N-terminus of Drosophila melanogaster MSL1 is critical for dosage compensation
Figures

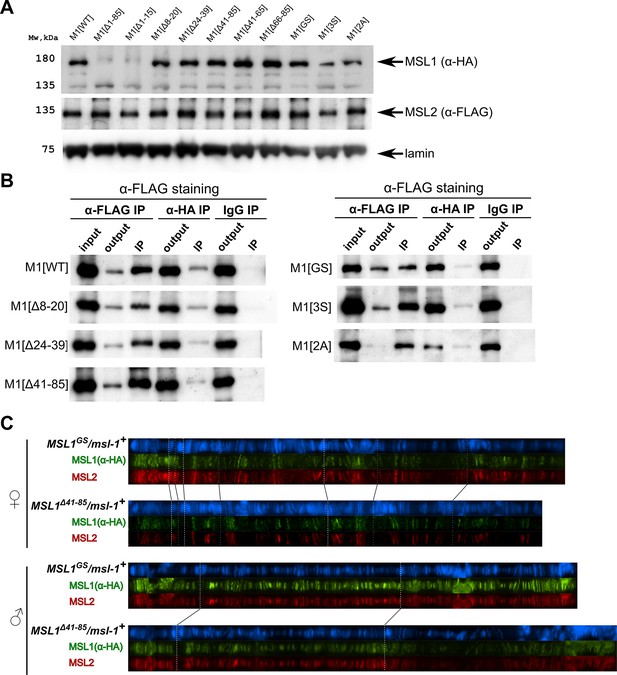
Testing the functional activity of male-specific lethal (MSL)1 mutants.
(A) Schematic presentation of the MSL1 deletion proteins expressed in flies. Alignment of the N-terminal domain (1–207 aa) of MSL1 among Drosophilidae. (B) Immunoblot analysis of protein extracts prepared from adult males y1w1118 (control) and msl-1−; Ubi:msl-1WT/ Ubi:msl-1WT(M1[WT]), and from adult females y1w1118 (control), –msl-1− (msl-1L60/msl-1γ269), M1[WT] (msl-1−; Ubi:msl-1WT/Ubi:msl-1WT), and M1[WT]+MSL2 (msl-1−; Ubi:msl-2WT-FLAG / Ubi:msl-1WT). Immunoblot analysis was performed using anti-MSL1, anti-MSL2, and anti-lamin Dm0 (internal control) antibodies. (C) Immunoblot analysis of protein extracts prepared from adult females expressing different MSL1 protein variants (MSL1WT, MSL1Δ1-85 MSL1Δ1-15, MSL1Δ8-20 MSL1Δ24-39, MSL1Δ41-85, MSL1Δ41-65, MSL1Δ66-85) and y1w1118 females (control). Immunoblot analysis was performed using anti-MSL1, and anti-lamin Dm0 (internal control) antibodies. (D) Viability of males expressing MSL1 variants in the msl-1-null background (marked as M1[*]). Viability (as a relative percentage) of msl-1−(msl-1L60/msl-1γ269); Ubi:msl-1*/ Ubi:msl-1* males to msl-1−; Ubi:msl-1*/Ubi:msl-1* females obtained in the progeny of crosses between females and males with the msl-1−/ CyO, GFP; Ubi:msl-1*/Ubi:msl-1* genotype. Ubi:msl-1* is any transgene expressing one of the tested MSL1 variants. The results are expressed as the mean of three independent crosses, with error bars showing standard deviations. (E) Immunoblot analysis of protein extracts prepared from adult female flies expressing MSL2-FLAG in combination with different MSL1 protein variants (MSL1WT, MSL1Δ1-85 MSL1Δ1-15, MSL1Δ8-20 MSL1Δ24-39, MSL1Δ41-85, MSL1Δ41-65, MSL1Δ66-85) and y1w1118 females (control). Immunoblot analysis was performed using anti-MSL1, anti-MSL2, and anti-lamin Dm0 (internal control) antibodies.
-
Figure 1—source data 1
Original files for the immunoblot blot analysis are in Figure 1B, C and E.
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig1-data1-v1.zip
-
Figure 1—source data 2
Files containing original immunoblots for Figure 1B, C and E, indicate the relevant bands.
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig1-data2-v1.zip
-
Figure 1—source data 3
The spreadsheet with numbers of males and females expressing male-specific lethal (MSL)1 variants in the msl-1-null background and obtained in three independent experiments.
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig1-data3-v1.xlsx

Alignment of the N-terminal region of male-specific lethal (MSL)1 among Diptera.
Orange box – Coiled coil domain.
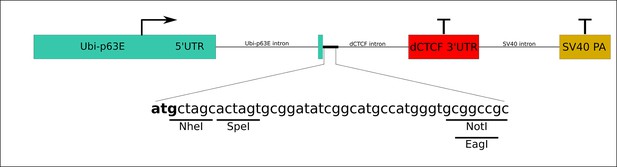
Scheme of the transgenic construct used to express male-specific lethal (MSL)1 variants.
Green boxes – promoter and 5’UTR of Ubi-p63E gene; red box – 3’UTR with polyadenylation signal from dCTCF gene; yellow box – polyadenylation signal from SV40 virus. Lines correspond to introns. Letters show the polylinker region used for cloning of MSL1 cDNAs.
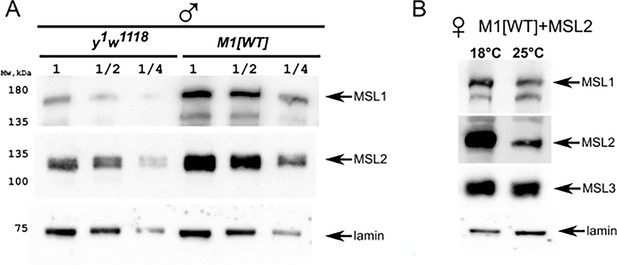
Immunoblot analysis of M1[wt] flies.
