92411-data1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/92411/elife-92411-mdarchecklist1-v1.docx






