Differential regulation by CD47 and thrombospondin-1 of extramedullary erythropoiesis in mouse spleen
Figures
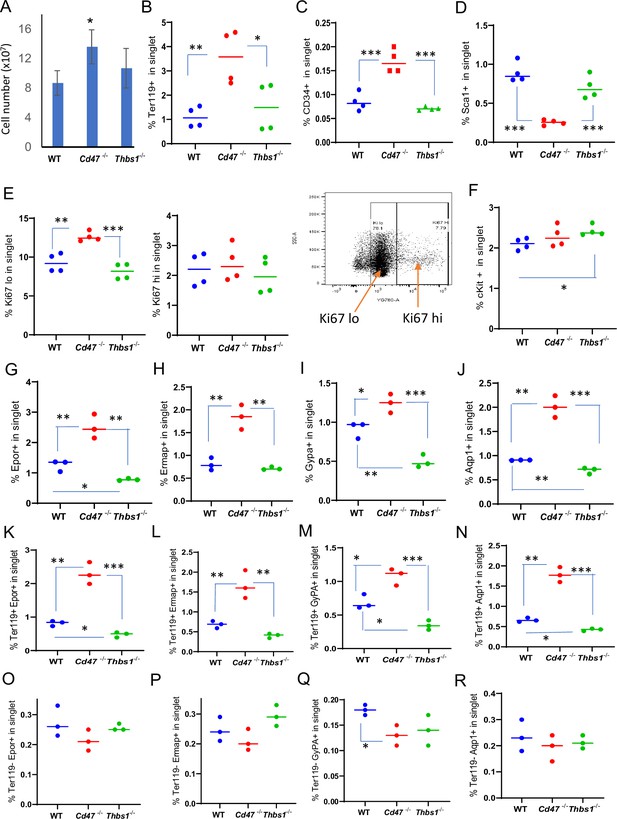
Effects of Cd47 or Thbs1 gene disruption on spleen cell numbers and content of cells expressing erythropoietic precursor markers or the proliferation marker Ki67.
(A) Total spleen cell numbers in WT, Cd47−/− and Thbs1−/− C57BL/6 mice determined after lysis of RBC (mean ± SEM, n=3). Flow cytometry was performed to analyze gated singlet spleen cells stained with Ter119 antibody (B), CD34 antibody (C), Sca1 antibody (D), Ki67 antibody with the indicated gating for high and low expression (E), cKit antibody (F), Epor antibody (G), Ermap antibody (H), Gypa antibody (I), or Aqp1 antibody (J). Further analysis of Epor (K, O), Ermap (L, P), Gypa (M, Q), and Aqp1 expression (N, R) was performed after gating for Ter119 expression. The percentages of cells positive for the indicated surface markers are presented (n=3 or 4 mice of each genotype). p-values were determined using a two-tailed t test for two-samples assuming equal variances in GraphPad Prism. *=p < 0.05, **=p < 0.01, ***=p < 0.001.
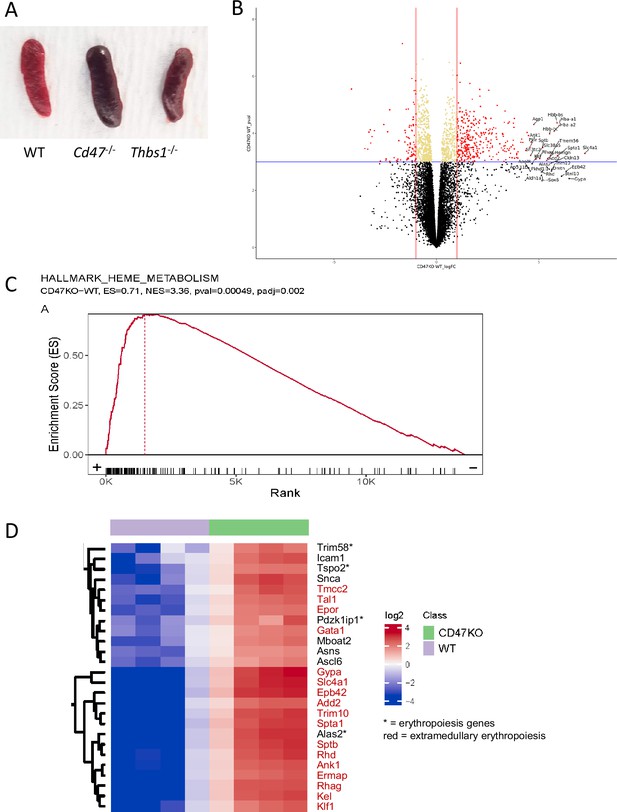
Enlargement of spleen in the absence of CD47 and bulk RNAseq analysis of lin− Cd47−/− vs WT spleen cells.
(A) Representative images of spleens from WT, Cd47−/−, and Thbs1−/− mice. (B) Volcano plot for differentially expressed genes between lineage-depleted Naive Cd47−/− vs WT CD8+-enriched spleen cells. Red dots represent significant genes with >2 fold change and p<0.001, and the top 30 genes were labelled. (C) GSEA plot showing RBC Heme Metabolism gene set enrichment. (D) Heat map visualization of the top differentially expressed genes in the RBC Heme Metabolism gene set.
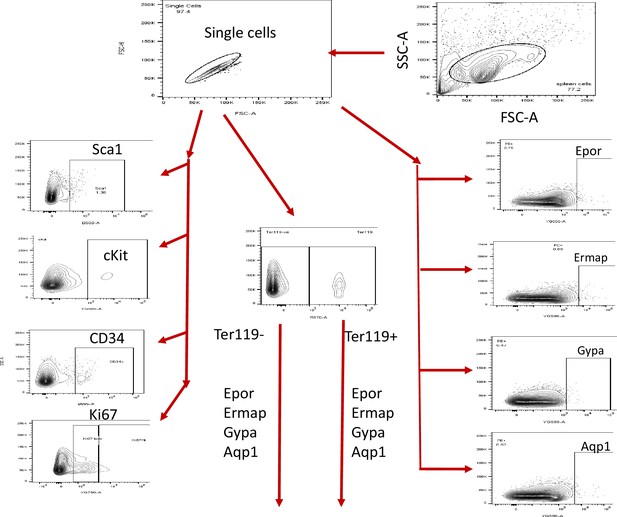
Flow cytometry analysis strategy.
