The physiological landscape and specificity of antibody repertoires are consolidated by multiple immunizations
Figures
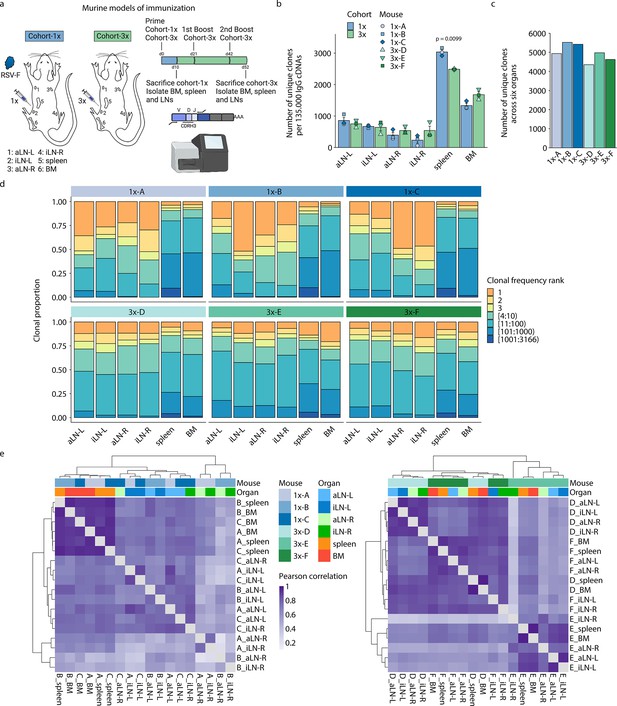
Profiling antibody repertoire diversity and clonal expansion across multiple lymphoid organs.
(a) Overview of mouse immunizations with RSV-F antigen: Mice (n = 3 per cohort) were subjected to single (cohort-1x) or multiple (cohort-3x) immunizations (subcutaneous injections into the left lateral flank), followed by deep sequencing of antibody repertoires (VH IgG) derived from six different lymphoid organs (BM, spleen, aLN-L, -R, and iLN-L, -R). (b) Shown is the average clonal diversity (mean ± SEM, n = 3) based on the total number of unique clones (clones were defined as antibody sequences possessing identical germline V- and J-genes and 100% CDRH3 amino acid [a.a.] identity and length) within each organ per mouse (point) and cohort (bar). p-Values represent the results from unpaired t-test, adjusted for multiple testing using the Benjamini–Hochberg method. The comparison was made across cohorts for the same organs, and only p-values <0.05 are depicted. (c) Bar plots represent the clonal diversity by quantifying the total numbers of unique clones across all organs within each mouse. (d) Clonal expansion profiles with each bar representing an organ and each colored section displaying frequency-ranked clones or clonal fractions and the corresponding percentage they occupy within each organ repertoire. (e) Hierarchical clustering of Pearson correlation coefficients of germline V-gene usage counts between organs. Cohort-1x and cohort-3x were separately clustered (left and right). Each tile represents the pairwise Pearson correlation coefficient of the germline V-gene repertoires of two organs. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes. Panel (a) created with BioRender.com.
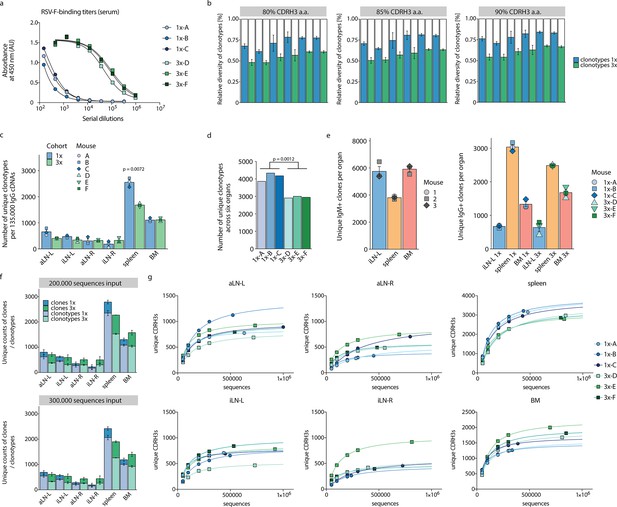
Serum titer measurements, evaluation of clonotyping, quantification of clonal diversities, and subsampling analysis.
(a) Serial dilutions of sera for the detection of RSV-F-specific titers 10 days after single or final booster immunizations for cohort-1x and cohort-3x, respectively, using ELISA. A four-parameter logistic curve was fitted to the data by nonlinear regression. (b) Comparison of different clonotyping thresholds using 80%, 85%, and 90% CDRH3 a.a. similarities by plotting average diversities of unique clonotypes (numbers of unique clonotypes) relative to average diversities of unique clones (numbers of unique clones) for each organ per cohort (mean ± SEM, n = 3). (c) Average clonotype diversity (mean ± SEM, n = 3) by quantification of total numbers of unique clonotypes within each organ per mouse (point) and cohort (bar). p-Value represents the result from unpaired t-test, adjusted for multiple testing using the Benjamini–Hochberg method. The comparison was made across cohorts for the same organs (only p-values <0.05 are shown). (d) Clonotype diversity by quantification of total numbers of unique clonotypes across all organs within each mouse. For statistical analysis, unpaired t-test was used on the average clonotype diversity of cohort-1x mice and cohort-3x mice. (e) Comparison of average clonal diversity within iLN-L, spleen, and BM between untreated Balb/c mice (n = 3, IgM+ clones, left graph) and immunized cohort-1x and cohort-3x Balb/c mice (n = 3/cohort, IgG+ clones, right graph). (f) Comparison of varying sequencing read input (2 × 105 and 3 × 105) into the MAF pipeline by quantification of both average clonal and average clonotype diversities for each cohort (mean ± SEM). (g) Yield of unique clonal sequences (defined by identical CDRH3) after subsampling sequence input into the MAF pipeline (5 × 104, 1 × 105, 2 × 105, 3 × 105 and maximal sequence number per sample). Curves were fitted to the data by nonlinear regression. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes.
