PfMORC protein regulates chromatin accessibility and transcriptional repression in the human malaria parasite, Plasmodium falciparum
Figures
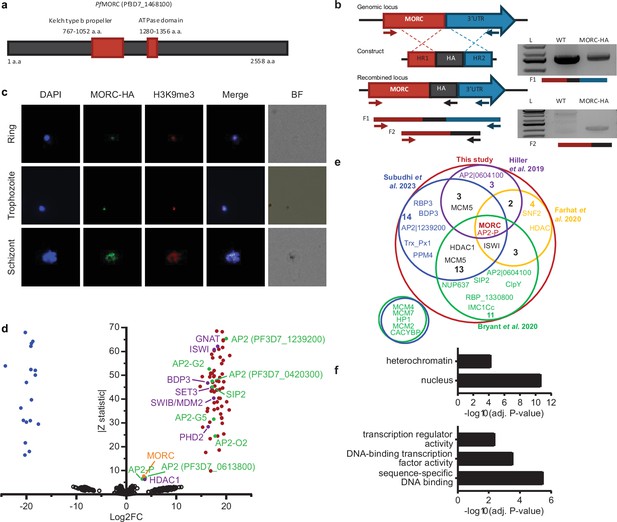
PfMORC-HA is associated with heterochromatin.
(a) Illustration of the PfMORC containing domains including Kelch type b propeller and ATPase domains using InterProScan. (b) Design strategy applied for PfMORC C-terminal HA tagging. PCR amplification of the genomic C-terminus end of Pfmorc region extending towards the 3’UTR (F1) as well as extension from the C-terminus towards the HA flanking sequence (F2) verifies the correct insertion site. NF54 genomic DNA was used as a negative control. (c) Immunofluorescence analysis (IFA) experiment: PfMORC foci (green) expressing co-localization with H3K9me3 marks (red). Cell nuclei are stained with DAPI (blue). BF: brightfield (d) Protein immunoprecipitation: Significance plot representing PfMORC interactome recovered through immunoprecipitation followed by mass spectrometry (IP-MS). Graph lists Microrchidia (MORC) (orange) bindings partners of the highest affinity associated with TF regulation (green) and chromatin remodelers, erasers, and writers (purple). Proteins enriched in the PfMORC-HA samples compared with controls were filtered with log2 FC ≥2 and Z statistic >5. (e) Venn diagram representing overlapping proteins identified among five publications. Values represent the total number of significant proteins identified as overlapping between two subsets. (f) Gene ontology enrichment analysis of the significantly enriched proteins. The top two terms of Cellular Component (top) and top three terms of molecular function (bottom) are represented as -Log10 (adjusted P-value) (Fisher’s exact test with Bonferroni adjustment). Figure 1b was created with BioRender.com.
-
Figure 1—source data 1
Original, unedited gel image for displayed PfMORC-HA results displayed in Figure 1.
- https://cdn.elifesciences.org/articles/92499/elife-92499-fig1-data1-v1.zip
-
Figure 1—source data 2
Original gel image for displayed PfMORC-HA results displayed in Figure 1 with marked ladder and described lanes.
- https://cdn.elifesciences.org/articles/92499/elife-92499-fig1-data2-v1.zip
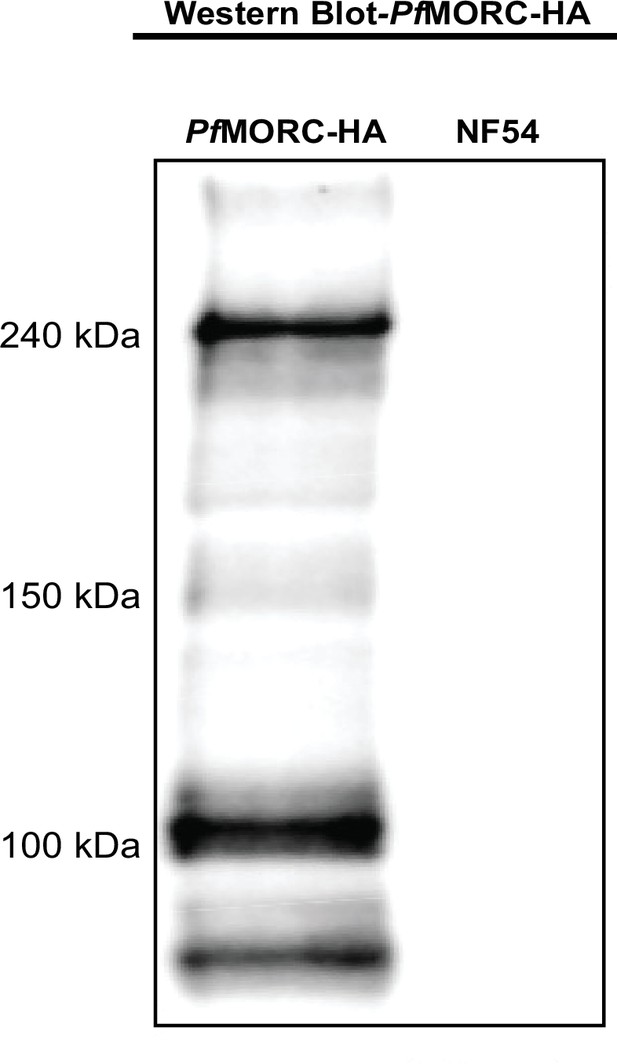
Expression of Microrchidia (MORC) in pDC2-MORC-HA lines.