The sequential flow cytometry gating strategy is illustrated.
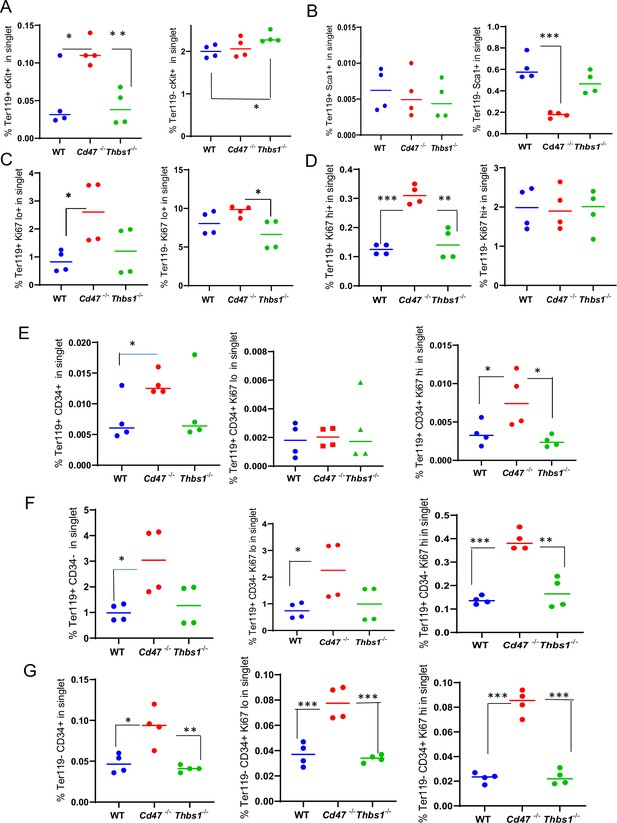
Effects of Cd47 or Thbs1 gene disruption on the percentage of Ter119+ and Ter119− spleen cells expressing markers of multipotent and committed erythroid precursors and cell proliferation.
Spleen cells isolated from WT, Cd47−/− and Thbs1−/− mice (2 male and 2 female of each genotype) were costained with Ter119 antibody along with cKit, Ki67, Sca1 Ermap, Gypa, Epor, or Aqp1 antibodies and acquired on an LSRFortessa SORP. After gating for singlet cells, the percentages of Ter119+ and Ter119− cells positive for stem cell markers cKit (A) and Sca1 (B), high or low levels of the proliferation marker Ki67 (C, D), were compared among WT, Cd47−/− and Thbs1−/− mouse spleens (n=3–4). The proliferation of CD34+ and CD34− populations of Ter119+ spleen cells from WT, Cd47−/−, Thbs1−/− mice was evaluated by staining with CD34, Ter119 and Ki67 antibodies. Ter119+CD34+ cells (E), Ter119+CD34− cells (F), and Ter119−CD34+ cells (G) were also quantified (left panels) and analyzed for the proliferation marker Ki67 (center and right panels). p-values were determined using a two-tailed t test for two-samples assuming equal variances in GraphPad Prism. *=p < 0.05, **=p < 0.01, ***=p < 0.001.
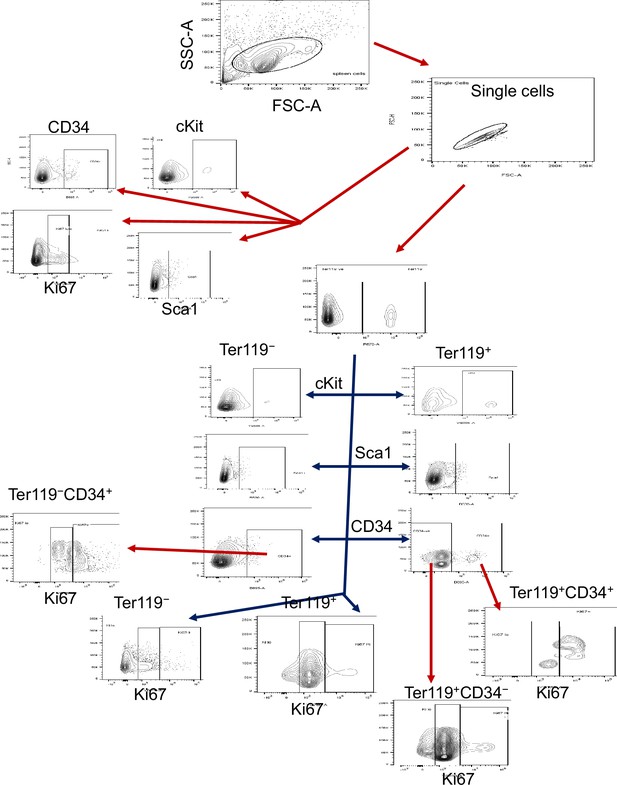
Flow cytometry analysis strategy.
The sequential flow cytometry gating strategy is illustrated.