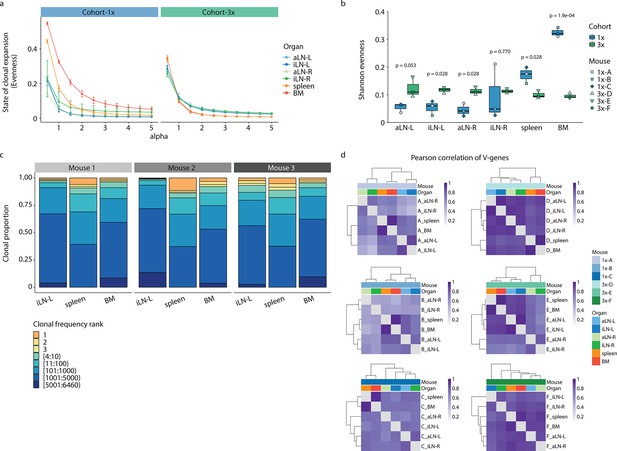
Average Hill-based evenness profiles, Shannon indices, IgM expansion profiles, and Pearson correlation of germline V-gene usage.
(a) Average Hill-based evenness profiles indicate the degree of clonal expansion within organ repertoires of cohort-1x and cohort-3x mice by using a continuum of diversity indices (mean ± SEM, n = 3 per cohort). The parameter α determines the impact of high-frequency clones, with increasing α values, higher-frequency clones are weighted more. Hill-based evenness values closer to 1 indicate uniform or even repertoires, while values tending to 0 indicate highly expanded (polarized) repertoires (Greiff et al., 2015; see ‘Materials and methods’ for more details). Each colored line depicts the antibody repertoire of one organ (see legend). (b) Boxplots representing the Shannon evenness (α=1E) of each lymphoid organ repertoire per cohort. p-Values represent the results from unpaired t-test, adjusted for multiple testing using the Benjamini–Hochberg method, comparing the same organs across both cohorts. (c) Clonal expansion profiles of three untreated mice 1–3. Each bar represents an organ and each colored section displays the repertoire space occupied by frequency-ranked IgM+ clones or clonal fractions. (d) Hierarchical clustering of pairwise Pearson correlations for the comparison of germline V-gene usage counts between organs for each mouse. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes.
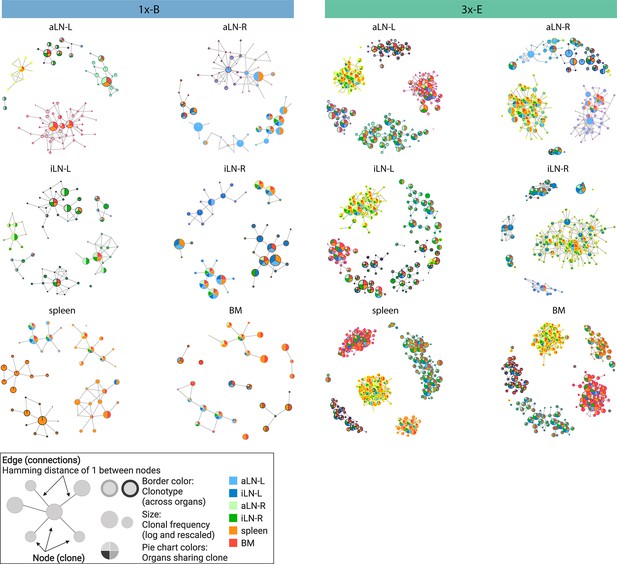
Sequence-similarity network analysis across lymphoid organs.
Sequence-similarity networks of the five most diverse clonotypes from a representative mouse from the single (1x-B) or multiple (3x-E) immunization cohorts. Within a network, each node represents a unique clone that is connected by edges to clones with a Hamming distance of 1 between CDRH3 a.a. Clones within each clonotype display the same border color throughout all organs in each mouse. The size of each clone represents the clonal frequency (in log) within the corresponding organ. Pie charts within each node depict organs sharing the corresponding clone (see legend). See Figure 2—figure supplement 1c for sequence similarity networks of the other cohort-1x and cohort-3x mice. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes. Legend created with BioRender.com.
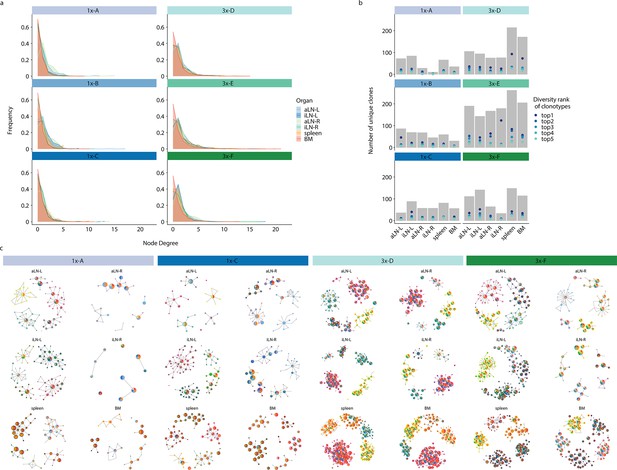
Quantification of clonal connectivity, clonal diversity and network-based analysis of the five most diverse clones.
