Bacteria are a major determinant of Orsay virus transmission and infection in Caenorhabditis elegans
Figures
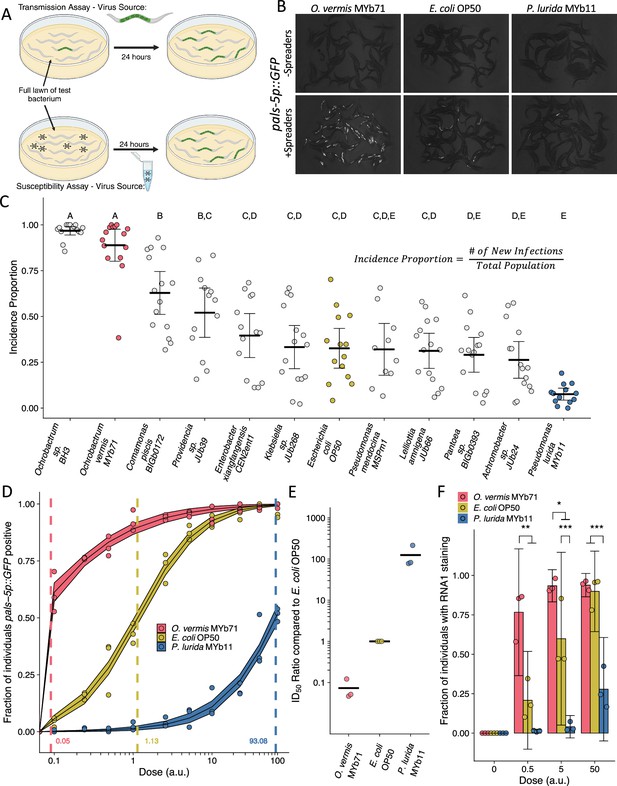
Ochrobactrum species and P. lurida MYb11 divergently modulate Orsay virus transmission and infection rates.
(A) Schematic representation of the transmission and susceptibility assays: Transmission can be assessed by combining infected spreader individuals (green nematode), uninfected reporter individuals (white nematodes), and various bacteria. Susceptibility can be assessed by combining uninfected reporter individuals with exogenous Orsay virus, and various bacteria. 24 hr later, infection is assessed. (B) Representative images of pals-5p::GFP expression among individuals exposed to spreaders or no spreaders in the presence of O. vermis MYb71, E. coli OP50, or P. lurida MYb11. (C) Incidence proportion, calculated as indicated, of Orsay virus transmission quantified on different bacteria from the environment of C. elegans. Data shown are from three experiments combined, each dot represents the incidence proportion from a single plate, and letters denote statistical significance. Treatments with the same letter are not significantly different from one another. (n=19,694 in total and n>42 for all dots). (D) Dose-response curves of C. elegans to exogenous Orsay virus. The dashed line indicates the calculated dose at which 50% of the population was infected, the ID50, the exact value which is given above the x-axis for each bacterium. The solid curves represent the 95% confidence interval of the modeled log-logistic function while in the presence of each bacterium. Data are from a single representative experiment and replicates can be found in Figure 1—figure supplement 3. Dots represent individual plates. (a.u., arbitrary units, see Materials and methods) (n=8334 in total and n>32 for all dots). (E) Ratios of the ID50 calculated on the indicated bacteria as compared to the ID50 calculated on E. coli OP50 from three separate experiments as in (D) and Figure 1—figure supplement 3. (F) The fraction of individuals with staining as assessed by fluorescence in situ hybridization targeting the RNA1 segment of Orsay virus. Data shown are from three experiments combined, each dot represents three pooled technical replicate plates from the susceptibility assays in (D) and Figure 1—figure supplement 3 (n=3446 in total and n>61 for all dots). For all plots the black bar is the mean and error bars are the 95% confidence interval (C.I.). p-Values determined using one-way ANOVA followed by Tukey’s honest significant difference (HSD) test (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001).
© 2024, BioRender Inc. Figure 1A was created using BioRender, and is published under a CC BY-NC-ND 4.0. Further reproductions must adhere to the terms of this license
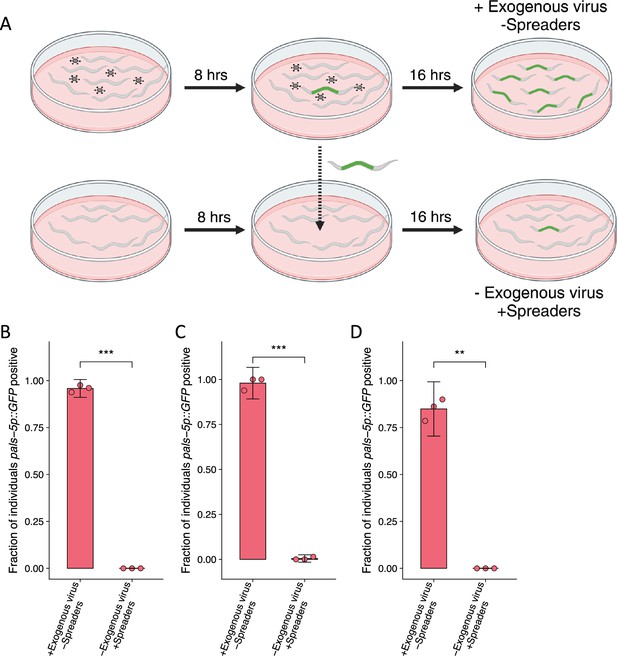
Transmission does not occur from individuals infected during the course of a transmission or susceptibility assay.