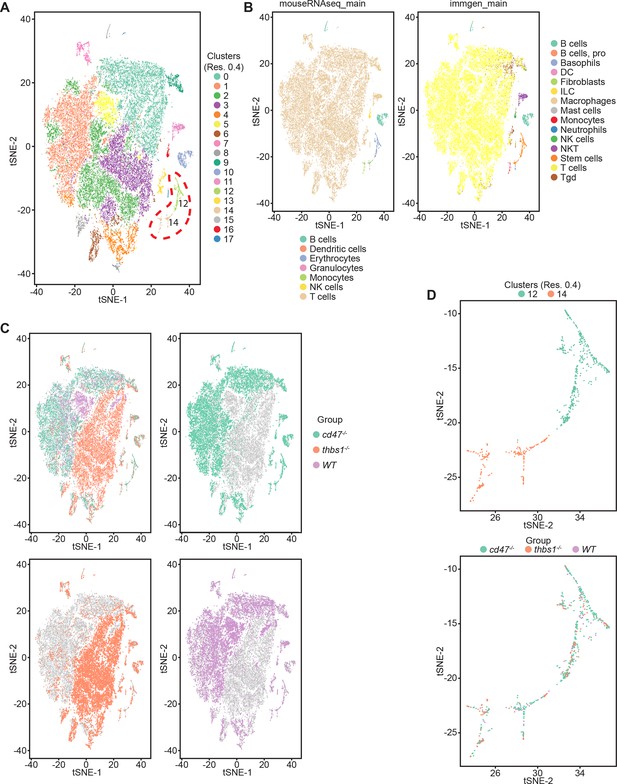
Effects of Cd47 and Thbs1 gene deletion on stem cell and erythroid precursor populations in mouse spleen identified using single cell RNA sequence analysis.
(A) tSNE clustering analysis of lineage-depleted spleen cells from WT, Cd47−/− and Thbs1−/−mice (n = 3). The encircled area contains erythroid cells (clusters 12) and stem cells (cluster 14). (B) Cell type analysis using Immgen and Mouse RNAseq and SingleR (v.1.0) databases. (C) Distribution of WT, Cd47−/− and Thbs1−/− spleen cells in each cluster of the tSNE plot. (D) Enlarged plots of clusters 12 and 14 and cells in these clusters colored by genotype.
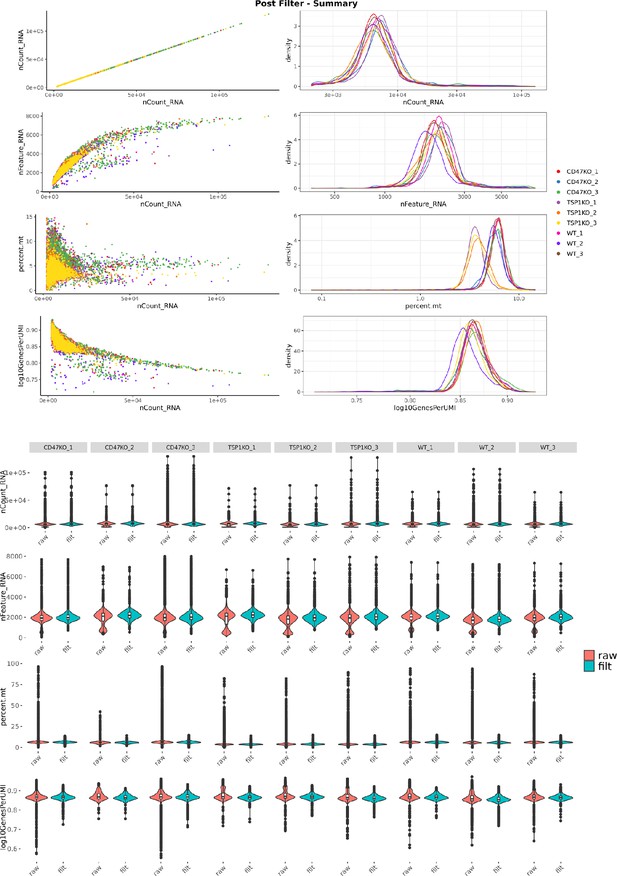
Single cell RNA sequence post-filter QC plots.
The scatter plots, histograms and violin plots of three samples lineage-depleted of WT, Cd47−/− and Thbs1−/− spleen cells are shown. The number of UMI per cell filter was set to exclude cells with <2000 UMI in nCount RNA. The Feature RNA plot shows the number of genes with non-zero expression detected per cell. Filtering was set to exclude cells having <15% mitochondrial gene expression as presented in the percent mt plot. The log10Genes per UMI plot represents the scores for complexity of the RNA library found in each cell. No filter was set in this row.
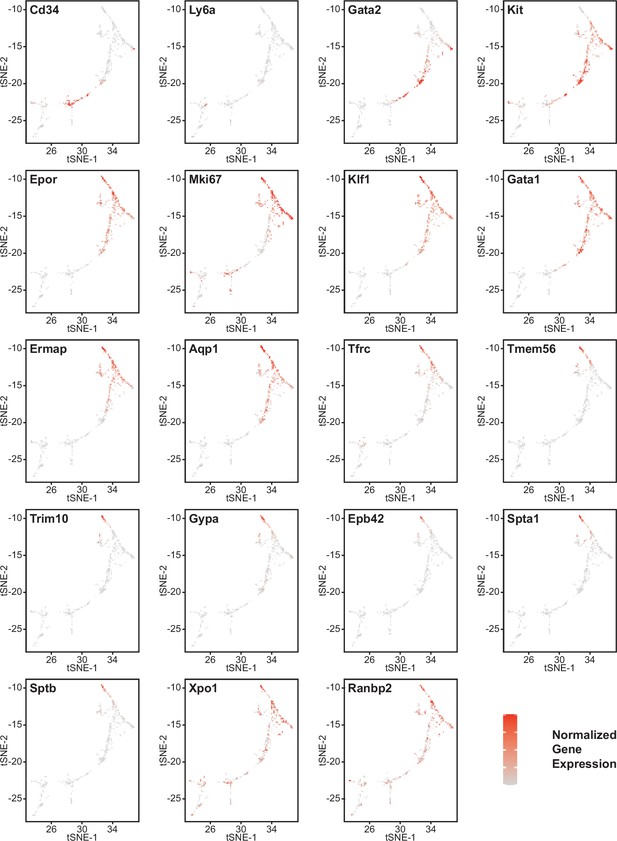
Effects of Cd47 and Thbs1 gene deletion on gene expression in stem cell and erythroid precursor clusters.