(A) Comparing of male-specific lethal (MSL)1 expression in y1w1118 and M1[wt] (msl-1−; Ubi:msl-1WT/ Ubi:msl-1WT) males. Male samples were titrated as twofold dilutions (‘1’, ‘1/2’, ‘1/4’). (B) Comparing of MSL1, MSL2, MSL3 expression in M1[wt]+MSL2 females (msl-1−; Ubi:msl-2WT-FLAG / Ubi:msl-1WT) growing at 18°C and 25°C. Immunoblot analysis was performed using anti-MSL1, anti-MSL2, anti-MSL3, and anti-lamin Dm0 (internal control) antibodies.
-
Figure 1—figure supplement 3—source data 1
Original files for the immunoblot blot analysis are in Figure 1—figure supplement 3.
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig1-figsupp3-data1-v1.zip
-
Figure 1—figure supplement 3—source data 2
Files containing original immunoblots for Figure 1—figure supplement 3 indicating the relevant bands.
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig1-figsupp3-data2-v1.zip
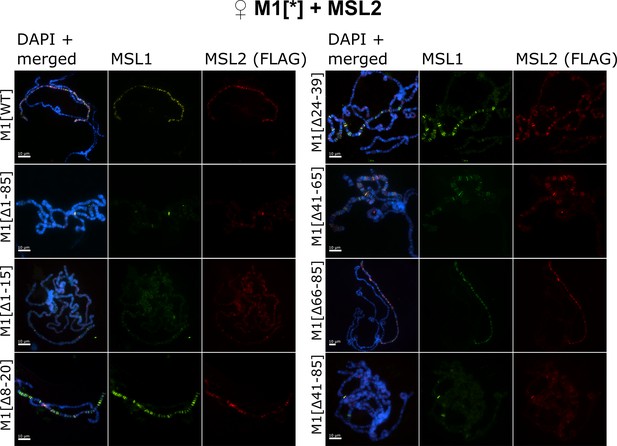
Testing the role of male-specific lethal (MSL)1 mutants in recruiting the MSL complex on the X chromosome in a female model system.
Distribution of the MSL complex on the polytene chromosomes from third day female larvae expressing both MSL1* variants and the MSL2WT-FLAG protein. Panels show the immunostaining of proteins using rabbit anti-MSL1 antibody (green) and mouse anti-FLAG antibody (red). DNA was stained with DAPI (blue). M1[WT](♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1WT); M1[Δ1–85] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ1-85); M1[Δ1–15] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ1-15); M1[Δ8–20] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ8-20); M1[Δ24–39] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ24-39); M1[Δ41–85] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ41-85); M1[Δ41–65] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ66-85); M1[Δ66–85] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ66-85).
-
Figure 2—source data 1
Raw files with images of polytene chromosomes.
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig2-data1-v1.zip
Testing the role of male-specific lethal (MSL)1 mutants in recruiting the MSL complex on the X chromosome in a female model system.
Distribution of the MSL complex on the polytene chromosomes from third day female larvae expressing both MSL1 variants and the MSL2-FLAG protein. Panels show the immunostaining of proteins using rabbit anti-MSL1 antibody (green) and mouse anti-FLAG antibody (red). DNA was stained with DAPI (blue). M1[WT] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1WT); M1[Δ1–85] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ1-85); M1[Δ1–15] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ1-15); M1[Δ8–20] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ8-20); M1[Δ24–39] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ24-39); M1[Δ41–85] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ41-85); M1[Δ41–65] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ66-85); M1[Δ66–85] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ66-85).
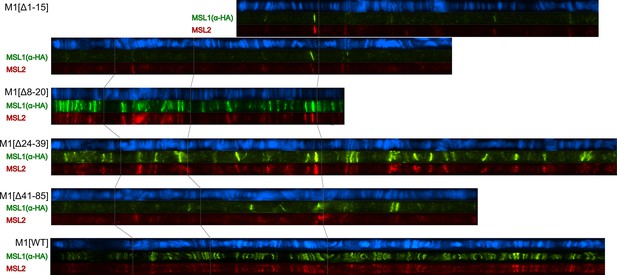
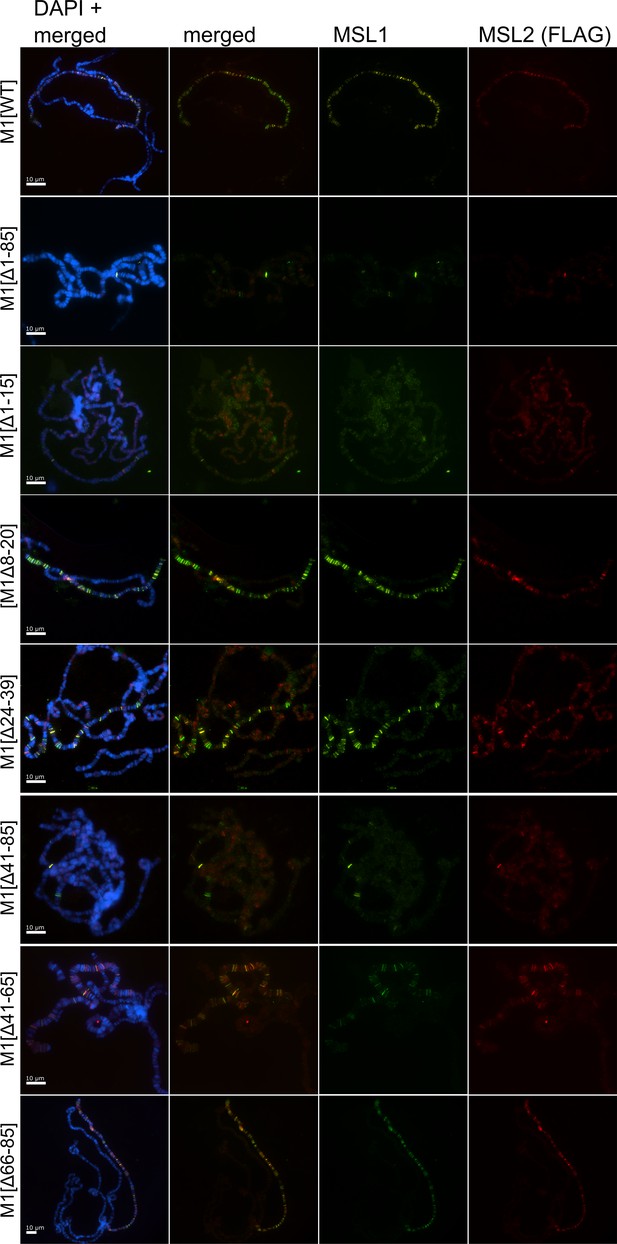
Cytological localization of male-specific lethal (MSL)1 variants and MSL2 on the straightened polytene chromosomes of the M1[Δ1–15] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ1-15); M1[Δ8–20] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ8-20); M1[Δ24–39] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ24-39); M1[Δ41–85] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ41-85); M1[WT] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1WT).