Western blot image reveals expression of PfMORC-HA or wild-type (WT) protein in samples extracted using anti-HA antibody.
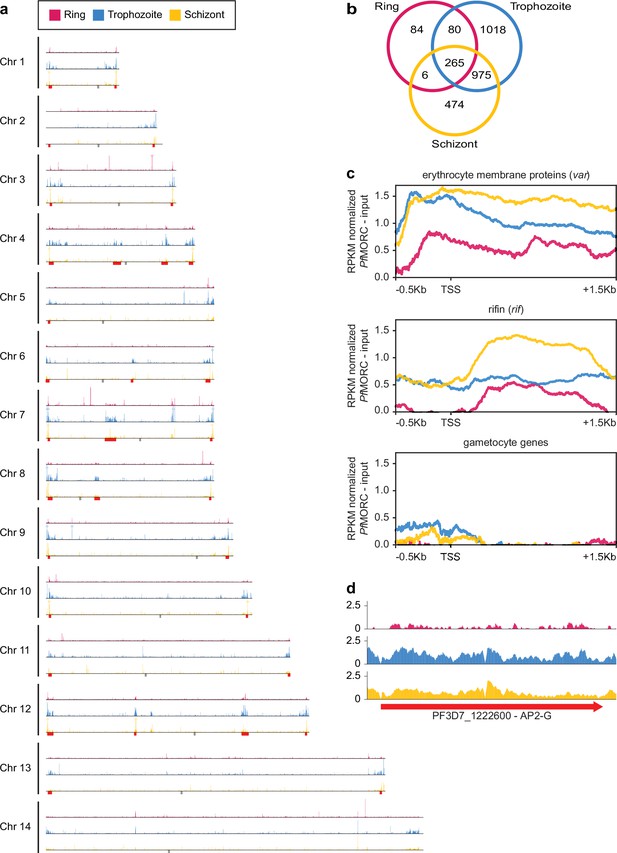
Genome-wide distribution of PfMORC proteins.
(a) Chromosome distribution plots of PfMORC binding show a predisposition for subtelomeric and internal var gene regions (red). Each track is input subtracted, and the per-million read count is normalized before normalizing the track height to allow for direct comparison between stages. Gray boxes indicate the position of centromeres. (b) Overlap of called peaks between time points. (c) Profile plots showing PfMORC coverage from 0.5 kb 5’ of the transcription start site (TSS) to 1.5 kb 3’ of the TSS in the ring, trophozoite, and schizont stages. Each plot includes per-million read count normalized coverage at 1 bp resolution for all genes within the var and rifin gene families as well as gametocyte-specific genes. (d) PfMORC coverage of the gametocyte-specific transcription factor ap2-g.
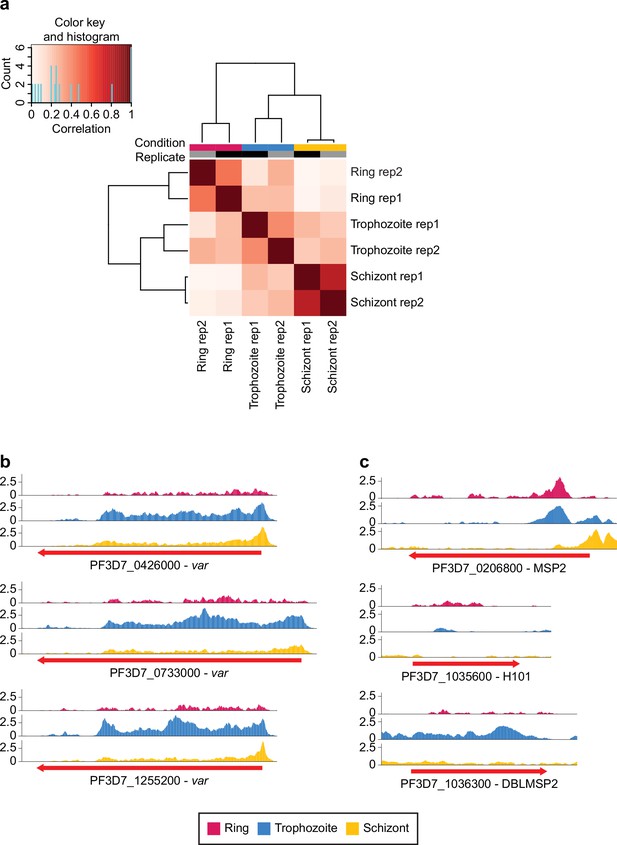
Correlation, peak calling, and gene family-specific coverage of PfMORC Chromatin immunoprecipitation followed by deep sequencing (ChIP-seq).
(a) Heatmap indicating the correlation between time points and replicates. (b) PfMORC coverage of select var genes from 500 bp 5’ of the transcription start site (TSS) to 500 bp 3’ of the coding region. (c) PfMORC coverage of three highly expressed msp genes from 500 bp 5’ of the TSS to 500 bp 3’ of the coding region.