High resolution tSNE plots showing the distribution of mRNAs encoding the multipotent stem cell markers CD34 and Ly6a (Sca1) and Gata2, the erythropoietic markers Kit and Epor, the proliferation marker Mki67, erythroid differentiation transcription factors Klf1 and Gata1, and erythroid differentiation and extramedullary erythropoiesis markers Ermap, Aqp1, Tmem56, Trim10, Gypa1, Spta1, Sptb, Ebp42, Xpo1, and RanBP2 in clusters 12 and 14. Expression levels were normalized to maximum expression of each mRNA in these clusters.
-
Figure 4—source data 1
Differential expression of erythropoietic, stem cell, and proliferation associated markers in cell clusters 12 and 14.
- https://cdn.elifesciences.org/articles/92679/elife-92679-fig4-data1-v1.docx
-
Figure 4—source data 2
Differential mRNA expression of the nuclear export protein Xpo1 and nuclear pore protein synthesis instructor Ranbp2 in cluster12 and CD34+ and CD34− subsets of cluster 12 cells.
- https://cdn.elifesciences.org/articles/92679/elife-92679-fig4-data2-v1.docx
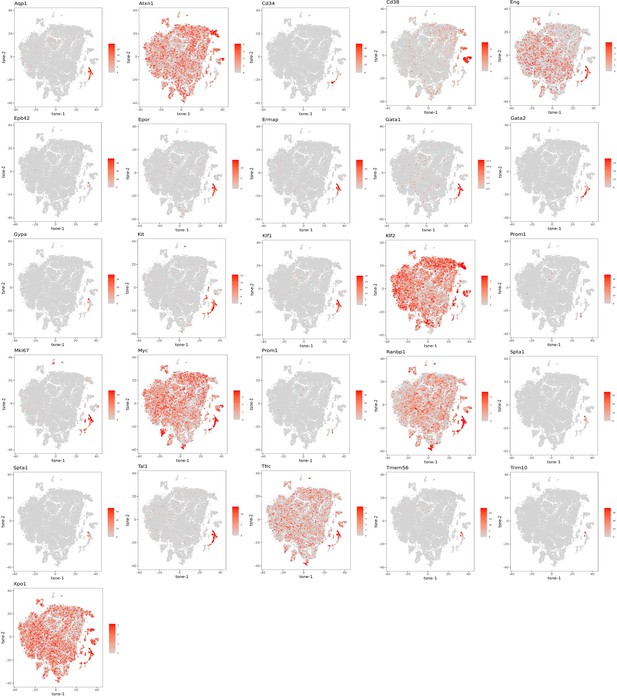
The distributions of mRNA expression of the indicated genes related to stem cells and erythropoietic cells are shown throughout the 18 clusters as a tSNE projection.
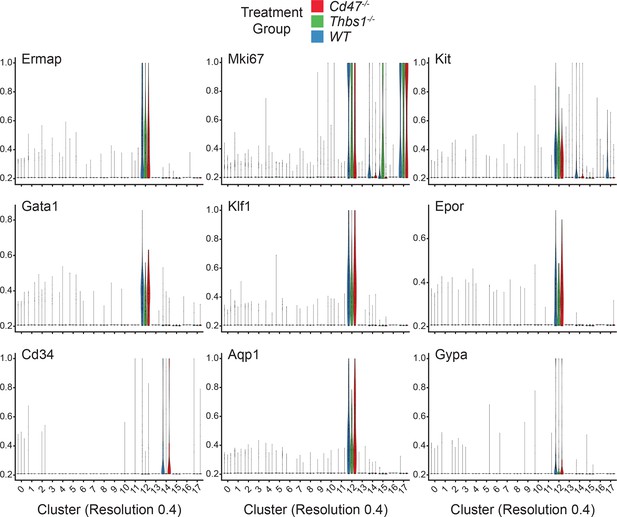
Violin plots of erythropoietic, stem cell, and proliferation associated marker mRNAs expressed in 18 cell clusters.
Data are from two female and one male mouse of each genotype.
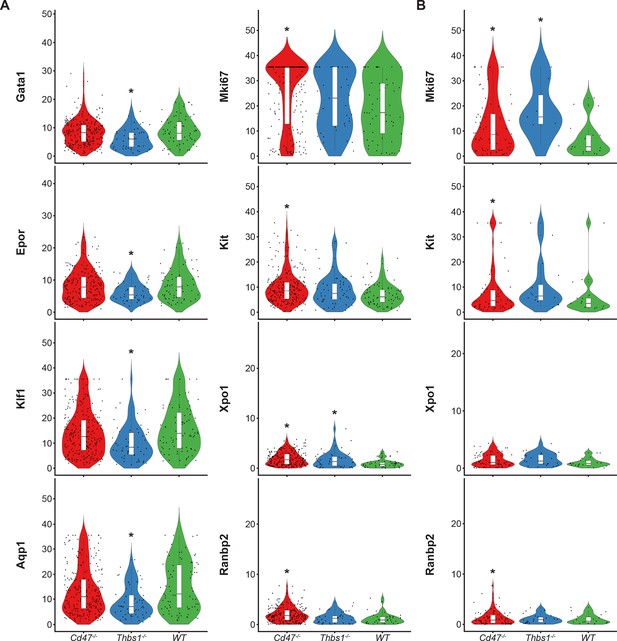
Differential effects of Cd47 and Thbs1 gene deletion on mRNA expression levels in erythroid precursor and stem cell clusters 12 and 14.
(A) Violin plots comparing mRNA expression levels of the indicated genes in Cd47−/−(red), Thbs1−/−(blue) and WT spleen cells (green) in cluster 12. (B) Violin plots comparing mRNA expression levels of the indicated genes in Cd47−/−, Thbs1−/−, and WT spleen cells in cluster 14. *=p < 0.05 relative to WT cells in the respective cluster. n = 2 female and 1 male mice of each genotype; *=p < 0.05.