Panels show the immunostaining of proteins using mouse anti-HA antibody (green) and rabbit anti-MSL2 antibody (red). DNA was stained with DAPI (blue). Polytene chromosomes were stretched and compared using ImageJ 1.54 f/Fiji 2.14.0 software.

Testing the functional role of N-terminal amino acids 3–7 of male-specific lethal (MSL)1.
(A) Schematic presentation of the mutations at the N-terminus of MSL1. (B) Immunoblot analysis of protein extracts prepared from adult female flies expressing different MSL1* variants in the msl-1− (msl-1L60/msl-1γ269) background (M1[WT]: MSL1WT; M1[2 A]: MSL1F5A W7A, M1[3 S]: MSL1K3S R4S K6S; M1[GS]: MSL1K3S R4S F5G K6S W7G). (C) Immunoblot analysis of protein extracts prepared from adult female flies expressing MSL2-FLAG in combination with different MSL1* variants. Immunoblot analysis was performed using anti-MSL1, anti-MSL2, anti-MSL3, and anti-lamin Dm0 (internal control) antibodies. (D) Viability of males expressing MSL1 variants (marked as M1[*]) in the msl-1− (msl-1L60/msl-1γ269) background. Viability (as a relative percentage) of msl-1−; Ubi:msl-1*/ Ubi:msl-1* males to msl-1−; Ubi:msl-1*/Ubi:msl-1* females obtained in the progeny of a cross between females and males with msl-1−/ CyO, GFP; Ubi:msl-1*/Ubi:msl-1* genotype. Ubi:msl-1* is any transgene expressing one of the tested MSL1* variants. (E) Viability of males expressing MSL2WT and MSL1* variants (marked as M1[*]) in the msl-1− background. Viability (as a relative percentage) of msl-1−; Ubi:msl-2WT-FLAG /Ubi:msl-1* males to msl-1−; Ubi:msl-2WT-FLAG /Ubi:msl-1* females obtained in the progeny of cross between males msl-1−/ CyO, GFP; Ubi:msl-1*/Ubi:msl-1* and females msl-1−; Ubi:msl-2/Ubi:msl-2. The results are expressed as the mean of three independent crosses, with error bars showing standard deviations. (F) Distribution of the MSL complex on the polytene chromosomes from third day female larvae expressing both MSL1* variants and the MSL2-FLAG protein. Panels show the merged results of immunostaining of MSL1 (green, rabbit anti-MSL1 antibody) and MSL2 (red, mouse anti-FLAG antibody). DNA was stained with DAPI (blue). Scale bar indicates 10 µm.
-
Figure 3—source data 1
Original files for the immunoblot blot analysis in Figure 3B and C.
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig3-data1-v1.zip
-
Figure 3—source data 2
Files containing original immunoblots for Figure 3B and C indicating the relevant bands.
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig3-data2-v1.zip
-
Figure 3—source data 3
The spreadsheet with numbers of males and females expressing male-specific lethal (MSL)1 variants in the msl-1-null background and obtained in three independent experiments.
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig3-data3-v1.xlsx
-
Figure 3—source data 4
The spreadsheet with numbers of males and females expressing MSL2WT and male-specific lethal (MSL)1 variants in the msl-1-null background and obtained in three independent experiments.
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig3-data4-v1.xlsx
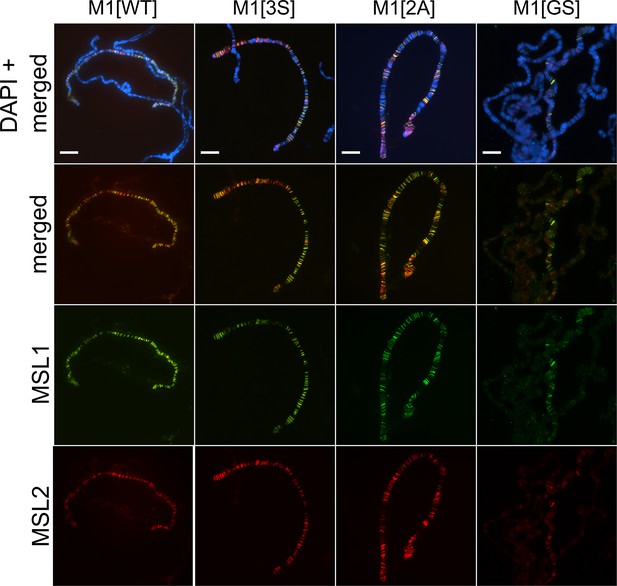
Distribution of the male-specific lethal (MSL) complex on the polytene chromosomes from third day female larvae expressing both MSL1* variants and the MSL2-FLAG protein.