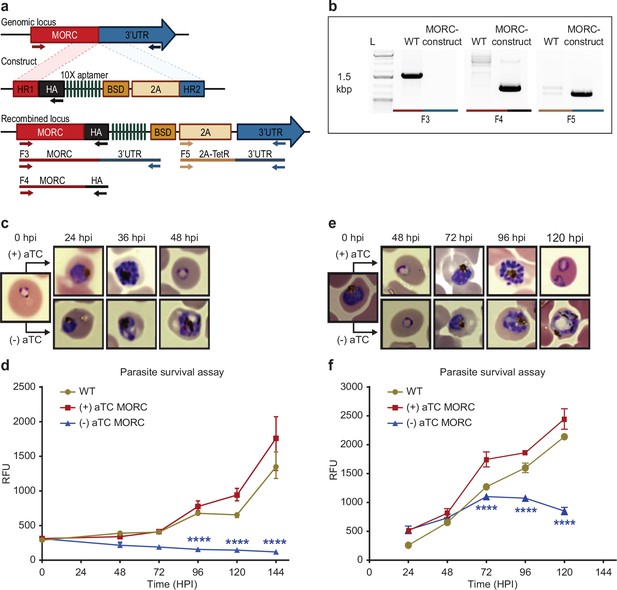
PfMORC is essential for cell survival.
(a) Diagram representation of PfMORC-HA-TetR-DOZI plasmid. (b). PCR amplification is used to verify genomic insertion using primers sets targeting 1.5 kbp of WT Pfmorc genome locus absent in transgenic line (Microrchidia, MORC construct) (F3) as well as verification of HA insertion (F4) and TetR-DOZI system extending along 3’ UTR of the construct (F5). (c) Phenotypic and (d) quantitative analysis of parasite cell progression after aTC withdrawal at the ring stage (0–6 hpi) (2-way ANOVA, n=3, p≤0.0001). (e) Phenotypic and quantitative (f) analysis of parasite cell progression after aTC removal at the trophozoite stage of cell cycle progression (24 hpi) (two-way ANOVA, n=3, p≤0.0001). Figure 3a was created with BioRender.com.
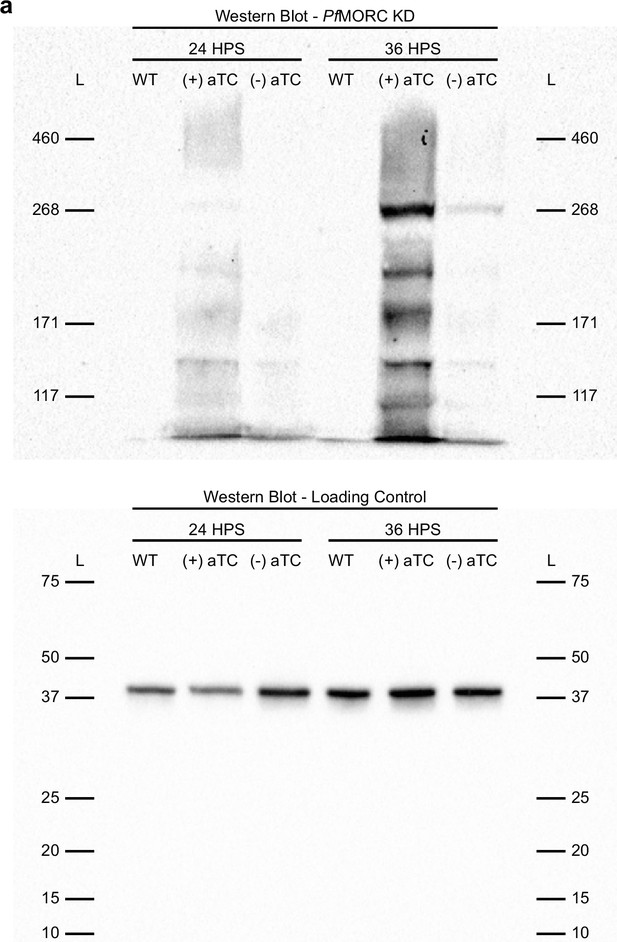
Expression of PfMORC-HA in PfMORC-HA-TetR-DOZI lines.
Western blot image reveals expression of PfMORC-HA (Top) or WT protein (Bottom) in samples extracted at 24 HPS or 36 HPS in +/-aTC conditions using anti-HA antibody. WT samples loaded with exactly 50% of the total protein obtained in PfMORC samples at each condition.
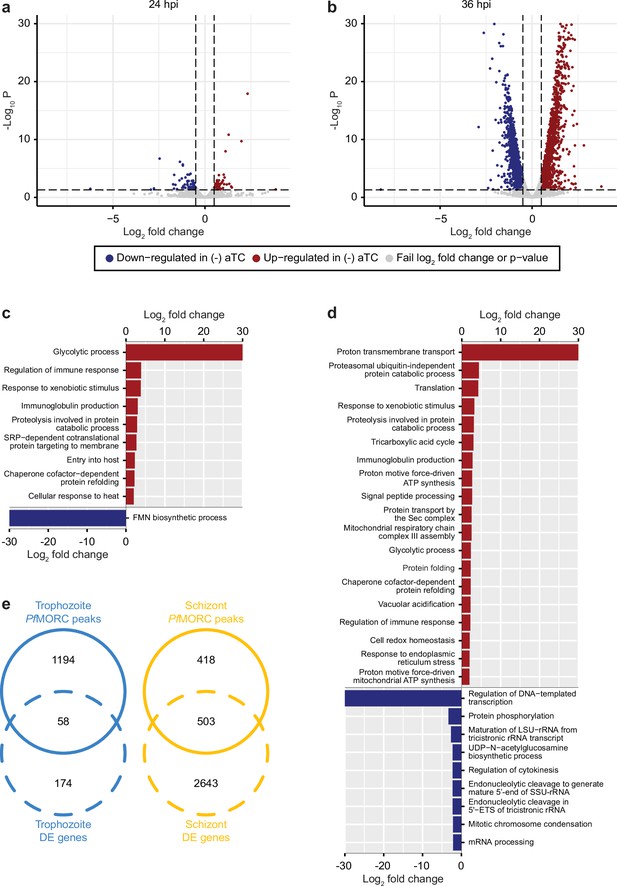
PfMORC knockdown (KD) on parasite transcriptome.