(A–D) Individuals were exposed to 0 a.u. or 5 a.u. exogenous Orsay virus in the presence of O. vermis MYb71. 8 hr post infection five infected individuals from the 5 a.u. plate were transferred to the 0 a.u. plate to assess whether transmission from these individuals would occur. Both plates were scored 16 hr after the transfer. Data for each plot are from a single representative experiment, bars represent mean, dots represent individual plates ((B) n=669 in total and n>83 for all dots, (C) n=377 in total and n>52 for all dots, (D) n=273 in total and n>33 for all dots). Error bars are the 95% confidence interval (C.I.). p-Value determined by Student’s t-test (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001) (a.u., arbitrary units).
© 2024, BioRender Inc. Figure 1—figure supplement 1A was created using BioRender, and is published under a CC BY-NC-ND 4.0. Further reproductions must adhere to the terms of this license

The range of all ID50 values calculated for each batch of Orsay virus.
(A) Dose-response curves as shown in Figure 1D and Figure 1—figure supplement 3 were measured for ZD2611(jyIs8[pals- 5p::GFP; myo-2p::mCherry];glp-4(bn2ts)) animals exposed to E. coli OP50 for each batch of Orsay virus used. The dose of virus required to infect 50% of the population, the ID50, was calculated and plotted. Bar represents the mean and individual dots represent the ID50 from a separate dose-response curve. Error bars are the 95% confidence interval (C.I.).
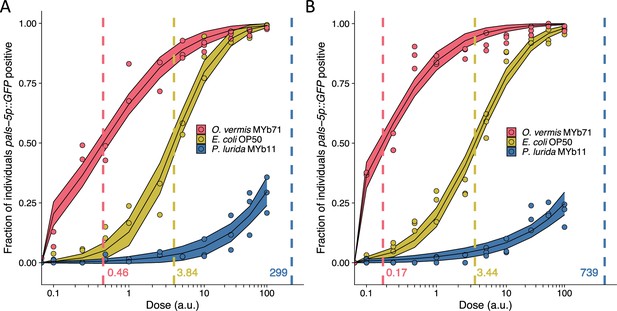
Replicate dose-response curves shown in Figure 1D.
(A–B) Dose-response curves of C. elegans to exogenous Orsay virus. The dashed line indicates the calculated dose at which 50% of the population was infected, the ID50, the exact value which is given above the x-axis for each bacterium. The solid curves represent the 95% confidence interval of the modeled log-logistic function while in the presence of each bacterium. Data are from a single representative experiment, dots represent individual plates ((A) n=11,350 in total and n>38 for all dots, (B) n=7753 in total and n>24 for all dots) (a.u., arbitrary units).
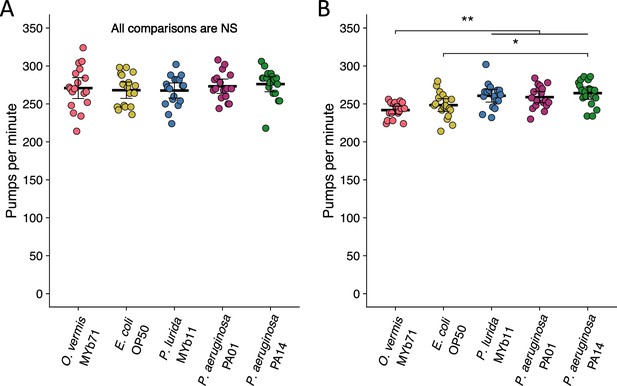
Pharyngeal pumping rates are unaltered at 6 hr and change only slightly by 24 hr after exposure to novel bacteria.
(A–B) Pharyngeal pumping rate of ZD2611 animals exposed to the indicated bacteria for (A) 6 hr or (B) 24 hr. Pumping rate was assessed via slow-motion video capture of animals feeding (Materials and methods). Data are from three separate experiments and each dot represents a single animal. The black bar is the mean and error bars are the 95% confidence interval (C.I.). p-Values determined using one-way ANOVA followed by Tukey’s honest significant difference (HSD) test (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001).