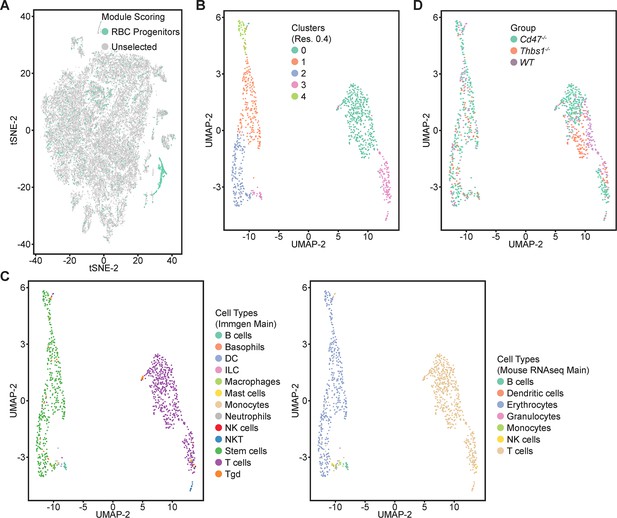
Re-clustering of lineage-depleted spleen cells selected for expression of erythroid signature genes.
(A) tSNE plot showing the distribution of cells selected for expressing threshold levels of Gypa, Ermap, Klf1, Gata1, and/or Aqp1. (B) Re-clustered cells expressing the erythroid progenitor signature are displayed in an UMAP projection. (C) Immgen and mouse RNAseq main cell type annotation of reclustered cells expressing the erythroid gene signature. (D) Distribution of WT (purple), Cd47−/− (green), and Thbs1−/− cells (orange) in the erythroid precursor and T cell clusters. Data are from two female and one male mouse of each genotype.
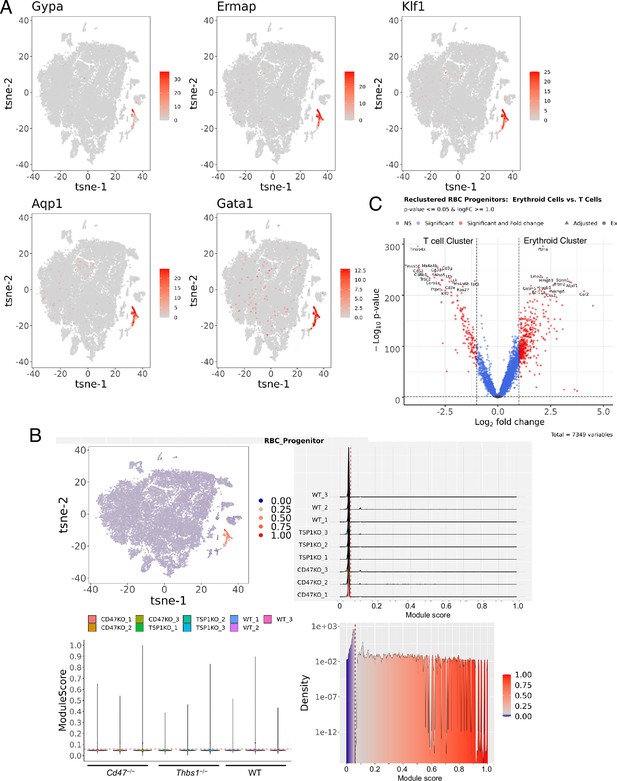
Strategy for reclustering spleen cells that express a gene signature for committed erythroid precursors.
(A) The left panels show the distribution of cells expressing threshold levels of the 5 gene signature in the original TSNE projection. (B) The RBC progenitor module scoring was performed, as module Scores (a.k.a. Signature Scores) calculated for each cell using expression of five genes (Gypa, Ermap, Klf1, Gata1, and Aqp1). The threshold was set manually (red dashed line on subplots) to separate high-scoring cells from low-scoring cells and expressed by the color on the TSNE plot (top left, high score cells are dark red and low scores cells are represented as light red). RBC Progenitor Module Scoring Plots show the distributions of module scores in lineage-depleted spleen cells from each mouse, with the threshold score indicated by the dotted red lines. (C) The volcano plot presents differentially expressed genes contrasting the reclustered erythropoietic progenitor cell clusters and the T cell cluster.
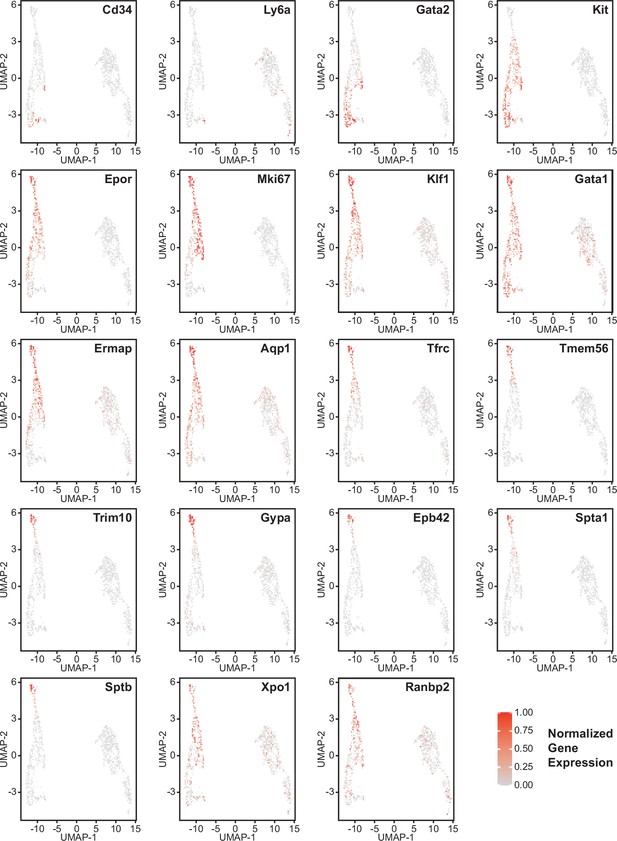
Gene expression in reclustered lineage-depleted spleen cells selected for expression of erythroid signature genes.