(a) Measurements of clonal connectivity degree distributions by quantifying the number of connections each node (clone) exhibits. Nodes belonging to the same clonotype with a Hamming distance of 1 were connected. Each line represents an organ. (b) Clonal diversities of the five most diverse clonotypes by quantifying the number of unique clones (clonal variants) within each clonotype in each organ. Each line represents one of the top 1–5 diversity ranked clones (see legend; the line indicates the diversity rank of the top 1–5 most diverse clonotypes for each organ, it does not indicate the identical clonotype across organs). Gray bars represent the sum of all clonal variants within top 1–5 most diverse clonotypes for each organ. (c) Graphs show sequence-similarity networks of the five most diverse clonotypes within each lymphoid organ for mice 1x-A, 1x-C, 3x-D, and 3x-F. Each node represents a unique clone and all clones belonging to the same clonotype with a Hamming distance of 1 are connected (edges). Clones within each clonotype display the same border color throughout all organs within each mouse. The size of each clone represents the clonal frequency (in log) within the corresponding organ. Pie charts within each node depict organs that share the corresponding clone. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes.
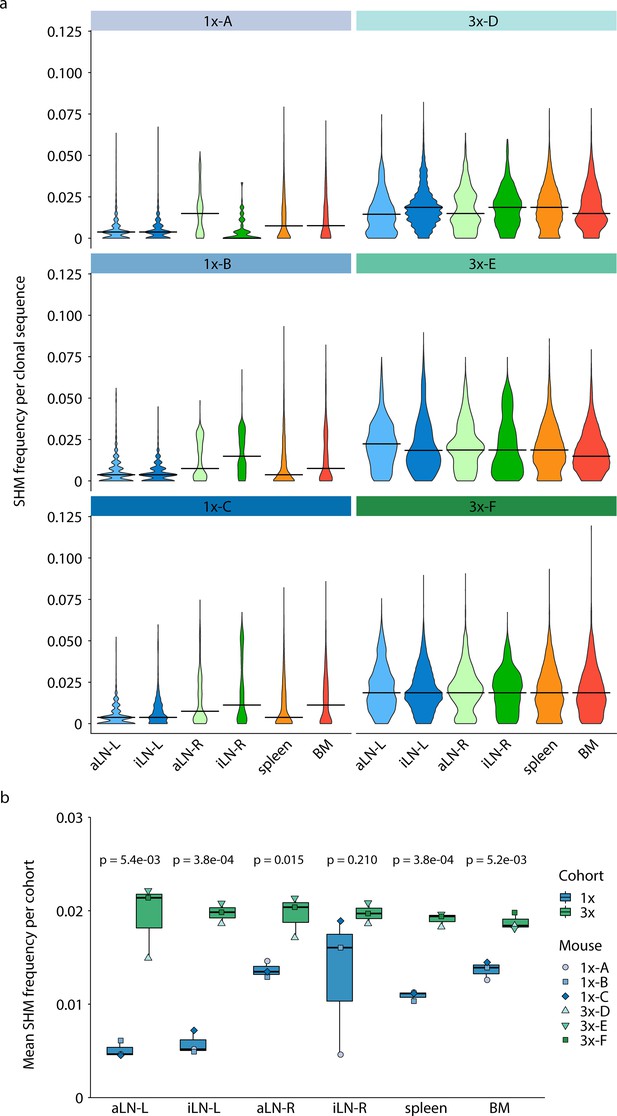
Quantification of somatic hypermutation (SHM).
(a) Quantification of SHM frequency for each organ repertoire per mouse (horizontal line indicates the median). (b) Boxplots indicating mean SHM clonal frequencies of organs per cohort. p-Values represent the results from unpaired t-test, adjusted for multiple testing using the Benjamini–Hochberg method, comparing the same organs across both cohorts. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes.
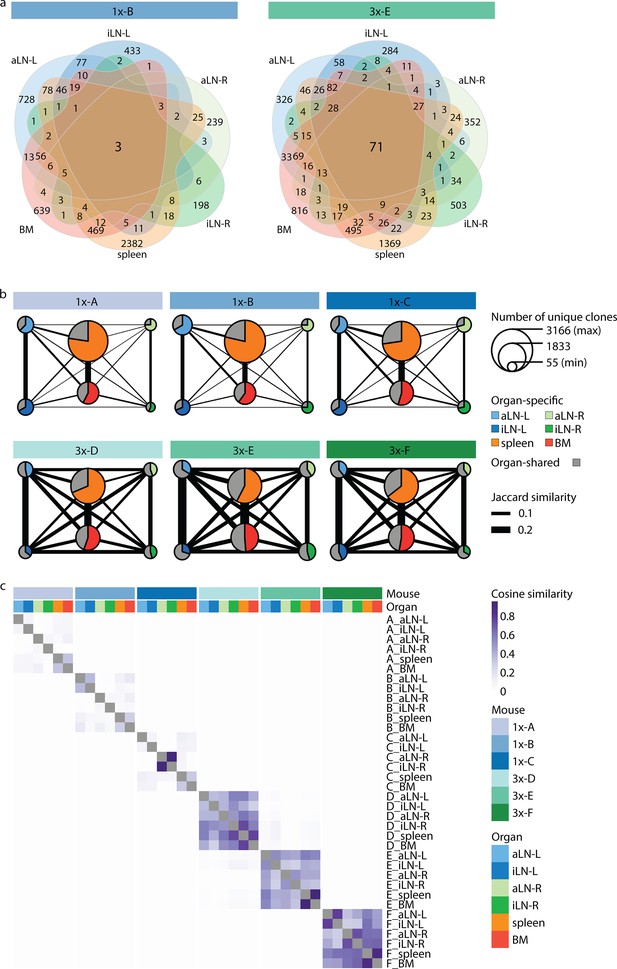
Strong humoral responses result in increased antibody repertoire similarities across multiple lymphoid organs.