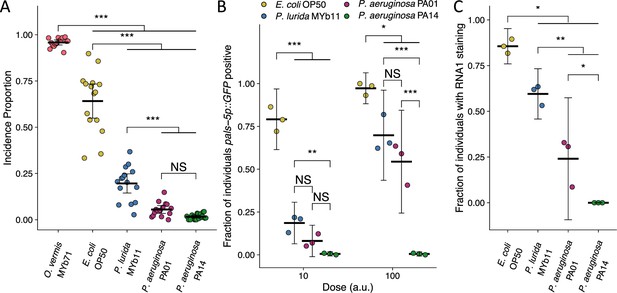
P. aeruginosa attenuates Orsay virus transmission and infection rates.
(A) Incidence proportion of Orsay virus transmission quantified on different bacteria from C. elegans’ environment and P. aeruginosa PA01 and P. aeruginosa PA14. Data shown are from three experiments combined, each dot represents the incidence proportion from a single plate (n=9214 in total and n>54 for all dots). (B) The fraction of individuals that became pals-5p::GFP positive following exposure to two doses of exogenous Orsay virus. Data are from a single representative experiment and replicates can be found in Figure 2—figure supplement 1. Dots represent individual plates (n=6539 in total and n>56 for all dots). (C) The fraction of individuals with staining following exposure to 100 a.u. Orsay virus as assessed by fluorescence in situ hybridization targeting the RNA1 segment of the Orsay virus genome. Data shown are from three experiments combined, each dot represents three pooled technical replicate plates from (B) and Figure 2—figure supplement 1 (n=2743 in total and n>116 for all dots). For all plots the black bar is the mean and error bars are the 95% confidence interval (C.I.). p-Values determined using one-way ANOVA followed by Tukey’s honest significant difference (HSD) test (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001) (a.u., arbitrary units).
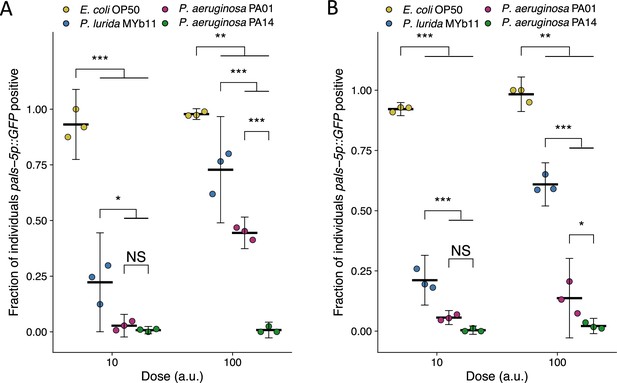
Replicate susceptibility assays shown in Figure 2B.
(A–B) The fraction of individuals that became pasl-5p::GFP positive following exposure to two doses of exogenous Orsay virus. Data are from a single representative experiment and dots represent individual plates ((A) n=2729 total and n>60 for all dots, (B) n=2051 total and n>48 for all dots). The black bar is the mean and error bars are the 95% confidence interval (C.I.). p-Values determined using one-way ANOVA followed by Tukey’s honest significant difference (HSD) test (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001).
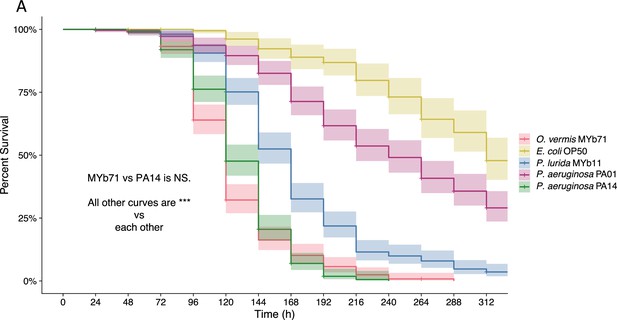
O. vermis MYb71, P. aeruginosa PA14, P. lurida MYb11, and P. aeruginosa PA01 reduce lifespan compared to E. coli OP50.
(A) Survival of ZD2611 animals following exposure to full lawns of the indicated bacteria on 5-fluorodeoxyuridine (FUdR) containing plates at 20°C. Hashes indicate censored animals. Lines represent the Kaplan-Meier curve, while shaded area represents the 95% confidence interval. Curve and confidence interval were calculated from three separate experiments with three replicate plates for each bacteria with approximately 30 individuals per plate. Statistics were calculated with the log-rank test implemented in the survdiff function in the survival package in R followed by the Bonferroni correction (***p<0.001).
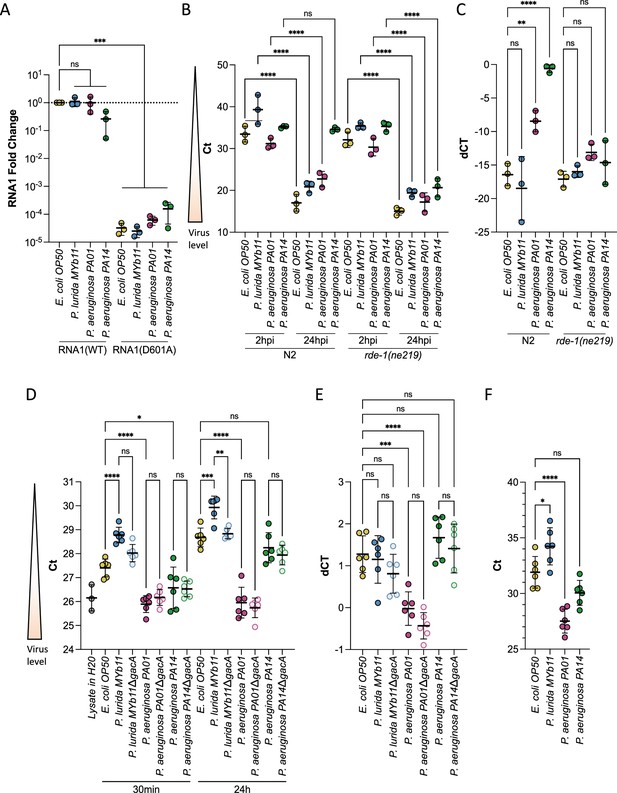
P. aeruginosa and P. lurida MYb11 do not eliminate Orsay virus replication, rapidly degrade Orsay virus, or eliminate Orsay virus shedding by infected spreader animals.