Panels show the results of immunostaining of MSL1 (green, rabbit anti-MSL1 antibody) and MSL2 (red, mouse anti-FLAG antibody). DNA was stained with DAPI (blue). M1[WT] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1WT); M1[2 A] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-12A); M1[3 S] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-13S); M1[GS] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1GS). Scale bar indicates 10 µm.
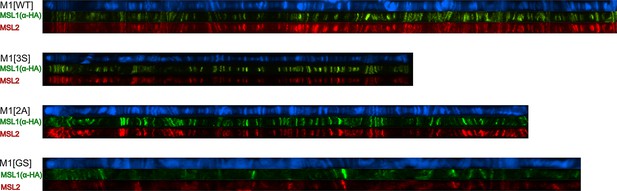
Cytological localization of male-specific lethal (MSL)1 variants and MSL2 on the straightened polytene chromosomes of the M1[WT] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1WT); M1[2 A] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-12A); M1[3 S] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-13S); M1[GS] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1GS). Panels show the immunostaining of proteins using mouse anti-HA antibody (green) and rabbit anti-MSL2 antibody (red). DNA was stained with DAPI (blue).
Testing the functional activity of mutant variants of the male-specific lethal (MSL)1 protein.
(A). Immunoblot analysis of protein extracts prepared from S2 cells expressing MSL2-FLAG and different MSL1-HAx3 protein variants (M1[WT]: MSL1WT; M1[Δ*]: MSL1Δ*; M1[2 A]: MSL12A, M1[3 S]: MSL13S; M1[GS]: MSL1GS). Immunoblot analysis was performed using anti-HA (MSL1-HAx3 variants), anti-FLAG (MSL2-FLAG), and anti-lamin Dm0 (internal control) antibodies. (B) Protein extracts from Drosophila S2 cells co-transfected with different MSL1-HAx3 variants and MSL2-FLAG were immunoprecipitated with antibodies against FLAG or HA or nonspecific mouse IgG as a negative control. The immunoprecipitates were analyzed by immunoblotting for the presence of FLAG-tagged proteins in immunoprecipitated samples. (C) Distribution of the MSL1*-HA and MSL2-FLAG on the polytene chromosomes from third day female or male larvae expressing one of MSL1*-HA variants and MSL2-FLAG at the wild-type background (msl-1+; Ubi:msl-2WT-FLAG /Ubi:msl-1*). Panels show the merged results of immunostaining of MSL1*-HA (green, mouse anti-HA antibody) and MSL2 (red, rabbit anti-MSL2 antibody). DNA was stained with DAPI (blue).
-
Figure 4—source data 1
Original files for the immunoblot blot analysis are in Figure 4A and B.
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig4-data1-v1.zip
-
Figure 4—source data 2
Files containing original immunoblots for Figure 4A and B indicating the relevant bands.
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig4-data2-v1.zip
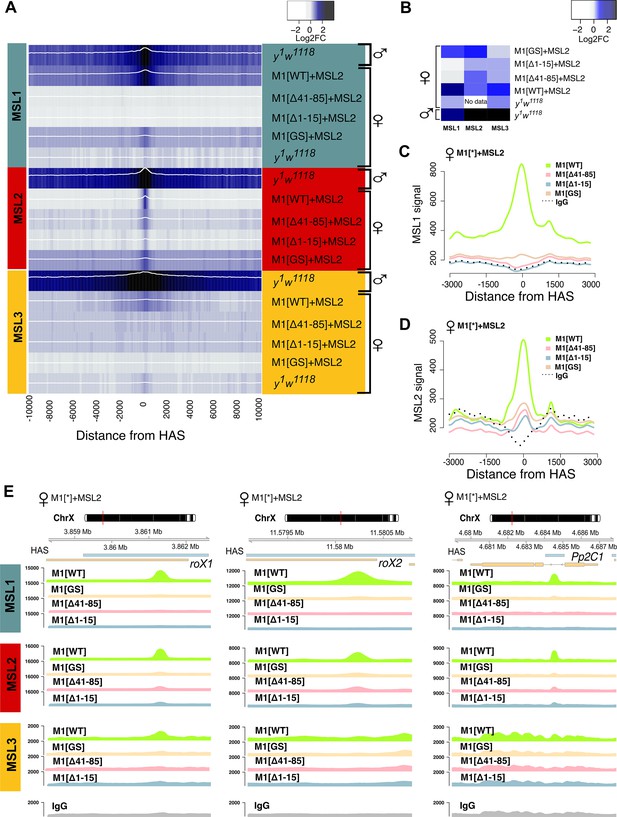
Role of the N-terminal regions in the recruitment of the dosage compensation complex to the high-affinity ‘entry’ sites (HAS).