(a) Venn diagrams depicting numbers of shared unique clones from a representative mouse from the single (1x-B) or multiple (3x-E) immunization cohorts. See Figure 3—figure supplement 1a for Venn diagrams of the other cohort-1x and cohort-3x mice. (b) Repertoire similarity networks are based on clonal overlap across all six lymphoid organs per mouse. Each node represents an organ, with the size of the node being proportional to the number of unique clones within the organ. Width of edges between the nodes depict the pairwise repertoire similarity based on the Jaccard index (see ‘Materials and methods’). The pie chart within each node represents the proportion of organ-specific and organ-shared clones for each organ. (c) Heatmap representing pairwise cosine similarity indices based on clonal overlap including clonal frequency information across all samples. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes.
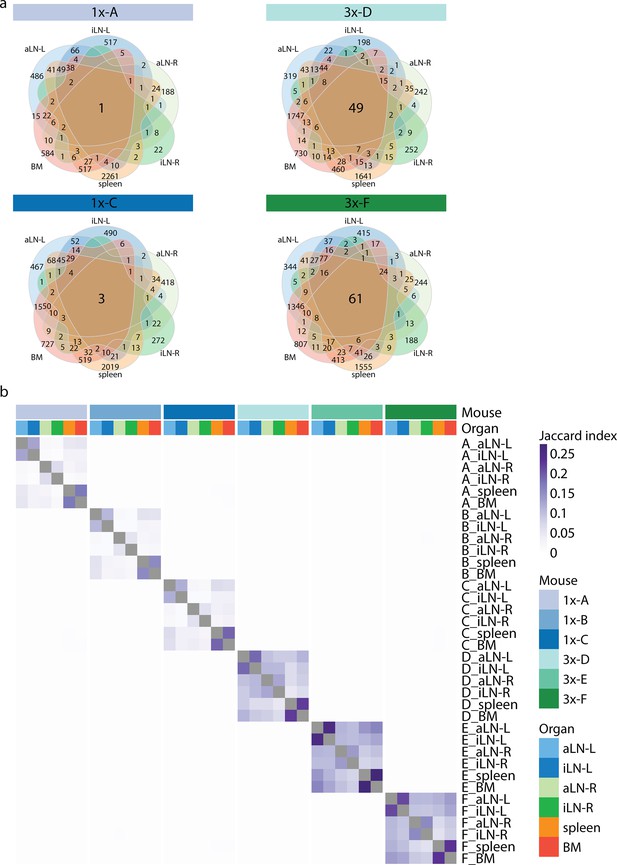
Repertoire similarity based on clonal overlap.
(a) Venn diagrams depicting numbers of shared unique clones within mice 1x-A, 1x-C, 3x-D, and 3x-F. (b) Heatmap depicting pairwise Jaccard index to assess repertoire similarity based on overlap of unique clones across all samples. The pairwise Jaccard index is calculated by quantifying the size of intersection between two samples and dividing it by the length of the union of the same sample. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes.
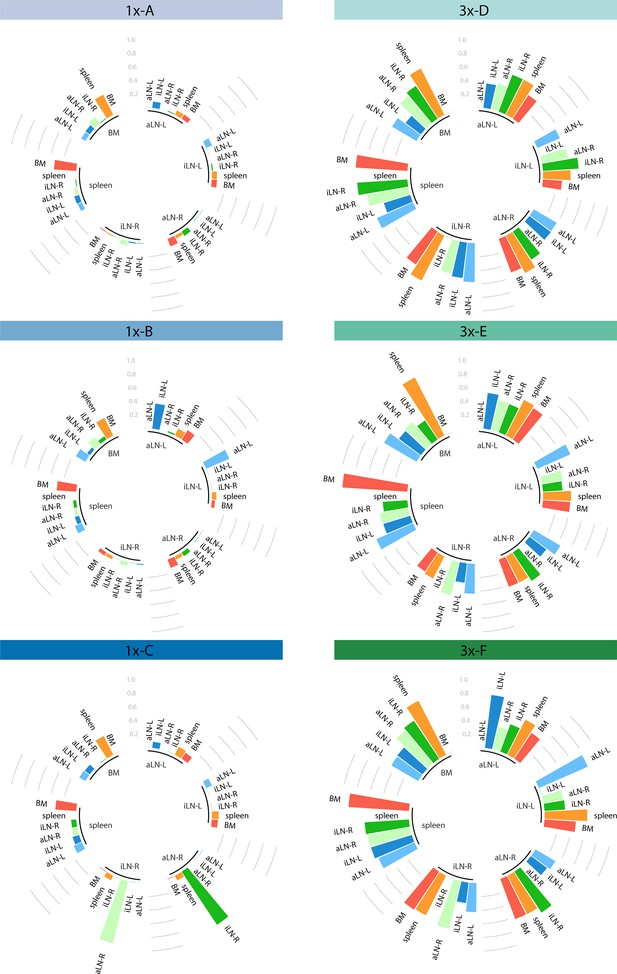
Repertoire similarity based on pairwise cosine similarities.
Graphs depicting cosine similarities of clones between organ pairs for each mouse per cohort. Cosine similarity measurements represent the degree of repertoire similarity between two organs by including both clonal overlap and frequency information (by giving weight to clonal frequencies), ranging from 0 to 1. Each section of the plot represents one organ and each bar depicts the cosine similarity of the given organ to another of the same mouse. Higher cosine values correspond to higher repertoire similarities between two organs. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes.