Distribution of the multipotent stem cell markers CD34 and Ly6a (Sca1), erythropoietic markers Gata2, Kit, and Epor, the erythroid differentiation transcription factors Klf1 and Gata1, and erythroid differentiation and extramedullary erythropoiesis marker the proliferation marker Mik67, and the erythroid markers Ermap, Tfrc, Aqp1, Tfrc, Tmem56, Trim10, Gypa1, Ebp42, Spta1, Sptb, Xpo1, and Ranbp2 in the erythroid lineage cluster (left) and T cell cluster (right). Expression levels were normalized to maximum expression of each mRNA in these clusters. Data are from two female and one male mouse of each genotype.
-
Figure 7—source data 1
Differential expression of erythropoietic, stem cell, and proliferation associated markers in reclustered erythroid and T cell clusters.
- https://cdn.elifesciences.org/articles/92679/elife-92679-fig7-data1-v1.docx
Tables
Expression of erythropoiesis-associated genes in lineage-depleted Cd47−/− versus WT spleen cells.
| Gene | Erythropoiesis expression/function | Fold change Cd47-/-/WT* | T statistic | p-value |
|---|---|---|---|---|
| Ermap | extramedullary erythropoiesis marker† | 21.5 | 6.04 | 9.35x10–5 |
| Tal1 | extramedullary erythropoiesis marker† | 13.9 | 5.13 | 3.55x10–4 |
| Gypa | extramedullary erythropoiesis marker† | 89.3 | 3.67 | 3.86x10–3 |
| Gata1 | extramedullary erythropoiesis marker† | 7.69 | 4.86 | 5.38x10–4 |
| Kel | extramedullary erythropoiesis marker† | 29.0 | 4.61 | 8.04x10–4 |
| Slc4a1 | extramedullary erythropoiesis marker† | 151.4 | 4.90 | 5.05x10–4 |
| Klf1 | extramedullary erythropoiesis marker† | 20.6 | 5.06 | 3.95x10–4 |
| Cldn13 | extramedullary erythropoiesis marker† | 49.2 | 4.42 | 1.09x10–3 |
| Trim10 | extramedullary erythropoiesis marker† | 49.7 | 4.11 | 1.83x10–3 |
| Epor | extramedullary erythropoiesis marker† | 12.3 | 4.91 | 5.01x10–4 |
| Sptb | extramedullary erythropoiesis marker† | 36.5 | 5.21 | 3.16x10–4 |
| Rhag | extramedullary erythropoiesis marker† | 33.0 | 4.64 | 7.68x10–4 |
| Hba-a1 | erythroblasts | 63.8 | 6.46 | 5.23x10–5 |
| Hbb-bs | erythroblasts | 59.2 | 6.60 | 4.34x10–5 |
| Gata1 | BFU-E through erythroblasts | 7.69 | 4.86 | 5.38x10–4 |
| Tfrc (CD71) | CFU-E through erythroblasts | 3.12 | 6.19 | 7.58x10–5 |
| Kit | Progenitors through CFU-E | 1.72 | 6.24 | 7.02x10–5 |
| Sox6 | Adult definitive erythropoiesis | 35.5 | 3.56 | 0.0046 |
| Aqp1 | Adult definitive erythropoiesis | 26.9 | 6.51 | 4.91x10–5 |
| Nr3c1 | Adult definitive erythropoiesis | 1.12 | 2.80 | 0.018 |
| Mki67 | Proliferation marker | 4.43 | 7.65 | 1.14x10–5 |
| Cd34 | Multipotent progenitors through CFU-E | 1.60 | 1.59 | 0.141 |
| Ly6a (Sca1) | Multipotent progenitors | 1.17 | 1.02 | 0.331 |
| Anpep (CD13) | Multipotent progenitors | 1.16 | 1.14 | 0.28 |
| Cd33 | Multipotent progenitors | –1.06 | –0.37 | 0.71 |
| Gata2 | Multipotent progenitors | 1.08 | 0.66 | 0.52 |
| Xpo1 | Stability of nuclear Gata1 | 1.23 | 3.86 | 2.7x10–3 |
-
*
Gene enrichment in naïve Cd47−/− vs WT spleen cells depleted for CD4, CD11b, CD11c, CD19, CD45R (B220), CD49b (DX5), CD105, MHC Class II, Ter-119, and TCRγ/δ.
-
†
Reported markers of stress-induced extramedullary erythropoiesis (Delic et al., 2020; Thompson et al., 2010).
Differential mRNA expression of erythropoietic, stem cell, and proliferation associated markers in WT, Cd47−/−, and Thbs1−/− cells in clusters 12 and 14.
| Cd47−/− vs WT | Thbs1−/− vs WT | ||||
|---|---|---|---|---|---|
| Cluster | Gene | p-value | Avg log2 FC | p-value | Avg log2 FC |
| 12 | Klf1 | 0.463 | –0.115 | 0.0023 | –0.510 |
| 12 | Aqp1 | 0.331 | –0.130 | 0.0011 | –0.472 |
| 12 | Tfrc | 0.017 | 0.531 | 0.93 | 0.009 |
| 14 | Tfrc | 0.64 | 0.071 | 0.922 | 0.016 |
| 12 | Epor | 0.506 | –0.078 | 4.75x10–5 | –0.576 |
| 12 | Ermap | 5.1x10–3 | 0.309 | 0.65 | –0.073 |
| 12 | Gata1 | 0.57 | 0.007 | 0.0011 | –0.377 |
| 12 | Mki67 | 9.16x10–6 | 0.997 | 0.22 | 0.456 |
| 14 | Mki67 | 0.0089 | 0.798 | 0.0017 | –0.062 |
| 12 | Kit | 0.0015 | 0.331 | 0.057 | 0.264 |
| 14 | Kit | 0.020 | 0.409 | 0.58 | 0.222 |
| 12 | Xpo1 | 3.31x10–8 | 0.472 | 0.0069 | 0.323 |
| 14 | Xpo1 | 0.079 | 0.211 | 0.19 | 0.148 |
| 12 | Ranbp1 | 0.24 | 0.108 | 0.13 | 0.079 |
| 14 | Ranbp1 | 0.91 | 0.069 | 0.076 | –0.486 |
| 12 | Ranbp2 | 1.98x10–14 | 0.754 | 0.092 | 0.269 |
| 14 | Ranbp2 | 0.0056 | 0.365 | 0.88 | 0.042 |
| 12 | Nr3c1 | 8.5x10–4 | 0.320 | 8.8x10–4 | 0.430 |
| 14 | Nr3c1 | 0.012 | 0.153 | 8.6x10–5 | 0.428 |
| 12 | Ddx46 | 3.04x10–8 | 0.530 | 2.68x10–10 | 0.779 |
| 14 | Ddx46 | 0.0014 | 0.385 | 0.0023 | 0.490 |
Co-expression of the indicated erythropoiesis related genes in WT, Cd47−/− and Thbs1−/− spleen cells in cluster 12 was quantified and expressed as a percentage of the total cell number of each genotype in cluster 12.
