Signatures of transposon-mediated genome inflation, host specialization, and photoentrainment in Entomophthora muscae and allied entomophthoralean fungi
Figures

A new Entomophthora muscae genome assembly and comparative entomophthoralean datasets.
(A) Fungal cladogram, based on Spatafora et al., 2016. All unshaded taxa are fungal phyla. Green-shaded branches are subphyla within phylum Zoopagomycota, purple-shaded branches are orders within subphylum Entomophthoromycotina. Branch lengths are not proportional to phylogenetic distance. (B) Overview of sequencing, assembly and annotation statistics for new E. muscae genome assembly. Asterisk indicates that this value is for reads that passed the base-calling threshold only, not all bases sequenced. (C) Cladogram presenting the evolutionary relationships of the Entomophthorales species considered in this study. Red-shaded branches are genera within the family Entomophthoraceae. Phylogenetic tree was constructed with FastTree using a concatenated set of conserved protein coding genes. Parentheses around Pandora formicae and Strongwellsea castrans indicate that transcriptomic datasets were used for these species; for all other species genomic datasets were used. Tree branch length is proportional to phylogenetic distance (substitution rate given in legend below). Species whose genomes comprised our core analysis set are colored. Classification of host specificity of each fungus (i.e. specialist or generalist) is denoted with a box to the right of the species name. Specialist species infect a narrow host range; generalists can infect a broad range of species. An example depiction of a host killed by each of these fungi is drawn to the right. Host tissues are gray, with fungal conidiophores depicted in black. Hosts (top to bottom): fruit fly (Drosophila melanogaster adult), spongy moth (Lymantria dispar larva), periodical cicada (Magicicada septendecim adult), ant (Formica exsecta adult), cabbage maggot fly (Delia radicum adult), Bagrada bug (Bagrada hilaris adult), aphid adult (Aphididae), and planthopper (Delphacodes kuscheli adult). (D) BUSCO completeness estimates for the predicted proteome corresponding to species listed in C, using fungi_odb10 BUSCO set (Simão et al., 2015).

Kmer distributions within E. muscae genome assembly.
(A) 33mers; (B) 29mers. Jellyfish was used to count kmers and plots were generated with GenomeScope. ‘Len’ indicates estimated assembly size (in bp); ‘uniq’: % of unique kmers observed; ‘kcov’: estimated coverage of assembly; ‘err’: % error kmers; ‘dup’: %duplicated kmers; ‘het’: indicates % of heterozygous bases; ‘k’: kmer size.
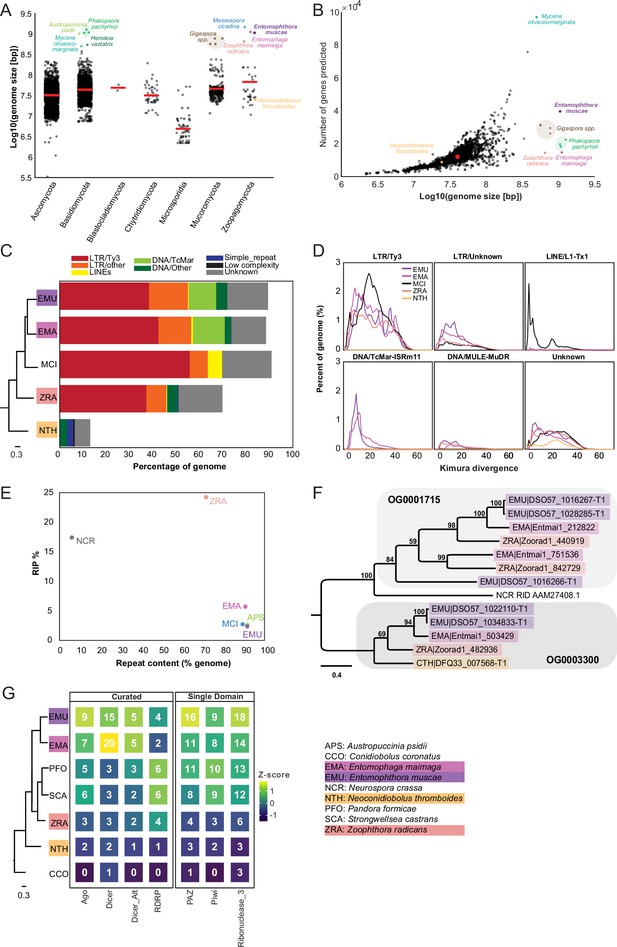
Entomophthoralean genomes are enlarged (relative to those of other fungi) due to proliferation of transposable elements.