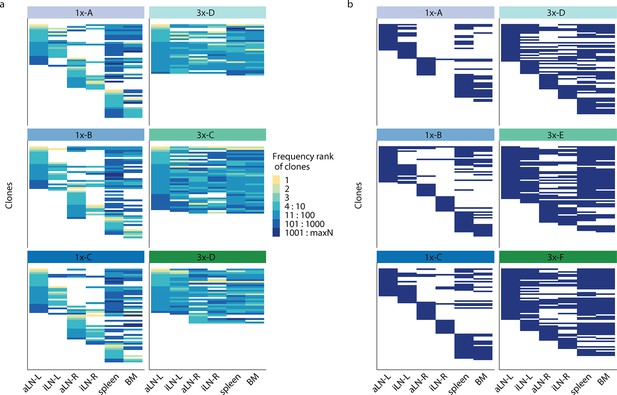
Tracking of diversity ranked clones across organs.
(a) String plots to determine if clones shared between lymphoid organs displayed similar frequencies. Tracking of the top 10 frequency-ranked clones of each lymphoid organ across all six organs within each mouse is shown. String plots are based on Boolean values (presence or absence of a clone in the corresponding organ) with each column representing one lymphoid organ and each line representing one clone (Rosenfeld et al., 2018). Each line (clone) that is in the same position across columns (organs) represents an identical shared clone. The color encodes the frequency rank of the clone within the specific organ (see color legend). (b) Plots for tracking the top 100–110 frequency-ranked clones of each lymphoid organ across all six organs within each mouse. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes.
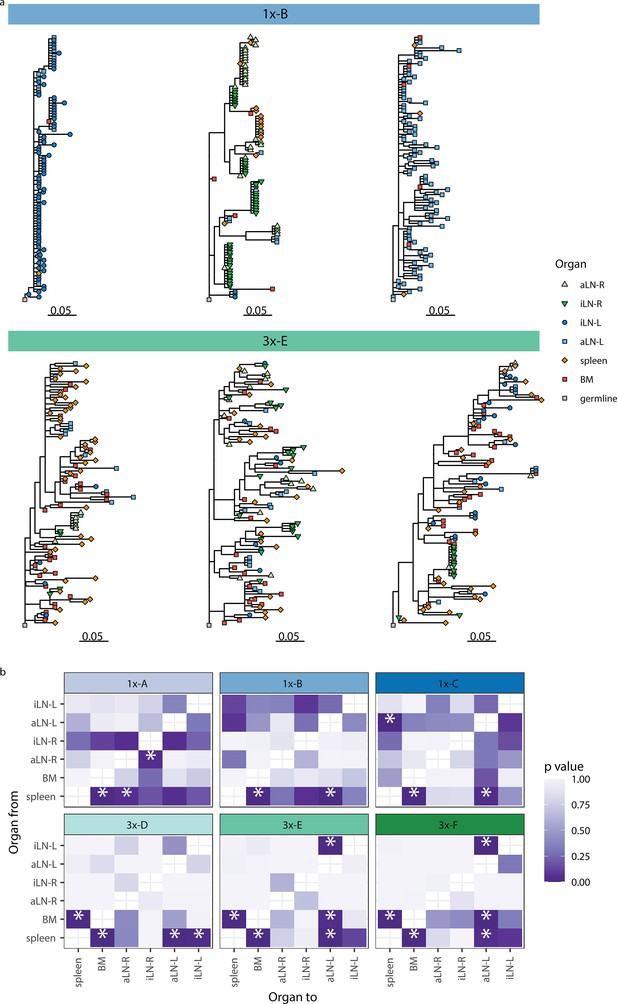
Phylogenetic analysis reveals physiological axes, diversification, and sharing of clonotypes across multiple lymphoid organs.
(a) Phylogenetic trees of the three most diverse clonotypes from a representative mouse from the single (1x-B) or multiple (3x-E) immunization cohorts. Color of nodes indicates the lymphoid organ origin of each tip with gray nodes depicting the clonotype’s unmutated germline sequence. Scale bar shows somatic hypermutations per codon site. See Figure 4—figure supplement 1a for phylogenetic trees of the other cohort-1x and cohort-3x mice. (b) Heatmaps displaying p-values derived from switch proportion (SP) tests, which quantify enrichment of transitions from one organ to another within lineage trees. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes. Asterisks indicate an SP test p<0.05.
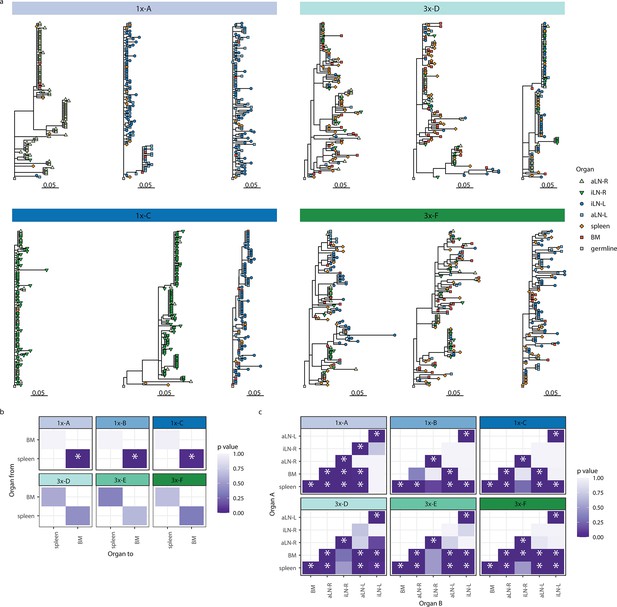
Phylogenetic analysis.