To study the functional role of the N-terminal region of male-specific lethal (MSL)1 for the recruitment of the dosage compensation complex, we compared the profiles of MSL1, MSL2, and MSL3 binding in 2–3 day females expressing MSL2 and one of four MSL1 variants (MSL1WT, MSL1GS, MSL1Δ1-15, MSL1Δ41-85). y1w1118 (wild-type males and females), ♀M1[WT]+MSL2 (msl-1−; Ubi:msl-2WT-FLAG/Ubi:msl-1WT females), ♀M1[Δ41–85]+MSL2 (msl-1−; Ubi:msl-2WT-FLAG/Ubi:msl-1Δ41-85 females), ♀M1[Δ1–15]+MSL2 (msl-1−; Ubi:msl-2WT-FLAG/Ubi:msl-1Δ1-15 females), ♀M1[GS]+MSL2 (msl-1−; Ubi:msl-2WT-FLAG/Ubi:msl-1GS females). (A) Average log fold-change between normalized (RPKM) test signals and nonspecific IgG signals in HAS (see Materials and methods). Average log fold-change was calculated after smoothing signals using the Daniell kernel with kernel size 50 and the addition of a pseudocount. Next, the tracks were aggregated using 100 bp bins. (B) Heatmap showing average log fold-change between normalized (RPKM) test signals and nonspecific IgG signals in region ±500 bps from HAS centers. (C and D) Average signal (RPKM) for (C) MSL1 protein and (D) MSL2 protein in different fly lines at HAS (see Methods). (E) Enrichment of MSL1, MSL2, and MSL3 signals associated with the X-linked genes encoding Pp2C1 and the non-coding RNAs roX1 and roX2 in females expressing MSL2 and one of four MSL1 variants (MSL1WT, MSL1GS, MSL1Δ1-15, MSL1Δ41-85).
-
Figure 5—source data 1
A list of high-affinity 'entry' site (HAS) regions.
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig5-data1-v1.xlsx
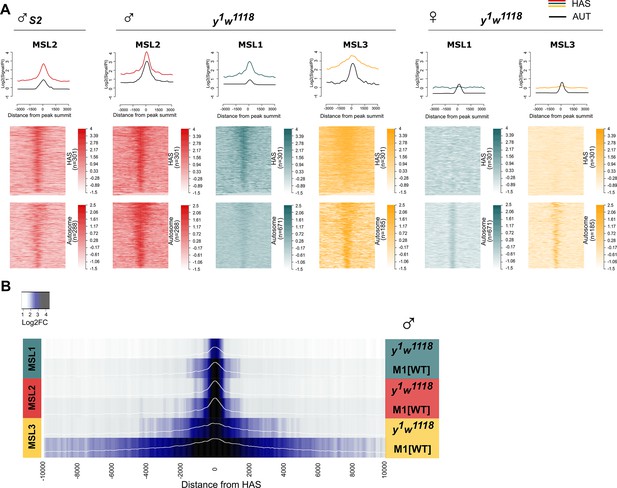
Comparison of MSL1, MSL2 and MSL3 binding in the flies of y1w1118 and M1[wt] lines.
(A) Comparison of male-specific lethal (MSL)1 (green), MSL2 (red), and MSL3 (yellow) binding in 2–3 day-old males (♂) and females (♀) of the y1w1118 line at HAS and autosomal sites for MSL proteins. Average signal (on the top) and heatmap (on the bottom) showing the log-fold change (FC) between normalized (Reads per kilobase of transcript, per million mapped reads, RPKM) test signals and nonspecific IgG signals in HAS and autosomal regions (see Materials and methods). MSL2 S2 track was obtained from Schauer et al., 2017. LogFC was calculated after smoothing signals using Daniell kernel, with kernel size 50 and the addition of a pseudocount. On the heatmaps, the peaks are ranked according to the average logFC in the ♂M1[wt] sample. (B) Comparing of MSL1, MSL2 and MSL3 binding to HAS in 2–3 day-old y1w1118 and M1[wt] (msl-1−; Ubi:msl-1WT / Ubi:msl-1WT) males. Average log fold-change between normalized (RPKM) test signals and nonspecific IgG signals in HAS (see Materials and methods). Average log fold-change was calculated after smoothing signals using the Daniell kernel with kernel size 50 and the addition of a pseudocount. Next, the tracks were aggregated using 100 bp bins.
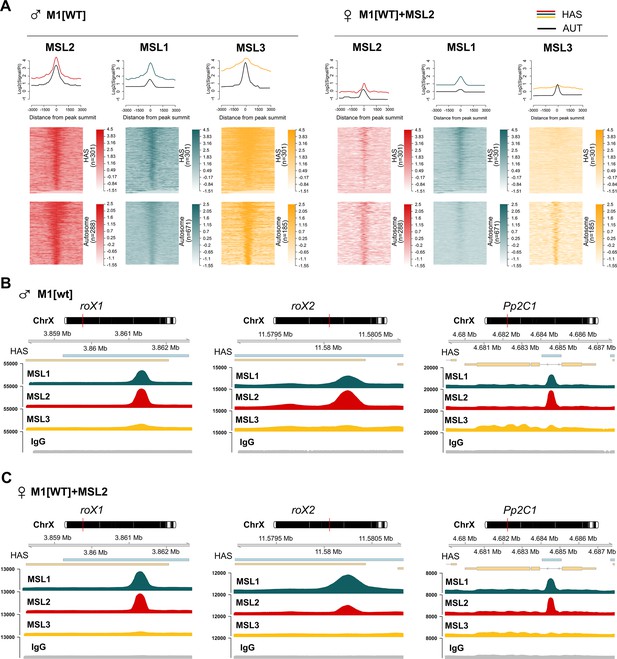
Comparisons of the binding patterns of male-specific lethal (MSL) proteins between 2–3 day adult males and females expressing MSL2.