(A) Genome assembly sizes of fungal phyla. Red lines indicate mean values per phylum. Species in the core Entomophthorales genomic analysis set are labeled (color-coded per Figure 1C). (B) Observed genome sizes versus predicted gene number in sequenced fungal genomes. Excluding genomes in excess of 500 Mb, the median genome size and number of genes across this dataset is 37.1 Mb and 11,843, respectively (red star). For both A and B, genomes exceeding 500 Mb are indicated by dots colored by species identity. Data for A and B available in Supplementary file 2. (C) Repeat element composition within E. muscae (EMU), E. maimaiga (EMA), M. cicadina (MCI), Z. radicans (ZRA), and N. thromboides (CTH) as determined by RepeatMasker (Smit et al., 2013). Only repeat elements that exceeded 0.1% of the genome for at least one species are shown. Cladogram modified from Figure 1C. (D) Landscapes of DNA repeat elements with those comprising less than 1% of DNA repeats in each genome binned as other. (E) Percentage of genome in which RIP was detected versus percent of genome comprised of repeat content. (F) Protein phylogeny representing relationships among two methyltransferase-containing orthogroups and RID from Neurospora crassa: a cytosine methyltransferase required for RIP (Freitag et al., 2002). Scale bar indicates 0.3 substitutions per site. (G) Counts of selected Pfams associated with RNAi pathway components across genomes. Curated counts include only candidates with the expected combination (and frequency) of Pfams for each of the listed RNAi pathway proteins. Cladogram modified from Figure 1C.

Comparison of domain architecture/gene content across Entomophthorales.
(A) Pfams significantly overrepresented in E. muscae (EMU) compared to other species analyzed E. maimaiga (EMA), Z. radicans (ZRA), S. castrans (SCA), P. formicae (PFO), N. thromboides (NTH), and C. coronatus (CCO). Bars represent the counts for E. muscae colored by fold-versus-the-median across all genomes. Point size represents the number of significant pairwise comparisons among other genomes and these are colored according to whether the value is above, below or equal to the median value across all genomes. Cladogram modified from Figure 1C. (B) Pfams significantly underrepresented in E. muscae compared to other species analyzed. Plot format as in A. (C) Combined UpSet plots showing the intersection among genomes for all domain categories (CAZy, Pfam and MEROPS; bar colors). (D) Counts of selected Pfams associated with circadian proteins across genomes. Curated counts include only candidates with the expected combination of Pfam domains for each of the listed circadian proteins. (For example, to be considered a curated wc-1 candidate, a gene needed to have one each of GATA, PAS_3 and PAS_9 domains.) (E) Venn diagram depicting intersections between predicted OGs among E. muscae, E. maimaiga, Z. radicans, and N. thromboides. Values only within a single ellipse indicate the abundance of species-specific genes. (F) Occupancy trends by species for 11,7111 OGs recovered from amalgamating E. muscae, Entomophaga maimaiga, Z. radicans and N. thromboides gene models. ‘Contain at least one gene’: percentage of OGs that contain at least one gene from a given species. ‘Species-specific’ is the percentage of OGs that only contain genes from one species. (G) Percentage of genes for each species found in each OG type as percentages of total genes annotated in the genome. ‘Assigned OG’ are genes that clustered with anOG; ‘Not assigned OG’ are genes that did not cluster with any OG. ‘Within species-specific OG’ reflects the percentage of genes that fall within an OG that is only populated by genes of the given species. ‘Potentially species-specific’ is the sum of genes ‘Not assigned OG’ and ‘Within species-specific OG’. The light purple bar marked with a black asterisk indicates the percentage of genes that are potentially species-specific with evidence of high expression in an in vivo dataset (expression <5, pooled dataset of 27 whole fly samples exposed with E. muscae).

Pfam UpSet plot analysis including E. muscae transcriptome.
UpSet plots displaying Pfam domain intersections including additional predictions from an E. muscae transcriptomic dataset (EMU-T; NCBI GSE111046), showing the intersection among included genomes and transcriptomes. This analysis compares the genome predictions (i.e. EMU, EMA, ZRA, CTH, and CCO) to the transcriptome predictions (i.e. EMU-T, SCA, and PFO). Blue highlights the Pfam domains uniquely shared by SCA and PFO. Orange highlights the Pfam domains uniquely shared by all transcriptomic datasets in this analysis.