Volcano plots denoting upregulated (red), and downregulated (blue) genes were discovered through differential expression analysis following PfMORC knockdown at (a) 24 hpi (b) 36 hpi. Gene ontology enrichment analysis for upregulated (red) and downregulated (blue) genes at (c) 24 hpi and (d) 36 hpi. (e) Overlap of differentially expressed genes and genes containing significant peaks called by PfMORC Chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) analysis at the trophozoite and schizont stages.
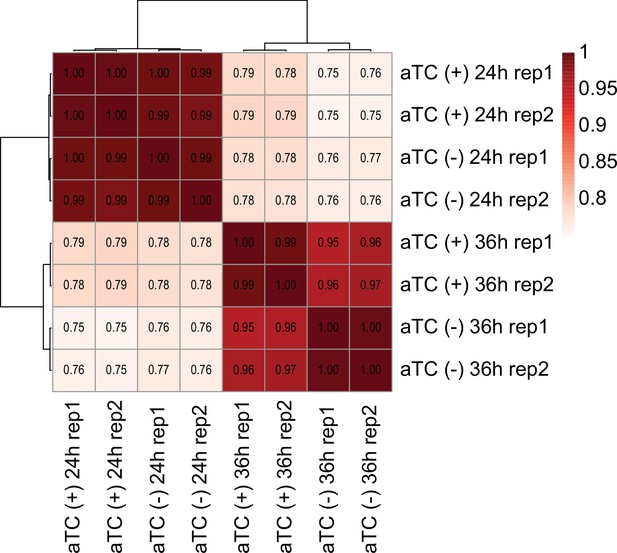
RNA-seq correlation heatmap.
Heatmap indicating the correlation between (+/-) aTC condition, time points, and replicates.
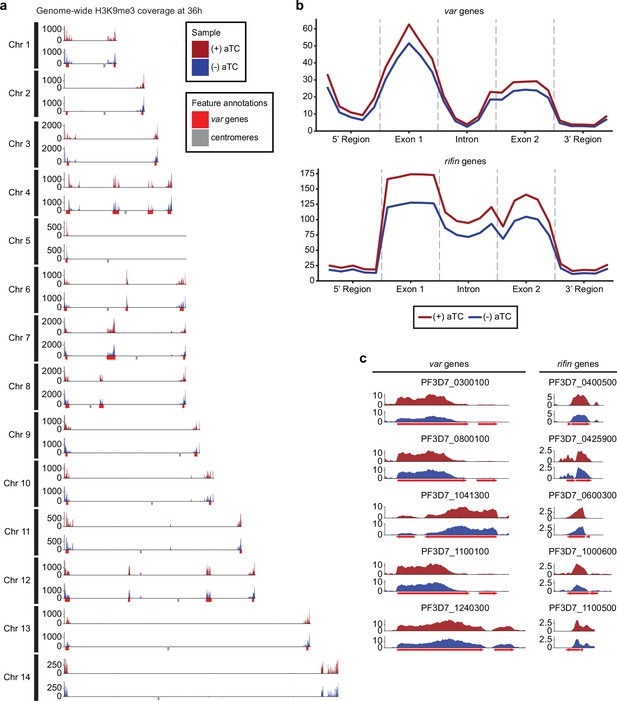
Impact of PfMORC knockdown (KD) on heterochromatin markers.
(a) Genome-wide H3K9me3 coverage of IGG subtracted and per-million normalized (+/-) aTC show similar distribution and concentration within telomeres and antigenic gene clusters highlighted in red. Replicates (n=2) are merged using the mean of the normalized read coverage per base pair. (b) Binned coverage of var and rifin genes from 1 kb upstream of the transcription start site (TSS) to 1 kb downstream of the end. The exons and intron of all genes within these families are split into five equal-sized bins and the 5’ and 3’ regions are binned into five 200 bp bins. Read counts within each bin are per million and bin length is normalized prior to plotting. (c) Coverage of the five most upregulated var and rifin genes as determined by the transcriptomic analysis which shows elevated coverage in (+) aTC cells.
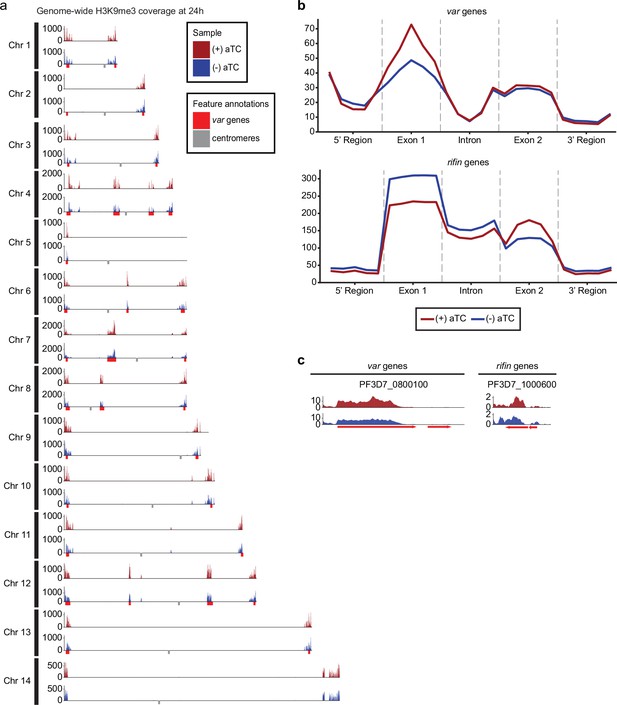
Impact of 24 hpi PfMORC knockdown (KD) on heterochromatin markers.