(A) Average signal (top) and heatmap (bottom) showing the log fold-change (FC) between normalized (Reads per kilobase of transcript, per million mapped reads, RPKM) test signals and nonspecific IgG signals for high-affinity 'entry' site (HAS) and autosomal regions (see Materials and methods). LogFC was calculated after smoothing signals using the Daniell kernel with kernel size 50 and the addition of a pseudocount. On the heatmaps, the peaks are ranked according to the average logFC in the M1[wt] male sample. ♂ M1[wt] indicates 2–3 day-old msl-1−; Ubi:msl-1WT/ Ubi:msl-1WT males. ♀M1[wt]+MSL2 indicates 2–3 day-old msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1WT females. (B and C) Enrichment of MSL1 (green), MSL2 (red), and MSL3 (yellow) signals associated with the X-linked genes encoding Pp2C1 and the non-coding RNAs roX1 and roX2 in (B) M1[WT] males and (C) M1[WT]+MSL2 females.
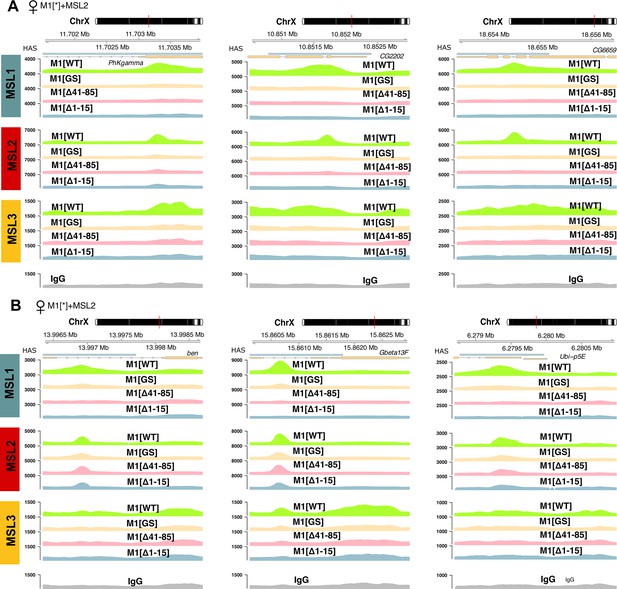
Examples of male-specific lethal (MSL)1, MSL2, and MSL3 signal enrichment associated with X chromosomal high-affinity ‘entry’ sites (HAS) in M1[WT] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1WT), M1[GS] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1GS), M1[Δ41–85] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ41-85), M1[Δ1–15] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ1-15) females.
MSL2 binds to HAS (A) only in M1[WT] females or (B) in M1[WT], M1[GS], M1[Δ41–85] and M1[Δ1–15] females.

Male-specific lethal (MSL)1 shows different scenarios of binding upon N-terminal region modification.
Regions clusterization with MSL1∩MSL2 overlapping peaks according to MSLs binding in different female fly lines expressing MSL2 and one of four MSL1 variants (MSL1WT, MSL1GS, MSL1Δ41-85, MSL1Δ1-15) (see Materials and methods). M1[WT] (msl-1−; Ubi:msl-2WT-FLAG/Ubi:msl-1WT females), M1[Δ41–85] (msl-1−; Ubi:msl-2WT-FLAG/Ubi:msl-1Δ41-85 females), M1[GS] (msl-1−; Ubi:msl-2WT-FLAG/Ubi:msl-1GS females), M1[Δ1–15] (msl-1−; Ubi:msl-2WT-FLAG/Ubi:msl-1Δ1-15 females). (A) Left - Heatmap of average logFC between MSL and nonspecific IgG signal in female fly lines, hierarchical clusterization, and cluster subdivision is drawn on the left corner. Right - Average logFC profiles of MSL signal in different clusters for female fly lines. Average log fold-change was calculated after smoothing signals using the Daniell kernel with kernel size 100 and the addition of a pseudocount. (B) Boxplot of average LogFC signal of MSL1 and MSL2 in cluster 2. All sites in cluster 2 were subdivided into PionX sites and other HAS, and the strength of MSL1 and MSL2 signals was tested.
-
Figure 6—source data 1
A list of unique male-specific lethal (MSL)1 peaks for females M1[WT]+MSL2 fly line (msl-1−; Ubi:msl-2WT-FLAG/Ubi:msl-1WT).
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig6-data1-v1.xlsx
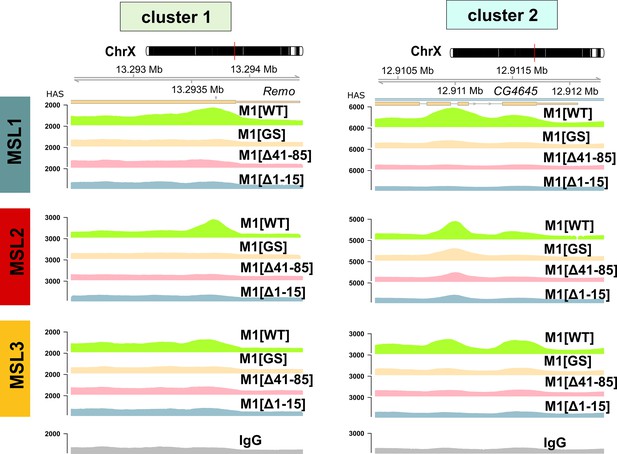
Examples of male-specific lethal (MSL)1, MSL2, and MSL3 signal enrichment in M1[WT] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1WT), M1[GS] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1GS), M1[Δ41–85] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ41-85), M1[Δ1–15] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ1-15) females.
Clusters 1 and 2 are associated with MSL2 binding to X chromosomal sites.

Regions with male-specific lethal (MSL)1 alone (no colocalization with MSL2) peaks (autosomes and X chromosome).