(A) Animals carrying either a wild-type (RNA1(WT)) or replication defective (RNA1(D601A)) transgenic viral RNA replicon system (Jiang et al., 2017) were used to assess Orsay virus replication efficiency independently of virus entry. p-Values determined using one-way ANOVA followed by Dunnett’s test. (B) Orsay virus RNA1 levels of N2 and rde-1(ne219) animals exposed to the indicated bacterium and exogenous Orsay virus 2 hr and 24 hr post infection (hpi). p-Values were determined using Welch’s t-test. For each plot, each dot represents five pooled technical replicates, the black bar is the mean, error bars are the standard deviation, RNA1 levels were quantified by qPCR, and the data shown are for three independent experiments. (C) Delta Ct comparing the 24 hr and 2 hr timepoints from (B). (D) Stability of Orsay virus in the presence of lawns of the indicated bacteria for the indicated time at 20°C. (E) Delta Ct comparing the 24 hr and 30 min timepoints from (D). (F) Orsay virus RNA1 levels recovered from the indicated lawns after infected ZD2610(rde- 1(ne219);jyIs8[pals-5p::GFP;myo-2p::mCherry];glp-4(bn2ts)) spreaders were allowed to shed for 24 hr mimicking a transmission assay (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001).
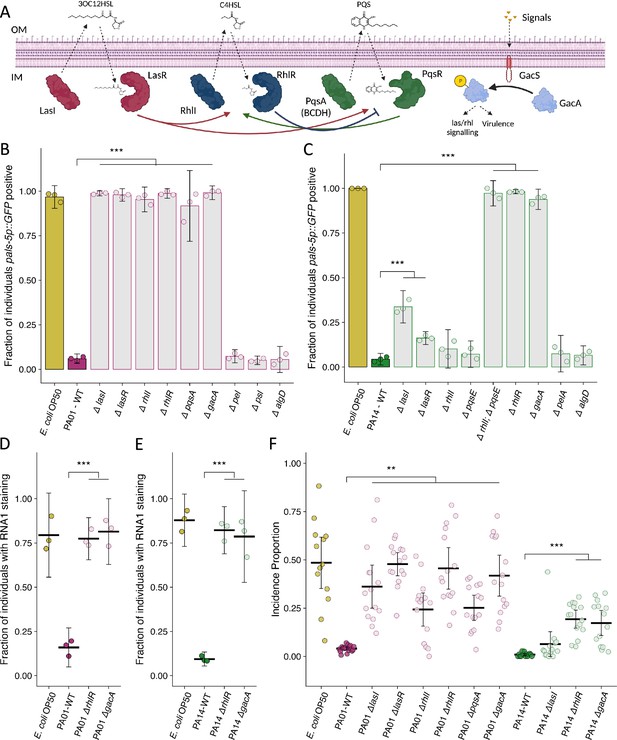
Mutation of P. aeruginosa quorum sensing regulators and the two-component response regulator gacA suppresses the attenuation of Orsay virus infection.
(A) Diagram demonstrating the three quorum sensing systems in P. aeruginosa. Each system encodes the enzyme(s) (LasI, RhlI, PqsA(BCDH)) to produce an autoinducer (3OC12HSL – 3-oxo-C12-homoserine lactone, C4HSL – butanoyl homoserine lactone, PQS – Pseudomonas quinolone signal) that is recognized by its cognate receptor (LasR, RhlR, PqsR) that influences gene transcription and the activity of the other quorum sensing systems. An additional level of regulation stems from the two-component gacS/gacA system. External signals are recognized by the histidine kinase GacS, which phosphorylates the response regulator GacA that indirectly influences quorum sensing processes. IM is inner membrane, OM is outer membrane. Arrows represent crosstalk between the various system. Adapted from Rutherford and Bassler, 2012 and Song et al., 2023. (B–C) The fraction of individuals that became pals-5p::GFP positive following exposure to exogenous Orsay virus in the presence of the indicated bacterium. Data are from a single representative experiment, bars represent mean, dots represent individual plates. (B) Wild-type P. aeruginosa PA01 compared to mutant P. aeruginosa PA01 strains using 10 a.u. of Orsay virus. Replicates can be found in Figure 4—figure supplement 1 (n=4480 in total and n>76 for all dots). (C) Wild-type P. aeruginosa PA14 compared to mutant P. aeruginosa PA14 strains using 100 a.u. of Orsay virus. Replicates can be found in Figure 4—figure supplement 2 (n=3320 in total and n>23 for all dots). (D–E) The fraction of individuals with staining following exposure to (D) 10 a.u. or (E) 100 a.u. Orsay virus as assessed by fluorescence in situ hybridization targeting the RNA1 segment of the Orsay virus genome. Data shown are from three experiments combined, each dot represents three pooled technical replicates from a single experiment. ((D) n=1632 in total and n>61 for all dots, (E) n=2000 in total and n>60 for all dots). (F) Incidence proportion of Orsay virus transmission in the presence of P. aeruginosa wild-type versus select P. aeruginosa mutants. Data are from three experiments, bars represent mean, dots represent individual plates (n=20,452 in total and n>37 for all dots). For all plots error bars represent 95% C.I. For B–E, p-values were determined using one-way ANOVA followed by Dunnett’s test (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001) (a.u., arbitrary units).