| Cluster 12 gene coexpression | WT | Cd47−/− | Thbs1−/− |
|---|---|---|---|
| Cd34_Ermap | 0.0% | 1.8% | 1.6% |
| Cd34_Klf1 | 6.5% | 8.1% | 3.1% |
| Cd34_Gata1 | 6.5% | 8.4% | 0.0% |
| Mki67_Cd34 | 2.2% | 6.3% | 0.0% |
| Mki67_Ermap | 55.4% | 63.0% | 54.7% |
| Mki67_Kit | 48.9% | 67.2% | 57.8% |
| Mki67_Ly6a | 37.0% | 48.2% | 43.8% |
| Mki67_Klf1 | 62.0% | 74.6% | 57.8% |
| Mki67_Epor | 54.4% | 62.7% | 53.1% |
| Klf1_Ermap | 67.4% | 68.0% | 62.5% |
| Klf1_Gata1 | 78.3% | 82.4% | 59.4% |
| Ermap_Gata1 | 62.0% | 65.1% | 56.2% |
| Kit_Ermap | 54.4% | 56.7% | 59.4% |
| Kit_Klf1 | 71.7% | 77.1% | 78.1% |
| Ly6a_Ermap | 42.4% | 37.7% | 46.9% |
| Ly6a_Klf1 | 64.1% | 59.2% | 67.2% |
Differential expression of mitochondrial encoded genes in cluster 12 cells from WT, Cd47−/−, and Thbs1−/− spleens.
| Gene | p-val Cd47−/−vs WT | Avg log2FC Cd47−/−vs_WT | p-val Thbs1-/- vs_WT | Avg log2FC Thbs1-/- vs_WT |
|---|---|---|---|---|
| Mt-Atp6 | 0.887 | –0.027 | 5.43x10–11 | –0.812 |
| Mt-Atp8 | 7.23x10–20 | 0.98 | 0.0168 | 0.377 |
| Mt-Co1 | 0.028 | 0.135 | 2.60x10–7 | –0.532 |
| Mt-Co2 | 0.226 | 0.068 | 3.37x10–10 | –0.693 |
| Mt-Co3 | 0.207 | 0.097 | 1.02x10–8 | –0.671 |
| Mt-Cytb | 0.338 | –0.006 | 9.35x10–10 | –0.790 |
| Mt-Nd1 | 0.074 | 0.15 | 6.78x10–6 | –0.636 |
| Mt-Nd2 | 0.135 | 0.113 | 1.00x10–7 | –0.748 |
| Mt-Nd3 | 7.67x10–9 | 0.531 | 1.36x10–7 | –0.813 |
| Mt-Nd4 | 0.152 | 0.093 | 5.84x10–6 | –0.558 |
| Mt-Nd4l | 3.50x10–10 | 0.658 | 6.49x10–8 | 0.787 |
| Mt-Nd5 | 4.63x10–8 | 0.612 | 0.0154 | 0.375 |
| Mt-Nd6 | 1.65x10–7 | 0.416 | 4.39x10–6 | 0.524 |
Differential mRNA expression of erythropoietic, stem cell, and proliferation associated markers in reclustered WT, Cd47−/−, and Thbs1−/− erythroid and T cell clusters.