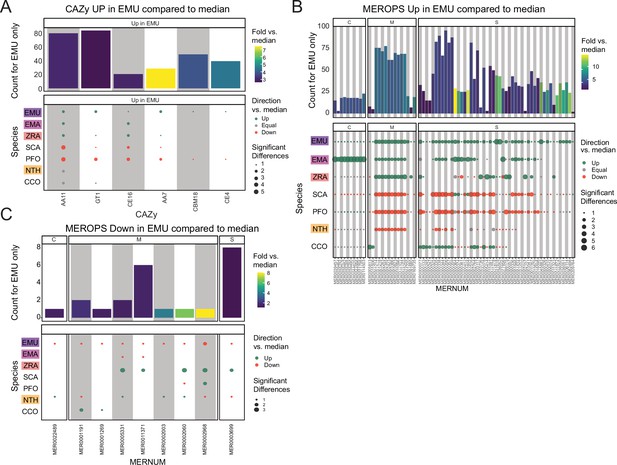
Additional domain analysis for CAZy and MEROPS databases.
(A) CAZy domains significantly overrepresented in E. muscae (EMU) compared to other species analyzed (E. maimaiga (EMA), Z. radicans (ZRA), S. castrans (SCA), P. formicae (PFO), N. thromboides (CTH), and C. coronatus (CCO)). Bars represent the counts for E. muscae colored by fold-versus-the-median across all genomes. Point size represents the number of significant pairwise comparisons among other genomes and are colored according to whether the value is above, below or equal to the median value across all genomes. (B) Plots following a similar format as panel A, displaying MEROPS domains that were found to be overrepresented in EMU by comparison, categorized by MEROPS peptidase category (C: cysteine peptidases, M: metallopeptidases, S: serine peptidases). (C) Plot following a similar format to panel B, displaying MEROPS domains that were found to be underrepresented in EMU by comparison.
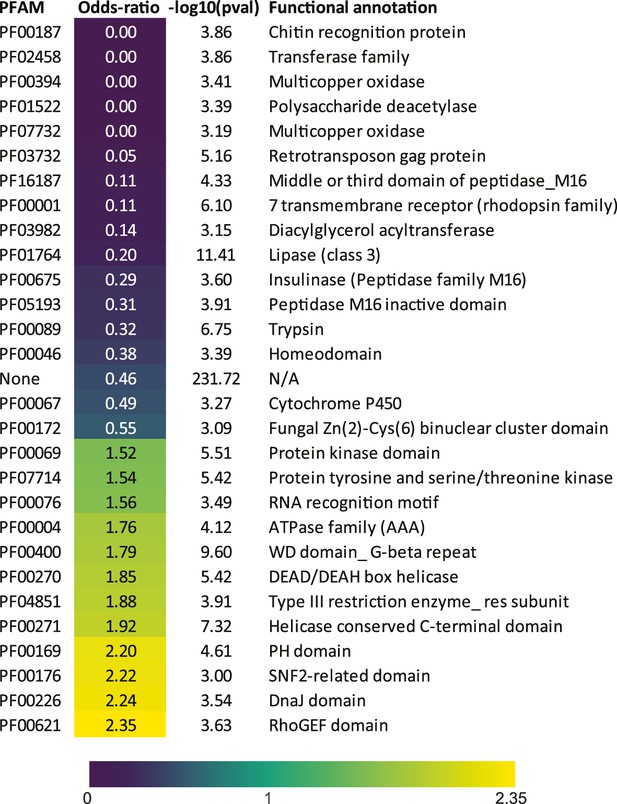
E. muscae core OG Pfam enrichment.
Enrichment among Pfam annotations for E. muscae genes belonging to core OG set. Odds-ratios are colored according to the scale bar below.