© 2024, BioRender Inc. Figure 4A was created using BioRender, and is published under a CC BY-NC-ND 4.0. Further reproductions must adhere to the terms of this license
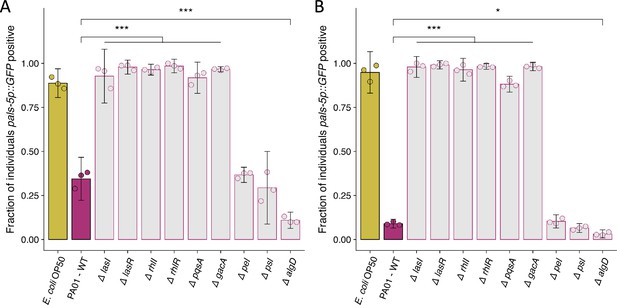
Replicate susceptibility assays shown in Figure 4B.
(A–B) The fraction of individuals that became pals-5p::GFP positive following exposure to 10 a.u. of exogenous Orsay virus. Data are from a single representative experiment and dots represent individual plates ((A) n=2627 in total and n>43 for all dots, (B) n=4068 in total and n>75 for all dots). The black bar is the mean and error bars are the 95% confidence interval (C.I.). p-Values determined using one-way ANOVA followed by Dunnett’s test (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001) (a.u., arbitrary units).
© 2024, BioRender Inc. Figure 4—figure supplement 1A was created using BioRender, and is published under a CC BY-NC-ND 4.0. Further reproductions must adhere to the terms of this license
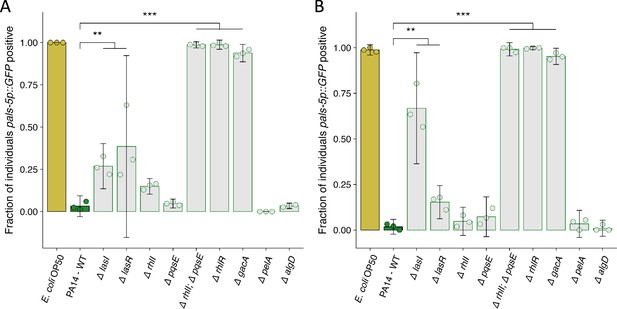
Replicate susceptibility assays shown in Figure 4C.
(A–B) The fraction of individuals that became pals-5p::GFP positive following exposure to 100 a.u. of exogenous Orsay virus. Data are from a single representative experiment and dots represent individual plates ((A) n=3672 in total and n>37 for all dots, (B) n=2728 in total and n>23 for all dots). The black bar is the mean and error bars are the 95% confidence interval (C.I.). p-Values determined using one-way ANOVA followed by Dunnett’s test (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001) (a.u., arbitrary units).
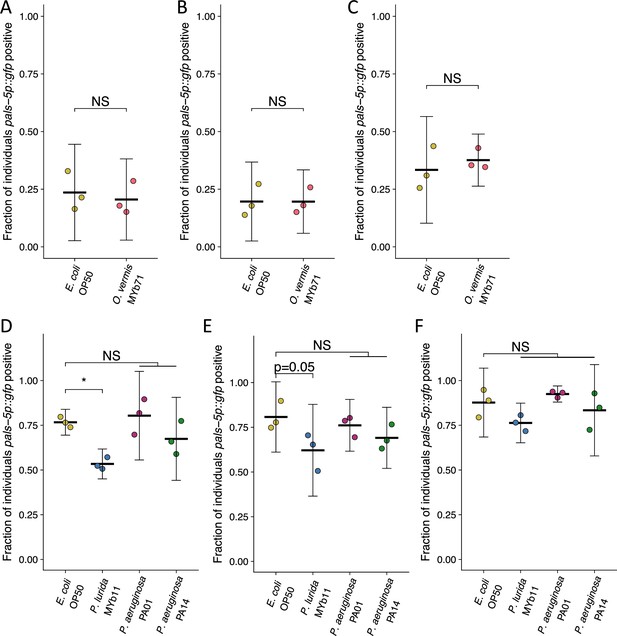
Supernatant from stationary phase cultures of different bacteria does not reliably attenuate or enhance Orsay virus infection in the presence of E. coli OP50.