(a) Genome-wide H3K9me3 coverage of IGG subtracted and per-million normalized (+/-) aTC show similar distribution and concentration within telomeres and antigenic gene clusters highlighted in red. Replicates (n=2) are merged using the mean of the normalized read coverage per base pair. (b) Binned coverage of var and rifin genes from 1 kb upstream of the transcription start site (TSS) to 1 kb downstream of the end. The exons and intron of all genes within these families are split into five equal-sized bins and the 5’ and 3’ regions are binned into five 200 bp bins. Read counts within each bin are per-million and bin length is normalized prior to plotting. (c) Coverage of the five most upregulated var and rifin genes as determined by the transcriptomic analysis which shows elevated coverage in (+) aTC cells.
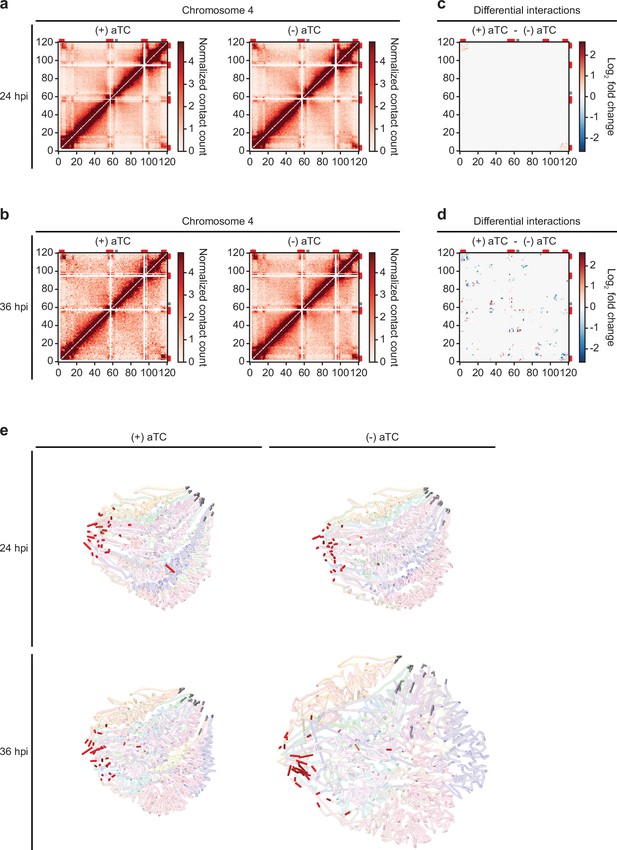
Loss of PfMORC expression correlates with heterochromatin expansion.
Intrachromosomal interaction heatmaps of (+/-) aTC PfMORC for chromosome 4 at (a) 24 hpi and (b) 36 hpi displaying heterochromatin clustering within antigenic (var, rifin, and stevor) gene-dense regions (red). Differential interaction heatmaps highlight changes in chromatin structure following removal of aTC and subsequent PfMORC knockdown at (c) 24 hpi and (d) 36 hpi. (e) Whole-genome 3D models of the chromatin structure at both time points (24 hpi and 36 hpi) and (+/-) aTC.
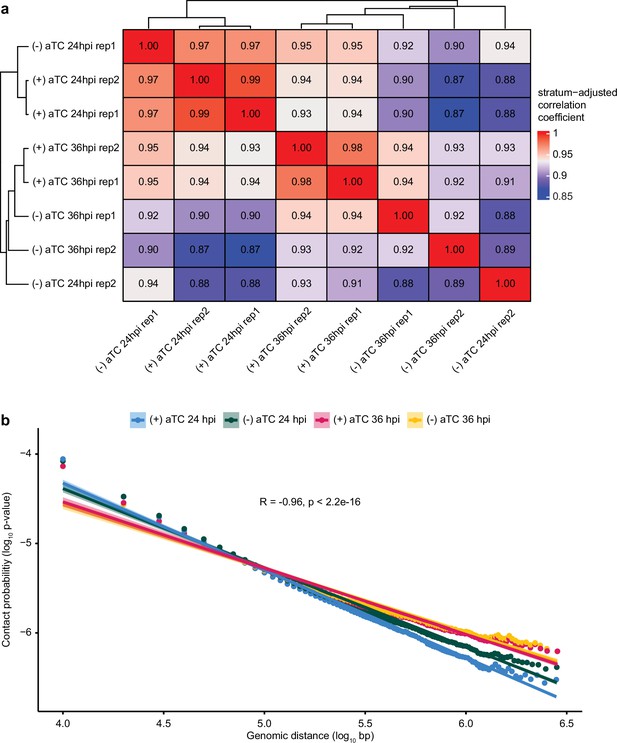
Chromatin conformation capture (Hi-C) correlation analyses.