(a) Phylogenetic trees of the three most diverse clonotypes from mice 1x-A, 1x-C, 3x-D, and 3x-F. Color of nodes indicates the lymphoid organ origin of each tip with gray nodes depicting the clonotype’s unmutated germline sequence. Scale bar shows somatic hypermutation per codon site. (b) Heatmaps displaying p-values of a switch proportion (SP) test, using only trees built from spleen and bone marrow sequences to quantify enrichments of transitions within clonotypes between spleen and bone marrow. (c) SP test permuting organ labels among trees and quantifying enrichment of transitions in either direction. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes. Asterisks indicate an SP test p<0.05.
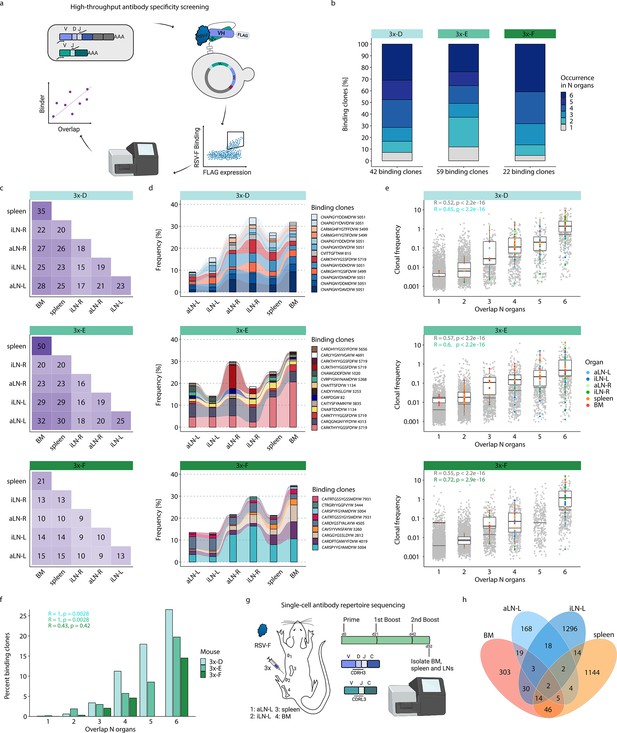
High-throughput antibody screening reveals that clonal overlap across lymphoid organs correlates with antigen-specificity.
(a) Workflow of high-throughput antibody screening by yeast display: antibody (scFv) libraries are generated by combinatorial pairing of variable heavy (VH) and variable light (VL) genes derived from BM of cohort-3x mice and expressed on the surface of yeast cells, followed by isolation of antigen-binding cells by fluorescence-activated cell sorting (FACS) and identification of clones by deep sequencing and repertoire analysis. (b) Bar plots represent proportions of binding clones occurring in N (1–6) organs within each corresponding mouse of cohort-3x (3x-D, 3x-E, and 3x-F). (c) Heatmaps depicting numbers of shared binding clones between two organs within each mouse. (d) Tracking of binding clones shared across all six lymphoid organs and their frequencies for each mouse. Legend displays CDRH3 of each clone with the number indicating the corresponding clonotype group. (e) Distribution of clonal frequencies per degree of organ overlap (1–6) for all clones (gray) and binding clones (colored). Gray horizontal lines represent the median frequency of all clones, black lines including box plots represent the median frequency of binding clones within the corresponding overlap sections (see color legend for organ origin). R and p-values of Spearman correlation analysis for all clones (gray) and binding clones only (green) are depicted. (f) Bar plots displaying ratios of binding clones to all clones with regard to their degree of organ overlap (1–6) for each mouse. R and p-values of Spearman correlation analysis for each mouse are depicted. (g) Workflow for RSV-F immunizations into the left lateral flank of one Balb/c mouse, followed by single-cell antibody repertoire sequencing (VH + VL) of lymphoid organs (BM, spleen, aLN-L, iLN-L). (h) Venn diagram of shared unique IgG+ clones (based on identical V- and J- genes and CDRH3+CDRL3) across lymphoid organs. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes. Panels (a) and (g) created with BioRender.com.
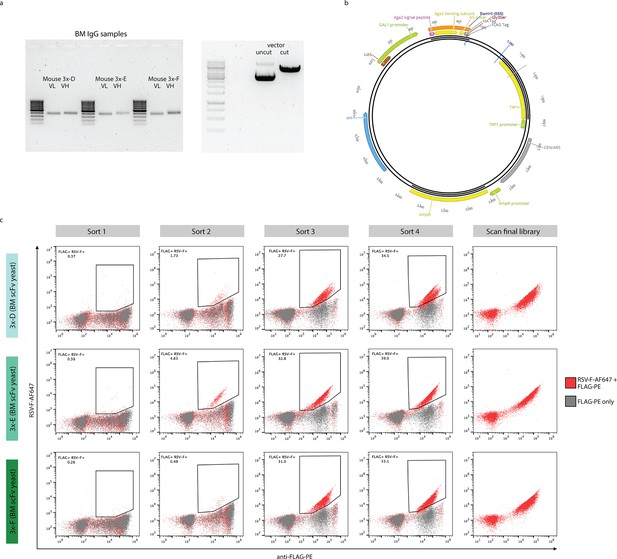
High-throughput antigen-specificity screening using scFv yeast display libraries.