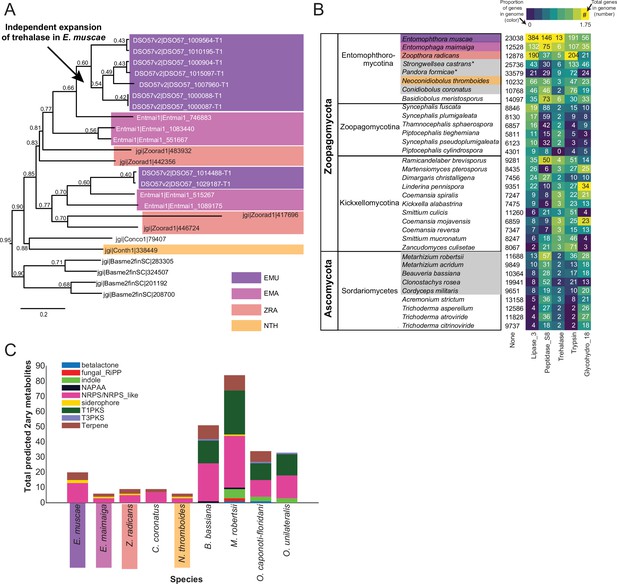
Gene family expansion and secondary metabolite production of E. muscae and other insect pathogens.
(A) A family of genes encoding extracellular trehalase enzymes (PF01204) is expanded in E. muscae (EMU) compared to other Zoopagomycetes: E. maimaiga (EMA), Z. radicans (ZRA), N. thromboides (NTH), C. conidiobolus (Conco1), and B. meristosporus (Basme2finSC). (B) Total number of genes and number of genes encoding Lipases (Lipase_3), Subtilisin-like serine peptidases (Peptidase_S8), Trehalases (Trehalase), Trypsins (Trypsin), and Chitinases (Glycohydro_18) in representative fungal species of Zoopagomycota and Ascomycota. Fungal species in gray are insect pathogens and the four Entomophthoromycotina species are outlined in the same colors as A. Numbers inside heatmap refer to the number of genes that encode a given Pfam domain, and color scale refers to proportion of genes with a given Pfam compared to the total number of genes in the genome (in percentages). (C) Predicted secondary metabolite production for select entomophthoralean genomes (E. muscae, E. maimaiga, Z. radicans, C. coronatus and N. thromboidies) and common ascomycete entomopathogens (B. bassiana, M. robertsii, O. caponoti-floridani, O. unilateralis), as predicted by AntiSMASH. Color indicates metabolite class.
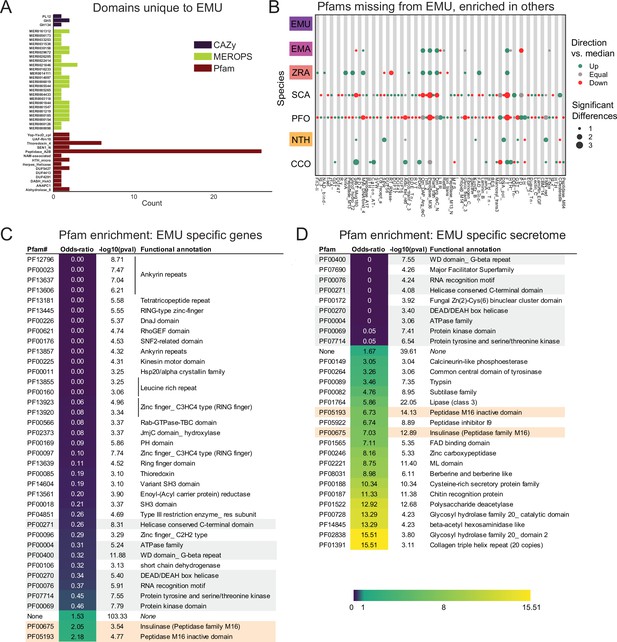
Unique features of E. muscae compared to E. maimaiga, Z. radicans and N. thromboides.
(A) Domains unique to E. muscae. (B) Pfam domains that are missing in E. muscae, but enriched in other entomophthoralean fungi. (C) Significantly enriched Pfam domains (p ≤ 0.001) within genes that are potentially E. muscae-specific (both genes that did not cluster with any orthogroup and genes that cluster with orthogroups that are species-specific; N=9,150 genes). (D) Significantly enriched Pfam domains (p-value ≤ 0.001) within potentially E. muscae-unique genes encoding proteins predicted to be secreted (N=1685 genes). Odds-ratios are colored according to the scale bar to bottom right. Two Pfam domains (PF00675 and PF05193; highlighted orange in C and D) are overrepresented in potentially E. muscae-specific E. muscae genes both genome-wide and within the predicted secretome. Pfam domains highlighted in gray are underrepresented across both of these sets.

CAZy and MEROPs domains missing in E. muscae and enriched in other fungi.
CAZy and MEROPS domains missing from E. muscae (EMU), but significantly underrepresented (red) or overrepresented (green) in other species analyzed (E. maimaiga (EMA), Z. radicans (ZRA), S. castrans (SCA), P. formicae (PFO), N. thromboides (CTH), and C. coronatus (CCO)).