(A–F) The fraction of individuals that became GFP positive following exposure to (A–C) 0.5 a.u. or (D–F) 5 a.u. of exogenous Orsay virus in the presence of E. coli OP50 saturated stationary phase supernatant from the indicated bacteria. Data from each plot are from a single experiment. The black bar is the mean, error bars are the 95% confidence interval (C.I.), and dots represent individual plates ((A) n=384 in total and n>35 for all dots, (B) n=525 in total and n>55 for all dots, (C) n=881 in total and n>103 for all dots, (D) n=843 in total and n>46 for all dots, (E) n=1040 in total and n>39 for all dots, (F) n=1603 in total and n>86 for all dots). (A–C) p-Values determined using Student’s t-test. (D–E) p-Values determined using a one-way ANOVA followed by Dunnett’s test (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001) (a.u., arbitrary units).
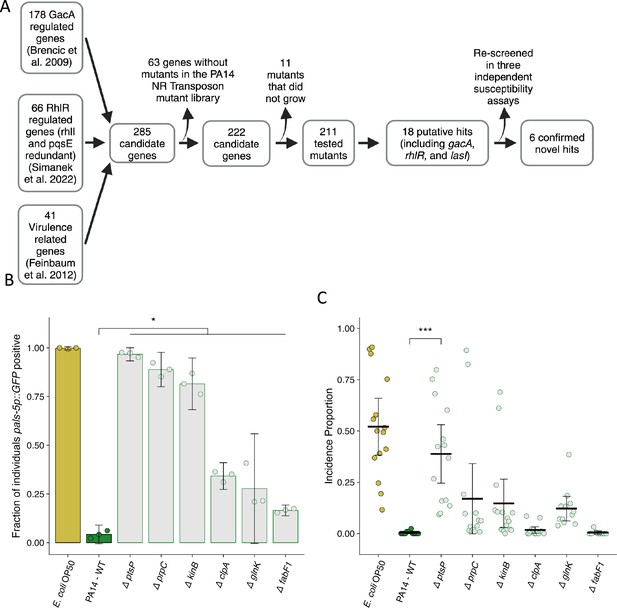
Mutation of P. aeruginosa PA14 genes related to virulence suppresses the attenuation of Orsay virus infection.
(A) Diagram describing the design and results of the screen to identify suppressors of P. aeruginosa PA14 Orsay virus attenuation. (B) The fraction of individuals that became pals-5p::GFP positive following exposure to 100 a.u. of exogenous Orsay virus. Data are from a single representative experiment. Replicates can be found in Figure 5—figure supplement 1 (n=3779 in total and n>55 for all dots). (C) Incidence proportion of Orsay virus transmission while individuals are present on P. aeruginosa PA14 wild-type compared to P. aeruginosa PA14 mutants. Data are from three experiments combined and dots represent individual plates (n=11,279 in total and n>35 for all dots). E. coli OP50 control shared with Figure 6F. For all plots the black bar is the mean and error bars are the 95% confidence interval (C.I.). p-Values determined using one-way ANOVA followed by Dunnett’s test (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001) (a.u., arbitrary units).
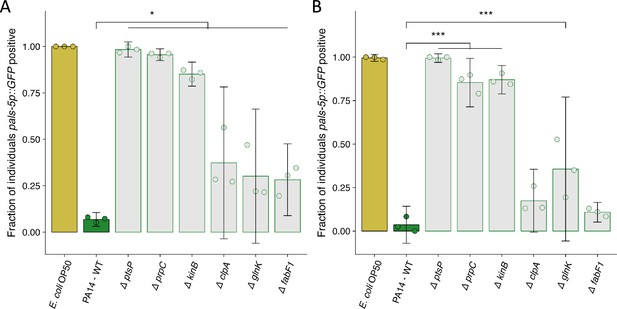
Replicate susceptibility assays shown in Figure 5B.
(A–B) The fraction of individuals that became pals-5p::GFP positive following exposure to 100 a.u. of exogenous Orsay virus. Data are from a single representative experiment and dots represent individual plates ((A) n=1957 in total and n>33 for all dots, (B) n=1730 in total and n>24 for all dots). The black bar is the mean and error bars are the 95% confidence interval (C.I.). p-Values determined using one-way ANOVA followed by Dunnett’s test (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001) (a.u., arbitrary units).
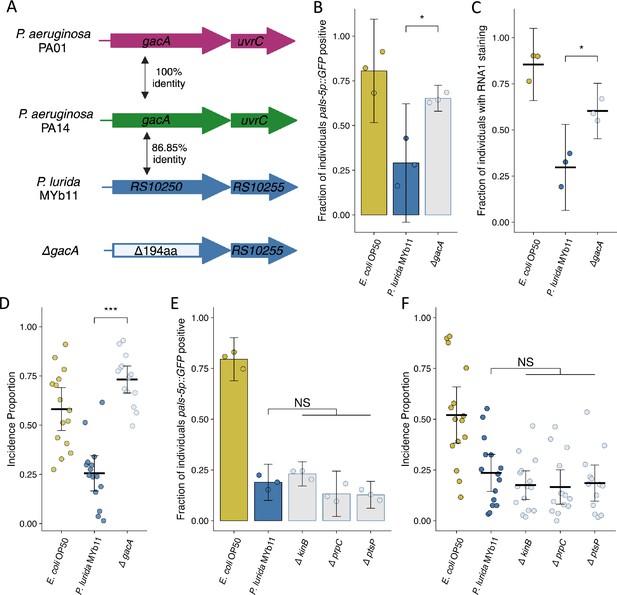
Mutation of P. lurida MYb11 gacA suppresses the attenuation of Orsay virus infection.