(a) Representative gel plots showing amplified variable light (VL) and variable heavy (VH) genes of BM samples for cohort-3x mice (3x-D, 3x-E, 3x-F), as well as both unrestricted and BamHI-restricted yeast surface display vector. (b) Schematic of yeast surface display vector used for generation of scFv yeast display libraries. (c) FACS enrichment of RSV-F-specific binders using scFv yeast display. Representative flow cytometry dot plots show scFv yeast display library screenings for BM samples 3x-D, 3x-E, 3x-F by four progressive enrichment steps for FLAG-PE+ (x-axis) and RSV-F-AF647+ (y-axis) double-positive cells (red, sort1-sort4). Negative controls containing cells stained for FLAG expression only (gray) were used to set the sorting gates. Representative flow cytometry plots are shown to illustrate successful enrichments. The final sorted populations of yeast cells were screened to confirm purity before sequencing libraries were prepared.
-
Figure 5—figure supplement 1—source data 1
Gel plot of amplified VL and VH genes.
Agarose gel electrophoresis (1%) of PCR-amplified VL and IgG VH genes from bone marrow (BM) samples of cohort-3x mice (3x-D, 3x-E, 3x-F), with 100 bp DNA size marker.
- https://cdn.elifesciences.org/articles/92718/elife-92718-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
Gel plot of yeast surface display vector.
Agarose gel electrophoresis (1%) of both unrestricted and BamHI-restricted pYD1 yeast surface display vectors, with 1 kb DNA size marker.
- https://cdn.elifesciences.org/articles/92718/elife-92718-fig5-figsupp1-data2-v1.zip
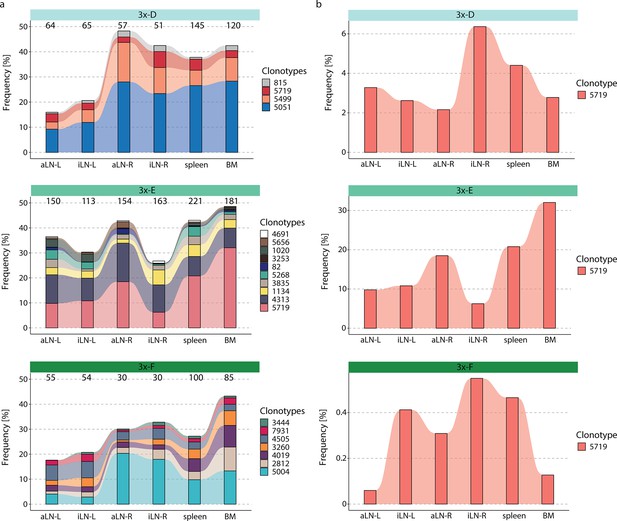
Tracking of clonotypes across all lymphoid organs.
(a) Tracking of clonotypes that are associated with binding clones across all six lymphoid organs and their frequencies for each mouse. Numbers above each column indicate the numbers of unique clones belonging to the clonotypes within the corresponding organ. (b) Frequency tracking of a public clonotype (containing binding clones) across all three cohort-3x mice (y-axis display different ranges). BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes.
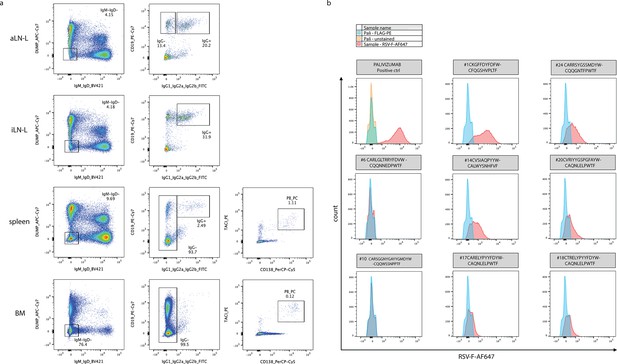
Isolation of B-cell subsets and screening of monoclonal yeast cells expressing single-chain variable fragments (scFvs).
(a) Overview of the FACS strategy used for the isolation of specific B-cell subsets for each organ as input for single-cell sequencing. (b) Flow cytometry histograms showing RSV-F-binding profiles of 9 representative monoclonal single-cell scFv-expressing yeast clones, selected among the 18 tested clones according to overlap analysis (including a positive control expressing RSV-F-binding monoclonal antibody palivizumab as scFv). Clones #1, 14, 17, 18, 20, and 24 were considered RSV-F-specific, whereas clones #6 and 10 are representatives for nonbinding profiles. Clones were pre-gated on FLAG-PE expression. BM: bone marrow; aLN-L: left axillary lymph node; iLN-L: left inguinal lymph node.
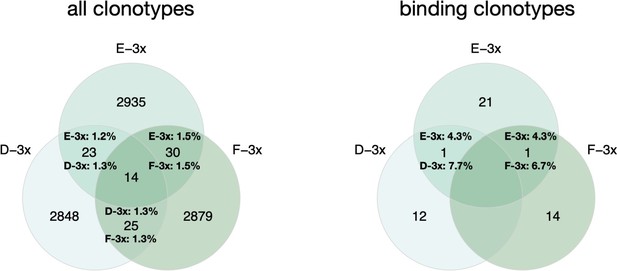
left: Venn diagram depicting numbers of shared clonotypes across cohort 3-x mice.
Percentages indicate the ratio of overlapping clones for the corresponding mouse repertoire. Right: Venn diagram depicting numbers of shared binding clonotypes across cohort 3-x mice. Percentages indicate the ratio of overlapping binding clones.
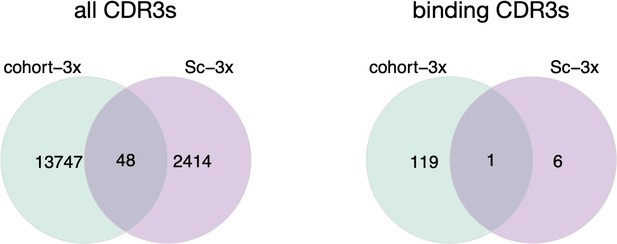
left: Venn diagram depicting numbers of shared CDR3s across cohort-3 mice (n=3, bulk sequenced) and one mouse (Sc-3x; immunized 3x, 10X single-cell sequenced).