(A) Heatmap of average logFC between MSL and nonspecific IgG signal in female fly lines. (B) Examples of MSL1, MSL2, and MSL3 signal enrichment at autosomes in M1[WT] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1WT), M1[GS] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1GS), M1[Δ41–85] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ41-85), M1[Δ1–15] (♀ msl-1−; Ubi:msl-2WT-FLAG/ Ubi:msl-1Δ1-15) females.

Testing role of the N-terminal region in recruiting of roX2 by male-specific lethal (MSL)1.
Extraction of RNA and RNP from adult flies, and immunoprecipitation followed by RNA extraction were prepared as described in the Materials and Methods section and briefly schematically explained in the drawing. (A) Expression levels of the roX2 RNA in females of the y1w1118, M1[WT] (msl-1−; Ubi:msl-1WT-HA/ Ubi:roX2), M1[GS] (msl-1−; Ubi:msl-1GS-HA/ Ubi:roX2), M1[Δ41–85] (msl-1−; Ubi:msl-1Δ41-85-HA/ Ubi:roX2) and msl1 –(msl-1−;Ubi:roX2/TM6,Tb). Individual transcript levels were determined by RT-qPCR with primers for the roX2 gene normalized relative to RpL32 for the amount of input cDNA. The error bars show standard deviations of triplicate measurements. This panel was created using BioRender.com. (B) RNP extracts from adult females were immunoprecipitated with antibodies against MSL1 (IP-MSL1) or nonspecific rabbit IgG (IP-IgG) as a negative control. The efficiency of immunoprecipitation was tested by immunoblot analysis for the presence of HA-tagged MSL1 protein in immunoprecipitate samples. (C) Total RNA was extracted from immunoprecipitates (α-MSL1 or IgG) and analyzed for the presence of roX2 RNA by RT-PCR. RpL32 was used as the negative control. The results of immunoprecipitations are presented as the percentage of input cDNA. ‘output’ – supernatant after immunoprecipitation; ‘IP’ – immunoprecipitated sample. The error bars indicate SDs from three independent biological samples.
-
Figure 7—source data 1
Original files for the immunoblot blot analysis are in Figure 7B.
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig7-data1-v1.zip
-
Figure 7—source data 2
Files containing original immunoblots for Figure 7B indicate the relevant bands.
- https://cdn.elifesciences.org/articles/93241/elife-93241-fig7-data2-v1.zip
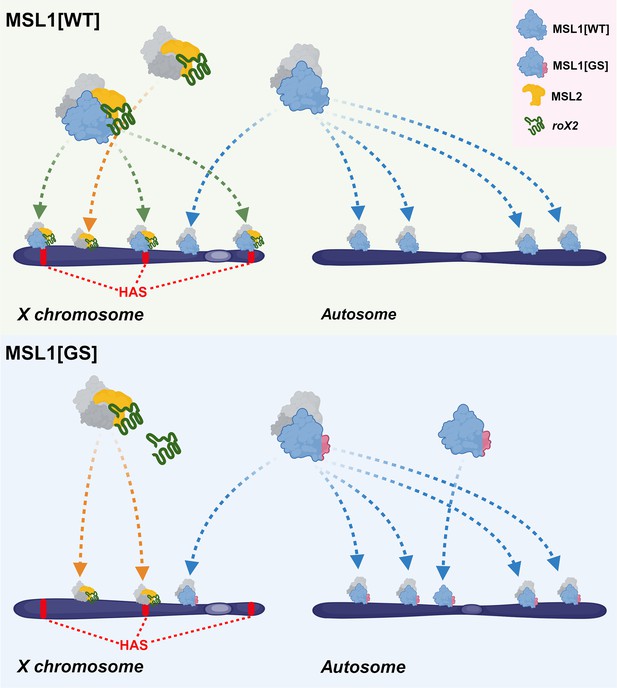
Schematic representation of male-specific lethal (MSL) complex recruiting to chromatin.
The difference in the principles of recruitment of a complex containing wild-type MSL1 protein (MSL1[WT]) and a complex containing MSL1 with a mutant N-terminus is shown (MSL1[GS]). This figure was created using BioRender.com.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Drosophila melanogaster) | 86Fb | Bloomington Drosophila Stock Center | BDSC: 23648 | P{ry[+t7.2]=hsp70-FLP}1, y[1] w[*]; M{3xP3-RFP.attP}ZH-86Fb; M{RFP[3xP3.PB] GFP[E.3xP3]=vas int.B}ZH-102D |
| Genetic reagent (D. melanogaster) | CyO, GFP | Bloomington Drosophila Stock Center | BDSC: 9325 | w[1118]; sna[Sco]/CyO, P[ActGFP.w-]CC2 |
| Genetic reagent (D. melanogaster) | y[1] w[1118] | Bloomington Drosophila Stock Center | BDSC: 6598 | y[1] w[1118] |
| Genetic reagent (D. melanogaster) | msl-2WT-FLAG | This lab (Tikhonova et al., 2019) | N/A | Ubi:msl-2WT-FLAG |
| Genetic reagent (D. melanogaster) | msl-1L60 | donated by M. Kuroda | N/A | msl-1L60 /CyO |
| Genetic reagent (D. melanogaster) | msl-1γ269 | donated by J. Lucchesi | N/A | |
| Genetic reagent (D. melanogaster) | Ubi-msl-1* | This paper | N/A | Ubi-msl-1* transgenes are described in Materials and Methods |
| Cell line (D. melanogaster) | S2 | Drosophila genomics resource center | DGRC Stock 181; https://dgrc.bio.indiana.edu//stock/181; RRID:CVCL_Z992 | FlyBase symbol: S2-DRSC. |
| Antibody | anti-MSL1 (rabbit polyclonal) | This lab | N/A | MSL1[423–1030] IF (1:500) ChIP (1:200) WB (1:500) IP (1:200) |
| Antibody | rabbit anti-MSL2 (rabbit polyclonal) | This lab | N/A | MSL2[421-540] IF (1:500) ChIP (1:100) WB (1:500) |
| Antibody | rabbit anti-MSL3 (rabbit polyclonal) | This lab | N/A | ChIP (1:200) WB (1:500) |