(A) Diagram depicting P. aeruginosa PA01, P. aeruginosa PA14, and P. lurida MYb11 gacA as well as the P. lurida MYb11 gacA deletion mutant. Percent identity of the encoded protein was assessed using Clustal Omega. The gacA deletion removed 194 amino acids from the protein leaving 10 amino acids from both the N and C termini. (B, E) The fraction of individuals that became pals-5p::GFP positive following exposure to 10 a.u. exogenous Orsay virus in the presence of wild-type P. lurida MYb11 compared to the indicated mutants. Data are from a single representative experiment. Replicates can be found in Figure 6—figure supplement 1. ((B) n=933 in total and n>46 for all dots, (E) n=1725 in total and n>78 for all dots). Bars represent mean and dots represent individual plates. (C) The fraction of individuals with staining following exposure to 10 a.u. Orsay virus as assessed by fluorescence in situ hybridization targeting the RNA1 segment of the Orsay virus genome. Data shown are from three experiments combined, each dot represents three pooled technical replicates from (B) and Figure 6—figure supplement 1 (n=1272 in total and n>61 for all dots). (D, F) Incidence proportion of Orsay virus transmission in the presence of wild-type P. lurida MYb11 versus the indicated mutants. Data are from three experiments combined, bars represent mean, dots represent individual plates ((D) n=4534 in total and n>42 for all dots, (F) n=8185 in total and n>58 for all dots). E. coli OP50 control of Figure 6F shared with Figure 5C. For all plots error bars represent 95% confidence interval (C.I.). (B–D) p-Values were determined using Student’s t-test. (E–F) p-Values determined using one-way ANOVA followed by Dunnett’s test (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001) (a.u., arbitrary units).
© 2024, BioRender Inc. Figure 6A was created using BioRender, and is published under a CC BY-NC-ND 4.0. Further reproductions must adhere to the terms of this license
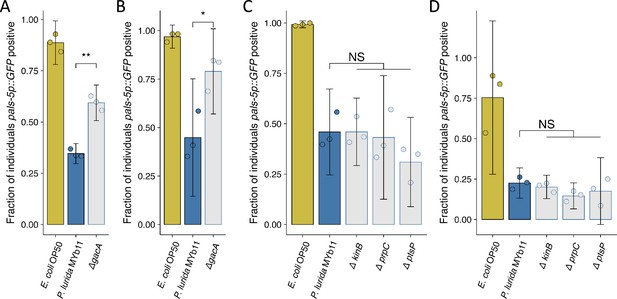
Replicate susceptibility assays shown in Figure 6B and E.
(A–D) The fraction of individuals that became pals-5p::GFP positive following exposure to 10 a.u. exogenous Orsay virus in the presence of wild-type P. lurida MYb11 versus the indicated mutants. Data are from a single representative experiment and dots represent individual plates ((A) n=681 in total and n>39 for all dots, (B) n=751 in total and n>41 for all dots, (C) n=2129 in total and n>106 for all dots, (D) n=1466 in total and n>28 for all dots). For all plots the bar is the mean and error bars are the 95% confidence interval (C.I.). (A–B) p-Values determined using Student’s t-test. (C–D) p-Values determined using one-way ANOVA followed by Dunnett’s test (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001) (a.u., arbitrary units).
© 2024, BioRender Inc. Figure 6—figure supplement 1A was created using BioRender, and is published under a CC BY-NC-ND 4.0. Further reproductions must adhere to the terms of this license