(a) Stratum-adjusted correlation between (+/-) aTC condition, time points, and replicates. (b) Negative log-linear relationship between contact probability and genomic distance.
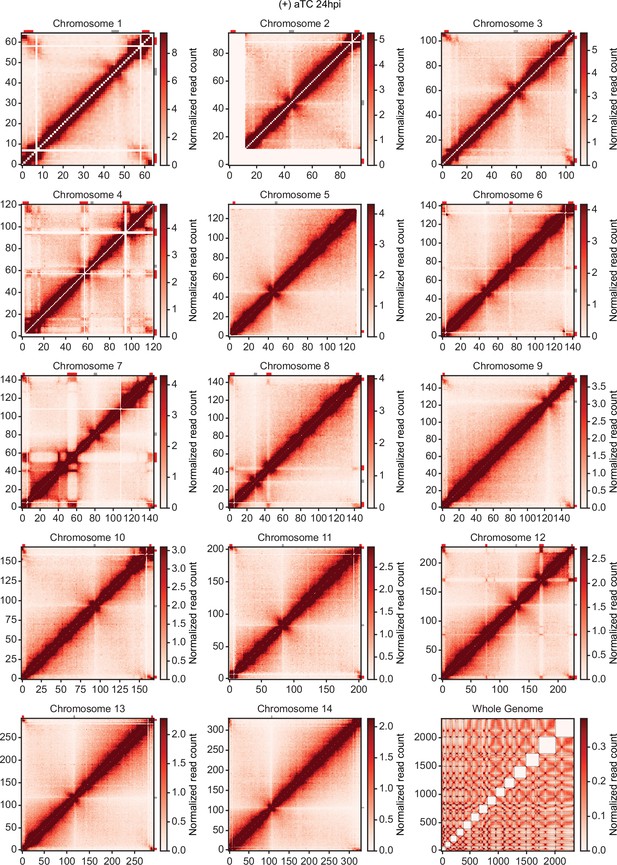
Chromatin conformation capture (Hi-C) interaction heatmaps binned at 10 kb resolution.
Each sample includes intrachromosomal contact count heatmaps for all 14 chromosomes within the P. falciparum genome and a genome-wide interchromosomal interaction heatmap at 24 hpi (+) aTC. Data for each heatmap is ICED and per-million read count normalized, and the maximum y-value for each heatmap is set to the highest value within the given chromosome for all samples. Antigenic gene-containing regions (red) and centromeres (gray) are highlighted.
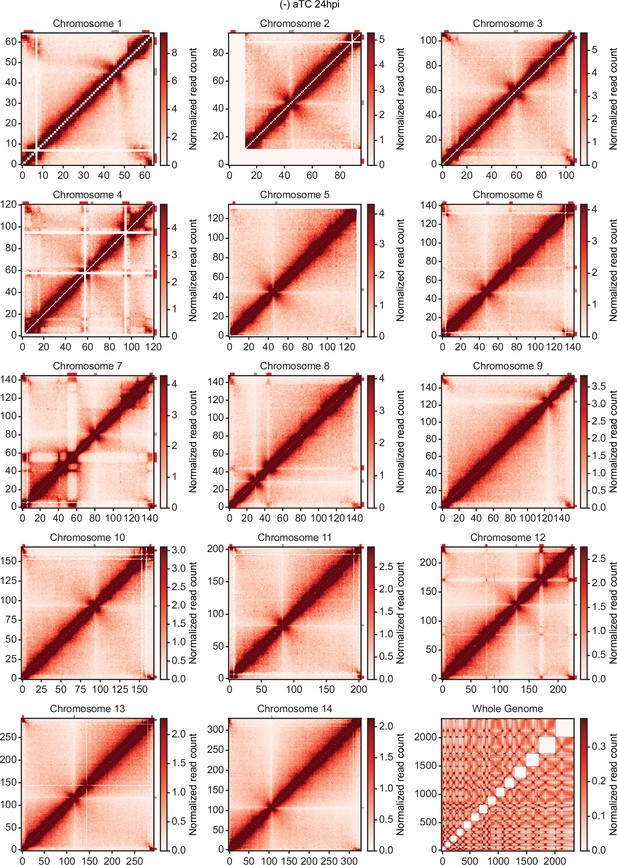
Chromatin conformation capture (Hi-C) interaction heatmaps binned at 10 kb resolution.
Each sample includes intrachromosomal contact count heatmaps for all 14 chromosomes within the P. falciparum genome and a genome-wide interchromosomal interaction heatmap at 24 hpi (-) aTC. Data for each heatmap is ICED and per-million read count normalized, and the maximum y-value for each heatmap is set to the highest value within the given chromosome for all samples. Antigenic gene-containing regions (red) and centromeres (gray) are highlighted.