| Antibody | anti-FLAG clone M2 (mouse monoclonal) | Sigma, USA | F3165 | IF (1:100) WB (1:1000) IP (1:200) |
| Antibody | anti-HA clone HA-7 (mouse monoclonal) | Sigma, USA | H9658 | IF (1:50) WB (1:500) IP (1:200) |
| Antibody | anti-lamin Dm0 clone ADL84.12 (mouse monoclonal) | DSHB, USA | ADL84.12 | WB (1:3000) |
| Antibody | anti-FLAG-HRP (mouse monoclonal) | Sigma, USA | A8592 | WB (1:1000) |
| Antibody | anti-mouse Alexa Fluor 555 (goat polyclonal) | Thermo Fisher Scientific, USA | A28180 | IF (1:2000) |
| Antibody | goat anti-rabbit Alexa Fluor 488 (goat polyclonal) | Thermo Fisher Scientific, USA | A11008 | IF (1:2000) |
| Chemical compound, drug | Pierce 16% Formaldehyde (w/v), Methanol-free | Thermo Fisher Scientific, USA | 28906 | |
| Chemical compound, drug | CNBr-activated Sepharose | Cytiva, UK | GE17-0430-01 | |
| Chemical compound, drug | Aminolink Plus Coupling Resin | Thermo Fisher Scientific, USA | 20505 | |
| Chemical compound, drug | TRI-reagent | MRC, USA | TR 118 | |
| Chemical compound, drug | Ribonucleoside Vanadyl Complex | NEB, USA | S1402S | |
| Chemical compound, drug | Nuclease-free BSA | Sigma, USA | 126609 | |
| Chemical compound, drug | Phase Lock Gel, QuantaBio - 2302830, Phase Lock Gel Heavy | VMR, USA | 10847–802 | |
| Chemical compound, drug | Ampure Xp beads | Beckman Coulter, USA | A63881 | |
| Chemical compound, drug | Diamant TaqA Hot-start polymerase | Belbiolab, Russia | E-TAP | |
| Chemical compound, drug | Dynabeads Protein A | Thermo Fisher Scientific, USA | 10008D | |
| Chemical compound, drug | DAPI | Applichem, Germany | A1001 | |
| Chemical compound, drug | MACSFectin | Miltenyi Biotec, USA | 130-098-410 | |
| Chemical compound, drug | SFX-Insect Cell Culture Media | HyClone, USA | SH30278 | |
| Chemical compound, drug | anti-HA magnetic beads | Thermo Fisher Scientific, USA | 88836 | |
| Chemical compound, drug | anti-FLAG magnetic beads | Sigma, USA | M8823 | |
| Chemical compound, drug | mouse IgG magnetic beads | NEB, USA | S1431 | |
| Chemical compound, drug | Halt Protease Inhibitor Cocktail | Thermo Fisher Scientific, USA | 78438 | |
| Chemical compound, drug | Calbiochem Complete Protease Inhibitor Cocktails V | Merck, USA | 539137 | |
| Chemical compound, drug | Calbiochem Complete Protease Inhibitor Cocktails VII | Merck, USA | 539138 | |
| Sequence-based reagent | Oligonucleotides used are listed in ‘Supplementary file 1' | Evrogen, Lytech, DNA synthesis | ||
| Commercial assay or kit | Crescendo Western Blotting substrate | Merck, USA | WBLUR0100 | |
| Commercial assay or kit | ChIP DNA Clean&Concentrator kit | Zymo Research, USA | D5205 | |
| Commercial assay or kit | NEBNext Ultra II DNA Library Prep Kit for Illumina | NEB, USA | E7645L | |
| Commercial assay or kit | Qubit dsDNA HS Assay Kit | Life Technologies Corporation | Q32851 | |
| Software, algorithm | ImageJ 1.54 f and Fiji bundle 2.14.0 | Schindelin et al., 2012 | N/A | fiji.sc |
| Software, algorithm | cutadapt | Martin, 2011 | N/A | |
| Software, algorithm | Bowtie version 2 | Langmead and Salzberg, 2012 | N/A | |
| Software, algorithm | deepTools | Ramírez et al., 2014 | N/A | |
| Software, algorithm | Picard | http://broadinstitute.github.io/picard/ | N/A | |
| Software, algorithm | MACS version 2 | Zhang et al., 2008 | N/A | |
| Software, algorithm | R version 4.2.1 | http://www.r-project.org | N/A | |
| Software, algorithm | ChIPpeakAnno package | Zhu et al., 2010 | N/A | |
| Software, algorithm | ChIPseeker package | Yu et al., 2015 | N/A | |
| Software, algorithm | pheatmap package. | https://github.com/raivokolde/pheatmap; Kolde, 2019 | N/A | |
| Software, algorithm | Gviz | Helaers et al., 2011 | N/A | |
| Other | 100 µm MACS SmartStrainers | Miltenyi Biotec, USA | 130-110-917 | |
| Other | 70 µm MACS SmartStrainers | Miltenyi Biotec, USA | 130-110-916 |
Additional files
-
Supplementary file 1
The list of oligonucleotides.
- https://cdn.elifesciences.org/articles/93241/elife-93241-supp1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/93241/elife-93241-mdarchecklist1-v1.docx