| Cd47−/− vs WT | Thbs1−/− vs WT | ||||
|---|---|---|---|---|---|
| Cluster | Gene | p-value | Avg log2 FC | p-value | Avg log2 FC |
| Erythroid | Klf1 | 0.507 | –0.095 | 0.0016 | –0.526 |
| Erythroid | Aqp1 | 0.196 | –0.088 | 8.1x10–4 | –0.390 |
| T cells | Aqp1 | 0.662 | 0.027 | 0.350 | –0.092 |
| Erythroid | Tfrc | 0.015 | 0.549 | 0.936 | 0.054 |
| T cells | Tfrc | - | - | - | - |
| Erythroid | Epor | 0.767 | –0.046 | 0.026 | –0.219 |
| Erythroid | Ermap | 0.0064 | 0.326 | 0.496 | –0.010 |
| T cells | Ermap | 0.335 | 0.076 | 0.697 | 0.063 |
| Erythroid | Gata1 | 0.38 | 0.009 | 0.0040 | –0.337 |
| T cells | Gata1 | 0.0044 | –.0.162 | 0.302 | 0.093 |
| Erythroid | Mki67 | 4.6x10–6 | 1.015 | 0.118 | 0.545 |
| T cells | Mki67 | - | - | - | - |
| Erythroid | Kit | 0.0066 | 0.293 | 0.059 | 0.275 |
| T cells | Kit | - | - | - | - |
| Erythroid | Xpo1 | 2.65x10–9 | 0.514 | 0.0025 | 0.373 |
| T cells | Xpo1 | 0.238 | 0.047 | 0.137 | 0.066 |
| Erythroid | Ranbp1 | 0.099 | 0.133 | 0.41 | 0.078 |
| T cells | Ranbp1 | 0.278 | –0.124 | 0.970 | 0.019 |
| Erythroid | Ranbp2 | 3.2x10–14 | 0.755 | 0.058 | 0.298 |
| T cells | Ranbp2 | 4.5x10–4 | 0.275 | 0.210 | 0.094 |
| Erythroid | Nr3c1 | 6.5x10–4 | 0.319 | 0.0011 | 0.447 |
| T cells | Nr3c1 | 0.0018 | 0.274 | 0.0018 | 0.233 |
| Erythroid | Ddx46 | 1.65x10–9 | 0.530 | 2.98x10–9 | 0.775 |
| T cells | Ddx46 | 2.3x10–5 | 0.348 | 7.5x10–6 | 0.446 |
| Erythroid | Hba-a1 | 0.906 | 0.034 | 0.0088 | –2.168 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Commercial assay or kit | CD8a+T Cell Isolation Kit | Miltenyi Biotec | Cat#: 130–104–075 | |
| Commercial assay or kit | CD8a (Ly-2) MicroBeads mouse | Miltenyi Biotec | Cat#: 130-117-044 | |
| Chemical compound, drug | EDTA solution | Sigma-Aldrich | Cat#: E8008 | 2 mM |
| Peptide, recombinant protein | bovine serum albumin (BSA) | Sigma-Aldrich | Cat#: A7906 | 0.5% |
| Chemical compound, drug | ACK Lysing Buffer, 100 mL | Quality Biologicals | Cat#: 118-156-721 | |
| Strain, strain background (Mus musculus, C57BL/6) | WT mice | Jackson Laboratories | WT C57BL/6 | |
| Strain, strain background (M. musculus, C57BL/6) | Cd47-/- mice | Jackson Laboratories | B6.129S7-Cd47tm1Fpl/J; Strain:003173 | Lindberg et al., 1996; PMID:8864123 |
| Strain, strain background (M. musculus, C57BL/6) | Thbs1-/- mice | Jackson Laboratories | B6.129S2-Thbs1tm1Hyn/J; Strain:006141 | Lawler et al., 1998; PMID:9486968 |
| Antibody | Anti-mouse Ter119-APC (Rat monoclonal) | Biolegend | Cat#: 116212; RRID:AB_313713 | IgG2b FACS (1 μg/100 μl/1 million cells) |
| Antibody | anti-mouse CD34-Percp/Cy5.5 (Rat monoclonal) | Biolegend | Cat#: 119327; RRID:AB_2728136 | IgG2a FACS (1 μg/100 μl/1 million cells) |
| Antibody | anti mouse Sca1 PE/Cy7 (Rat monoclonal) | Biolegend | Cat#: 108114; RRID:AB_493596 | IgG2a FACS (0.5 μg/100 μl/1 million cells) |
| Antibody | anti-mouse cKit PE (Rat monoclonal) | Biolegend | Cat#: 105808; RRID:AB_313217 | IgG2b FACS (0.1 μg/100 μl/1 million cells) |
| Antibody | anti-mouse Ki67- PE/Cy7 (Rat monoclonal) | Biolegend | Cat#: 652425; RRID:AB_2632693 | IgG2a FACS (0.5 μg/100 μl/1 million cells) |
| Antibody | IgG2b, κ APC Isotype Control Antibody (Rat monoclonal) | Biolegend | Cat#: 400611; RRID:AB_326555 | IgG2b FACS (1 μg/100 μl/1 million cells) |
| Antibody | IgG2a, κ PerCP/Cyanine5.5 Isotype Control Antibody (Rat monoclonal) | Biolegend | Cat#: 400531; RRID:AB_2864286 | FACS (1 μg/100 μl/1 million cells) |
| Antibody | IgG2a, κ PE/Cyanine7 Isotype Control Antibody (Rat monoclonal) | Biolegend | Cat#: 400521; RRID:AB_326542 | FACS (0.25 μg/100 μl/1 million cells) |
| Antibody | IgG2b, κ PE Isotype Control Antibody (Rat monoclonal) | Biolegend | Cat#: 400608; RRID:AB_326552 | FACS (0.1 μg/100 μl/1 million cells) |
| Antibody | IgG2a, κ Alexa Fluor 488 Isotype Control Antibody (Rat monoclonal) | Biolegend | Cat#: 400525; RRID:AB_2864283 | FACS (0.25 μg/100 μl/1 million cells) |
| Antibody | anti-Rabbit IgG (H+L) Cross-Adsorbed, Alexa Fluor 594 (Goat polyclonal) | Thermo Fisher | Cat#: A-11012 | FACS (0.2 μg/100 μl/1 million cells) |
| Antibody | Anti-ERMAP (Rabbit polyclonal) | Thermo Fisher | Cat#: BS-12333R | FACS (1:100 dilution) |
| Antibody | Anti-GYPA (Rabbit polyclonal) | Thermo Fisher | Cat#: BS-2575R | FACS (1:100 dilution) |
| Antibody | Anti-Aquaporin 1 (Rabbit polyclonal) | Thermo Fisher | Cat#: PA5-78806 | FACS (1 μg/100 μl/1 million cells) |
| Antibody | Anti-EPOR (Rabbit polyclonal) | Bioss; Thermo Fisher | Cat#: BS-1424R | FACS (1 μg/100 μl/1 million cells) |
| Extramedullary | CD47KO/WT_FC | CD47KO/WT_pval | DEG rank |
|---|---|---|---|
| erythropoiesis Gene | |||
| Ermap | 3.202941868 | 0.035938856 | 2087 |
| Gypa | 23.09289299 | 0.000169485 | 163 |
| Gata1 | 2.809105664 | 0.044897197 | 2273 |
| Slc4a1 | 4.352812248 | 0.005732723 | 1641 |
| Klf1 | 30.09543812 | 0.000442139 | 76 |
| Trim10 | 10.8936042 | 0.003179996 | 672 |
| Sptb | 4.443091877 | 0.00032532 | 1607 |
| Rhag | 25.90869436 | 8.45 E-05 | 121 |