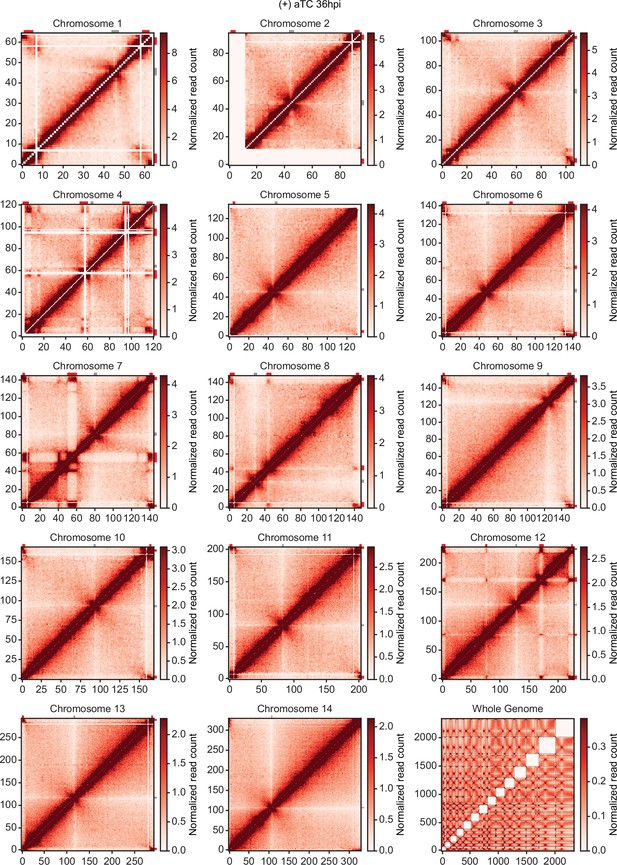
Chromatin conformation capture (Hi-C) interaction heatmaps binned at 10 kb resolution.
Each sample includes intrachromosomal contact count heatmaps for all 14 chromosomes within the P. falciparum genome and a genome-wide interchromosomal interaction heatmap at 36 hpi (+) aTC. Data for each heatmap is ICED and per-million read count normalized, and the maximum y-value for each heatmap is set to the highest value within the given chromosome for all samples. Antigenic gene-containing regions (red) and centromeres (gray) are highlighted.
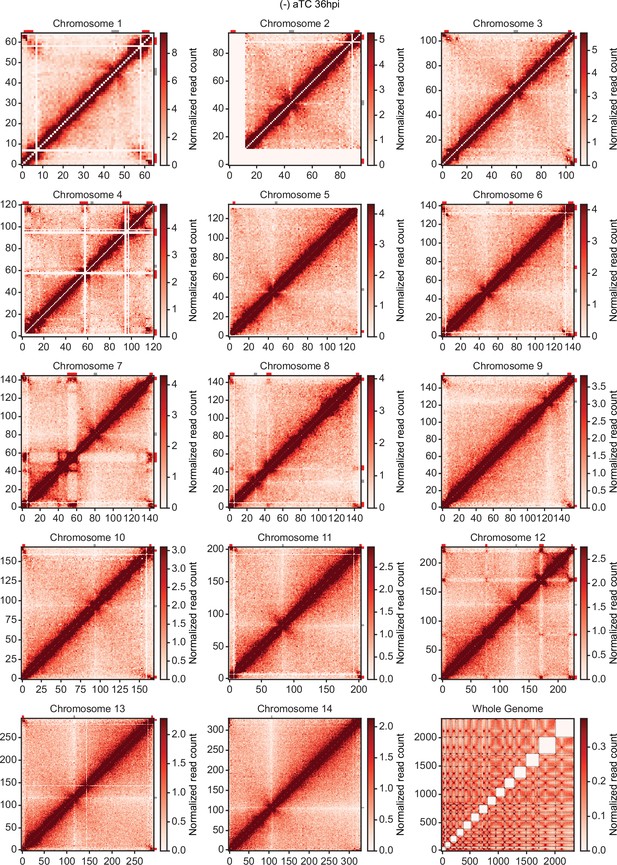
Chromatin conformation capture (Hi-C) interaction heatmaps binned at 10 kb resolution.
Each sample includes intrachromosomal contact count heatmaps for all 14 chromosomes within the P. falciparum genome and a genome-wide interchromosomal interaction heatmap at 36 hpi (-) aTC. Data for each heatmap is ICED and per-million read count normalized, and the maximum y-value for each heatmap is set to the highest value within the given chromosome for all samples. Antigenic gene-containing regions (red) and centromeres (gray) are highlighted.
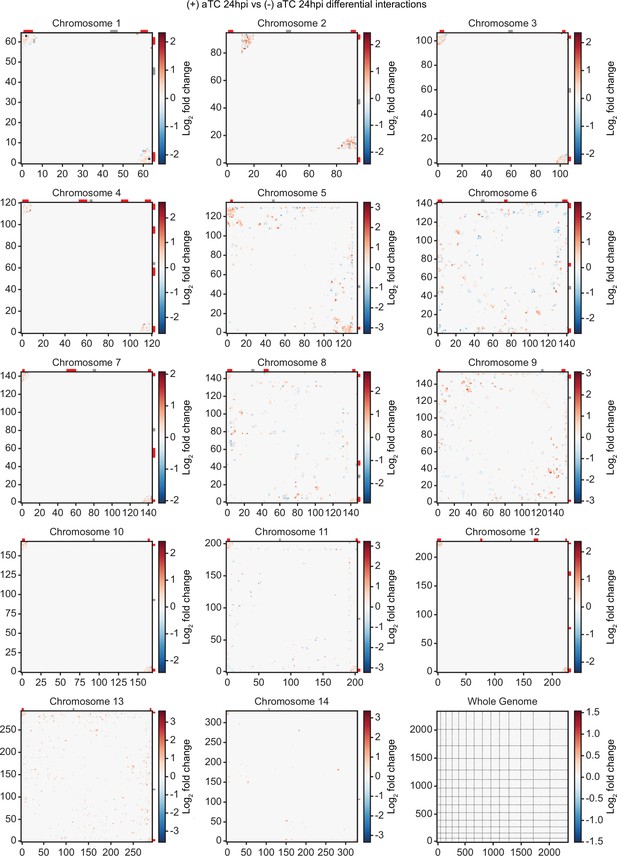
Differential chromatin conformation capture (Hi-C) interaction heatmaps binned at 10 kb resolution.