Right: Venn diagram depicting the numbers of shared binding CDR3s across cohort-3x mice and Sc-3x mouse.
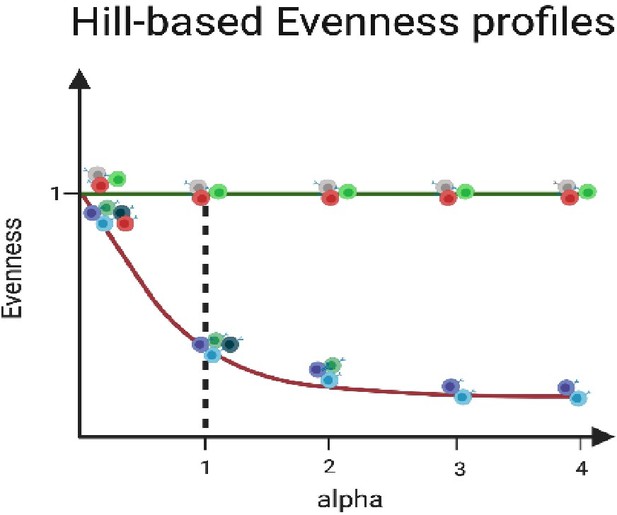
Exemplary Hill-based evenness profiles.
Evenness profiles allow visualization of the state of clonal expansion within antibody repertoires. The green line depicts an even (uniform) repertoire in which all clones are equally abundant, while the purple line represents a clonally expanded (polarized) antibody repertoire with one or few highly expanded clones. The dotted line indicates the Shannon evenness (α = 1); with the repertoire indicated in purple showing a low Shannon Evenness α=1E value, resembling a polarized repertoire compared to the green repertoire.
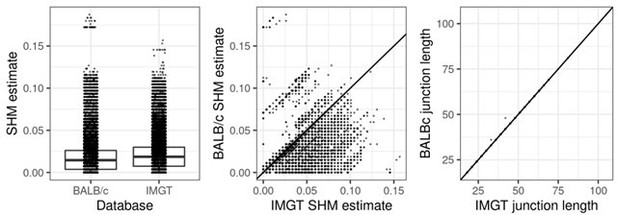
SHM and junction length estimates using the BALB/c reference versus the IMGT reference databases.
Using the BALB/c reference produces overall lower SHM estimates without affecting junction lengths. This shows the BALB/c reference is a better match to the data than the IMGT reference. Sequences above the diagonal line in the middle panel represent 1% of sequences that had higher SHM under the BALB/c database, and were excluded.
Additional files
-
Supplementary file 1
Sequence input into molecular amplification fingerprinting (MAF) pipeline.
Numbers of quality-processed and length-trimmed sequencing reads as input into the MAF pipeline for error and bias correction for mice 1x-A, 1x-B, 1x-C, 3x-D, 3x-E, and 3x-F. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes.
- https://cdn.elifesciences.org/articles/92718/elife-92718-supp1-v1.docx
-
Supplementary file 2
Clonal output after MAF processing.
Numbers of unique clones (and clonotypes; defined by antibody sequences possessing identical germline V- and J-genes and 90% CDRH3 a.a. identity and identical length) obtained after MAF processing. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes.
- https://cdn.elifesciences.org/articles/92718/elife-92718-supp2-v1.docx
-
Supplementary file 3
Overlap of the five most diverse clonotypes across lymphoid organs.
Table indicates overlap of the five most diverse clonotypes (top 1–top 5) across lymphoid organs within each mouse. The numbers represent organs (1: aLN-L; 2: iLN-L; 3: iLN-R; 4: aLN-R; 5: spleen; 6: BM) that share the same clonotypes, being among the top five in all indicated organs. BM: bone marrow; aLN-L, -R: left and right axillary lymph nodes; iLN-L, -R: left and right inguinal lymph nodes.
- https://cdn.elifesciences.org/articles/92718/elife-92718-supp3-v1.docx
-
Supplementary file 4
B-cell sort and 10× cell yield.
Numbers of FACS-isolated B-cell subsets per organ, as well as the yield of all cells and IgG+ B cells after single-cell V(D)J sequencing of antibody repertoires.
- https://cdn.elifesciences.org/articles/92718/elife-92718-supp4-v1.docx
-
Supplementary file 5
Overlap analysis for the identification of antigen-specific antibody sequences.
CDRH3 and CDRL3 information of IgG+ B cells shared among at least three organs, including cell counts for each organ. Black check marks indicate antibodies tested for RSV-F-binding as scFv format in yeast cells. Green check marks indicate confirmed RSV-F-binding of single-cell clones, whereas gray symbols indicate presumptive binding/non-binding of clones belonging to the corresponding clonotype. BM: bone marrow; aLN-L: left axillary lymph node; iLN-L: left inguinal lymph node.
- https://cdn.elifesciences.org/articles/92718/elife-92718-supp5-v1.docx
-
Supplementary file 6
Primers used for yeast display screening and NGS library preparation.
VL-binding primers p3-26 were adapted from Reddy et al., 2010 and IgG- and VH-binding primers p1, 2, 27–45 were adapted from Khan et al., 2016.
- https://cdn.elifesciences.org/articles/92718/elife-92718-supp6-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/92718/elife-92718-mdarchecklist1-v1.docx