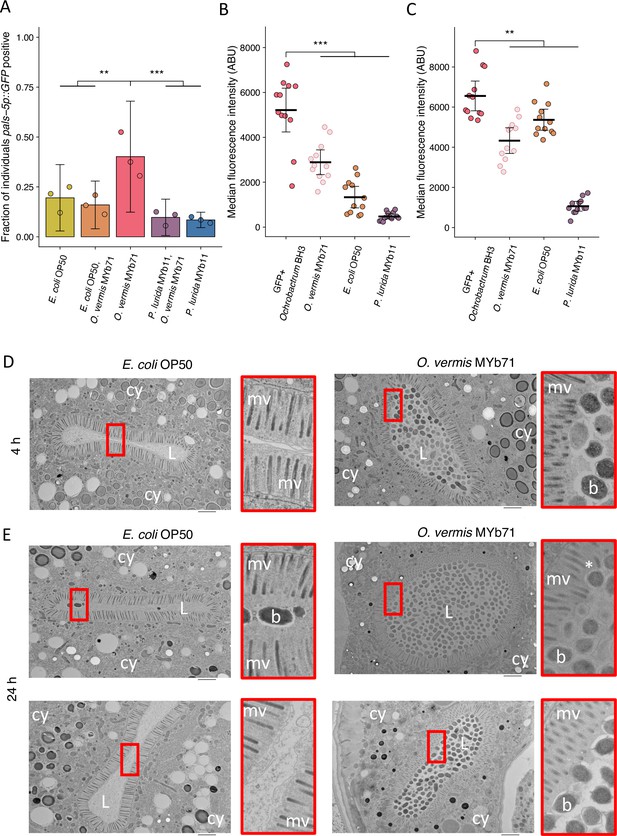
O. vermis MYb71 closely interacts with the brush border of C. elegans intestinal cells.
(A) The fraction of individuals that became pals-5p::GFP positive following exposure to 1 a.u. exogenous Orsay virus in the presence of pure lawns of the indicated bacteria or mixed lawns consisting of 50% of the indicated bacteria mixed with 50% O. vermis MYb71 (vol:vol, Materials and methods). Data are from a single representative experiment. Replicates can be found in Figure 7—figure supplement 1. Bars represent mean and dots represent individual plates (n=908 in total and n>36 for all dots). (B–C) The median fluorescence observed in the intestinal lumen of individuals following exposure for (B) 4 hr or (C) 24 hr to pure GFP expressing Ochrobactrum BH3 lawns or mixed lawns consisting of 50% of the indicated bacteria mixed with 50% GFP expressing Ochrobactrum BH3 (vol:vol, Materials and methods). Data are from a single representative experiment. Replicates can be found in Figure 7—figure supplement 1. Bars represent mean, dots represent individual animals. (D–E) Electron micrographs of animals exposed to the indicated bacterium for either (D) 4 hr or (E) 24 hr. Scale bar is 2 µm. cy = cytoplasm, L=lumen, mv = microvilli, b=bacterium, and * denotes a bent microvilli. For all plots error bars represent 95% confidence interval (C.I.). p-Values determined using one-way ANOVA followed by Dunnett’s test (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001) (a.u., arbitrary units).
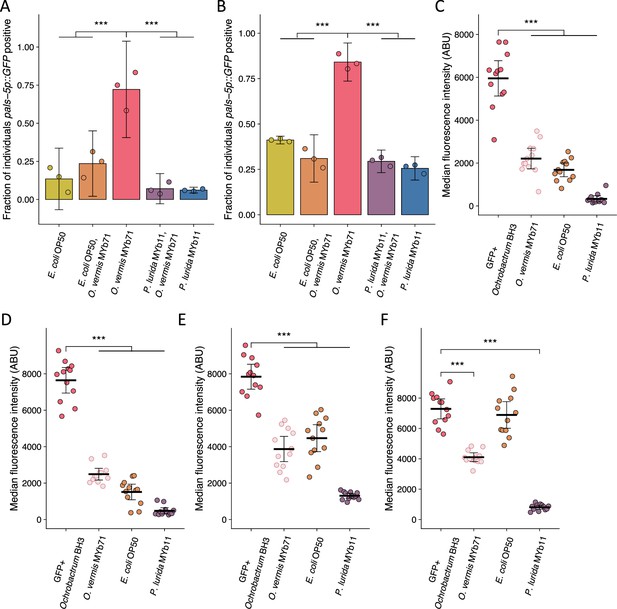
Replicate assays shown in Figure 7A–C.
(A–B) The fraction of individuals that became pals-5p::GFP positive following exposure to 1 a.u. exogenous Orsay virus in the presence of pure lawns of the indicated bacteria or mixed lawns consisting of 50% of the indicated bacteria mixed with 50% O. vermis MYb71 (vol:vol, Materials and methods). Data are from a single representative experiment. Bars represent mean and dots represent individual plates ((A) n=886 in total and n>43 for all dots, (B) n=833 in total and n>30 for all dots). (C–F) The median fluorescence observed in the intestinal lumen of individuals following exposure for (C–D) 4 hr or (E–F) 24 hr to pure GFP expressing Ochrobactrum BH3 lawns or mixed lawns consisting of 50% of the indicated bacteria mixed with 50% GFP expressing Ochrobactrum BH3 (vol:vol, Materials and methods). Data are from a single representative experiment. Bars represent mean, dots represent individual animals. For all plots error bars represent 95% confidence interval (C.I.). p-Values determined using one-way ANOVA followed by Dunnett’s test (NS, non-significant, *p<0.05, **p<0.01, ***p<0.001) (a.u., arbitrary units).
Additional files
-
Supplementary file 1
C. elegans strains, bacterial strains, and Pseudomonas aeruginosa PA14 transposon mutants used in this study.
- https://cdn.elifesciences.org/articles/92534/elife-92534-supp1-v1.docx
-
Supplementary file 2
Oligonucletides used in this study.
- https://cdn.elifesciences.org/articles/92534/elife-92534-supp2-v1.xlsx
-
Source data 1
Raw data included in this study.
- https://cdn.elifesciences.org/articles/92534/elife-92534-data1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/92534/elife-92534-mdarchecklist1-v1.docx