Heatmaps generated from differential interaction matrices, identifying regions with a positive (red) or negative (blue) log2 fold change between (-) aTC and (+) aTC at 24 hpi. Data for each heatmap is ICED and per-million read count normalized, and the maximum y-value for each heatmap is set to the highest value within the given chromosome for all samples. Antigenic gene-containing regions (red) and centromeres (gray) are highlighted.
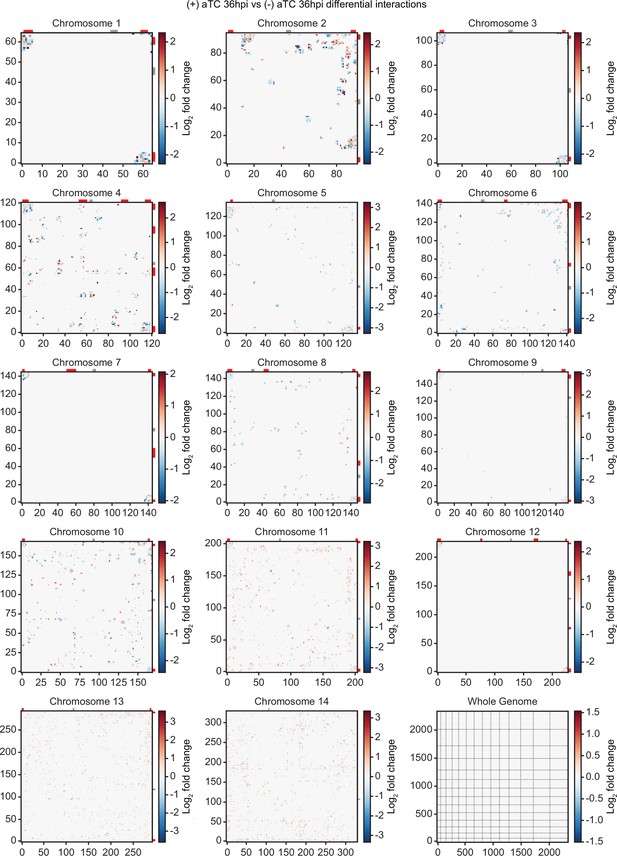
Differential chromatin conformation capture (Hi-C) interaction heatmaps binned at 10 kb resolution.
Heatmaps generated from differential interaction matrices, identifying regions with a positive (red) or negative (blue) log2 fold change between (-) aTC and (+) aTC at 36 hpi. Data for each heatmap is ICED and per-million read count normalized, and the maximum y-value for each heatmap is set to the highest value within the given chromosome for all samples. Antigenic gene-containing regions (red) and centromeres (gray) are highlighted.
Additional files
-
Supplementary file 1
PfMORC plasmid constructs to generate transgenic lines.
All constructs were performed through listed primers, restriction sites, and gRNAs.
- https://cdn.elifesciences.org/articles/92499/elife-92499-supp1-v1.xlsx
-
Supplementary file 2
Mapped whole genome sequencing results of PfMORC transfectants.
- https://cdn.elifesciences.org/articles/92499/elife-92499-supp2-v1.xlsx
-
Supplementary file 3
MORC-HA associated proteins identified via MudPIT analysis.
(a) Proteins identified by MudPIT analysis after MORC-HA immunoprecipitation. PfMORC (PF3D7_1468100) is highlighted in yellow and significantly purified proteins are in red. The filters used are QPROT log2 fold change >2 and Z statistic >5. Detailed protein list is provided in an additional sheet. (b) Raw data showing proteins identified by MudPIT analysis after PfMORC-HA immunoprecipitation.
- https://cdn.elifesciences.org/articles/92499/elife-92499-supp3-v1.xlsx
-
Supplementary file 4
Peak calling the result of PfMORC Chromatin immunoprecipitation followed by deep sequencing (ChIP-seq).
Results were obtained at (a) ring stages, (b) trophozoite stages, and (c) schizont stages of cell progression.
- https://cdn.elifesciences.org/articles/92499/elife-92499-supp4-v1.xlsx
-
Supplementary file 5
Parasite Survival Assay illustrating the effect of PfMORC down-regulation on parasites at the ring stage (top) or trophozoite stage (bottom) cell cycle.
Experiments were conducted in triplicates with parasitemia quantified via relative fluorescence units (RFUs) obtained by SYBR green assay for each time point. Significance derived through two-way ANOVA.
- https://cdn.elifesciences.org/articles/92499/elife-92499-supp5-v1.xlsx
-
Supplementary file 6
Effect of PfMORC KD on transcriptomic profile at 24 and 36 hr.
(a) RNA-seq read counts of PfMORC at 24 and 36 hr. DEseq2 analysis of differentially expressed genes of PfMORC at (b) 24 hr and (c) 36 hr.
- https://cdn.elifesciences.org/articles/92499/elife-92499-supp6-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/92499/elife-92499-mdarchecklist1-v1.docx