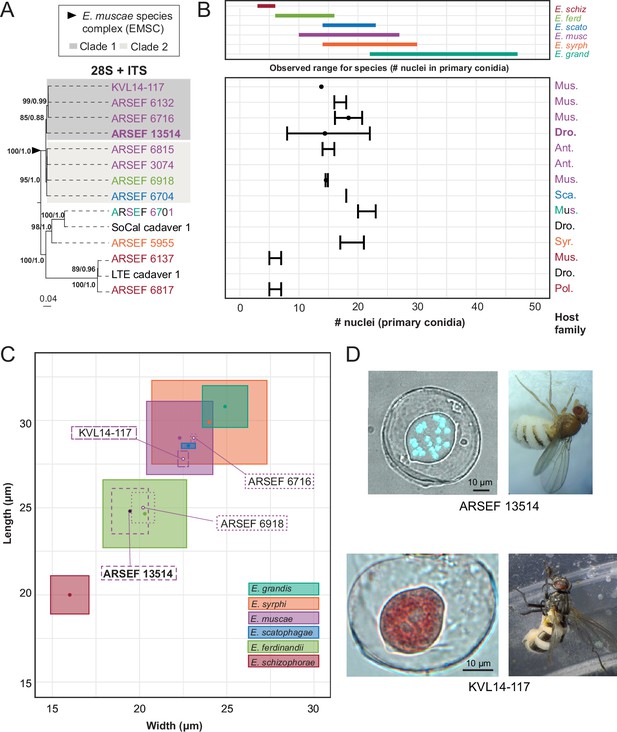
Phylogenetic and morphological data for E. muscae ARSEF 13514, members of the Entomophthora muscae species complex, and closely allied fly-infecting Entomophthora spp.
Across all panels, species are designated by color (see key in B). (A) Concatenated ITS +28 S phylogenetic tree of representative Entomophthora spp. including diverse strains across the EMSC. Gray boxes indicate distinct well-supported clades within the EMSC. Topology and branch lengths shown are from the ML analysis. Bootstrap support and posterior probabilities are indicated near each node (ML / BI), and only nodes with >50% support are labeled. ARSEF 6701 is denoted by three colors to indicate that this strain has multiple identifications (E. grandis (teal), E. muscae (purple) and E. sp. (black)). (B) Nuclei number of primary conidia among strains (bottom) relative to the known ranges for each of six described species included in this study as defined by Keller, 2002 (top). Fly family is noted at the far right for each strain (Mus.=Muscidae, Dro.=Drosophilidae, Ant.=Anthomyiidae, Sca.=Scathophagidae, Syr.=Syrphidae, and Pol.=Polleniidae). For panel A, ITS and 28 S sequence data for ARSEF 13514 (ITS and 28 S), ARSEF 6918, SoCal cadaver 1 and LTE cadaver 1 are original to this study, while data for the remainder are from the literature (see Supplementary file 1c). (C) Primary conidial length and width for strains with published spore measurements, overlaid atop measurements reported for each of the six species. Isolate in bold is E. muscae described in this study. (D) Primary conidia (left) and fly hosts (right) for E. muscae strains ARSEF 13514 (above) and KVL14-117 (below). The E. muscae ARSEF 13514 primary conidium is stained with Hoechst 33342 to visualize multiple nuclei contained within conidium. The E. muscae KVL 14–117 conidium is stained with aceto-orcein. Scale bars: 10 μm.
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/92863/elife-92863-mdarchecklist1-v1.docx
-
Supplementary file 1
Summary data for gene counts using different annotation methods, tables of fungal isolates, and RNA-seq datasets used in this study.
(a) Variance in predicted gene models using different annotation pipelines does not explain the large gene count predicted in the E. muscae genome. (b) Summary of fungal isolates and data used in this manuscript. (c) Information about strains used in phylogenetic and morphologic studies (related to Figure 6). (d) SRA accession numbers of E. muscae RNAseq data (NCBI GSE111046) used for pooled expression analysis (related to Figure 3).
- https://cdn.elifesciences.org/articles/92863/elife-92863-supp1-v1.xlsx
-
Supplementary file 2
Genome sizes and gene counts across Fungi (related to Figure 2).
- https://cdn.elifesciences.org/articles/92863/elife-92863-supp2-v1.xlsx





