The effect of combining antibiotics on resistance: A systematic review and meta-analysis
Figures
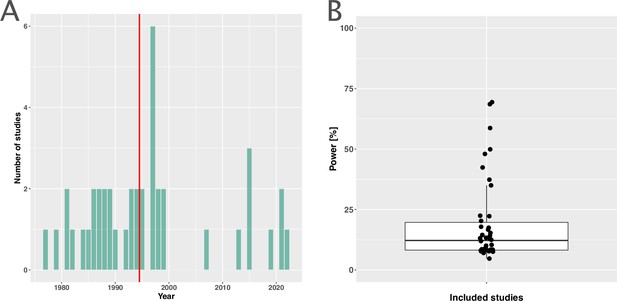
Measuring antibiotic resistance is not the current main objective of randomised controlled trials (RCTs).
(A) Distribution of the publishing year of included studies. The number of studies published per year is shown, with the red vertical line indicating the median of the distribution. (B) Calculated statistical power [%] of included studies to detect an odds ratio of 0.5. The power calculations were based on equal treatment arm sizes. For the calculations, the treatment arm with the higher number of patients of the respective studies was used.
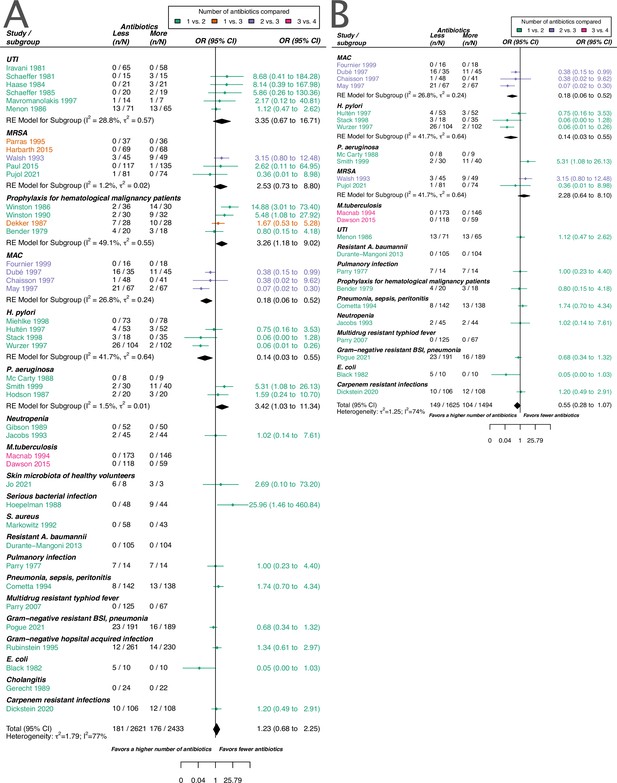
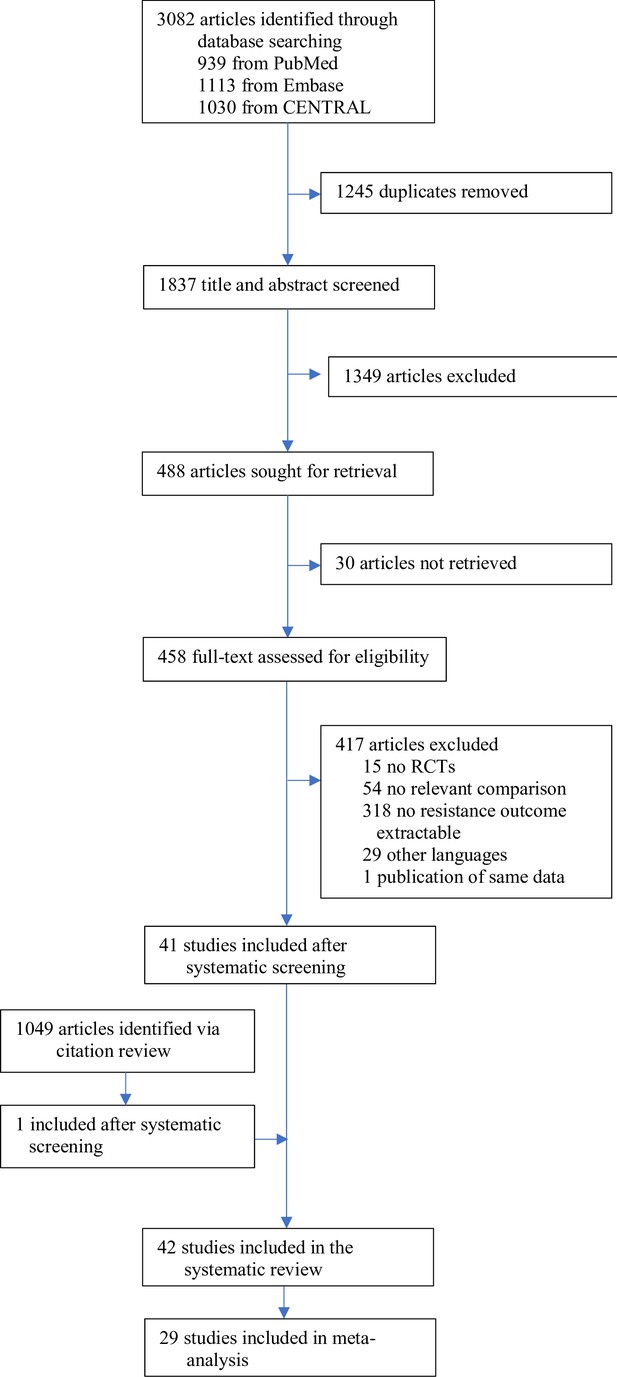
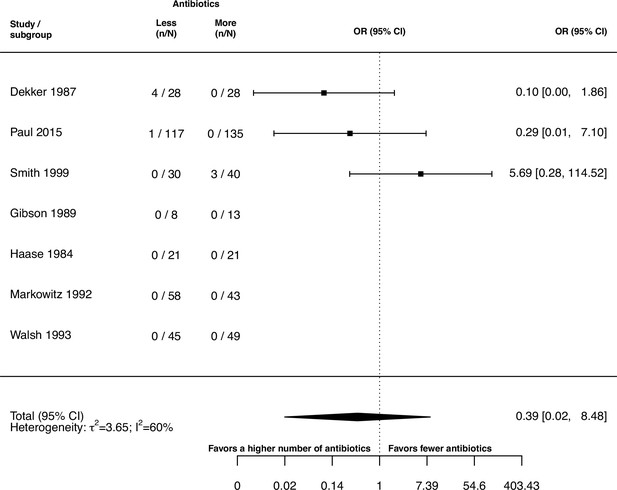
Forest plot of acquisition of bacterial resistance stratified by the reason antibiotics were administered.
The colouring indicates the number of antibiotics that were compared in each study. (A) The overall pooled OR of all included studies. (B) The pooled OR of studies with at least one antibiotic in common in the treatment arms. UTI stands for urinary tract infection, methicillin-resistant Staphylococcus aureus (MRSA) for methicillin-resistant Staphylococcus aureus, MAC for Mycobacterium avium complex, and BSI for blood stream infection.
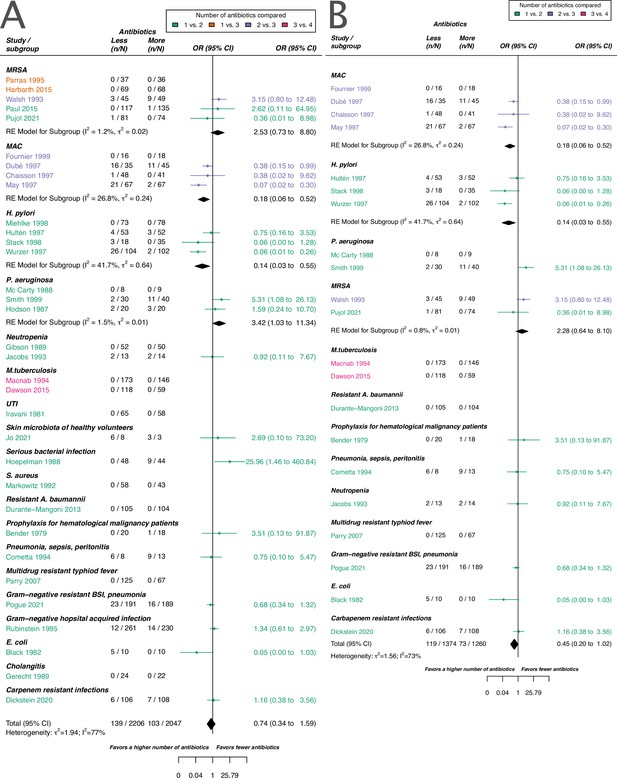
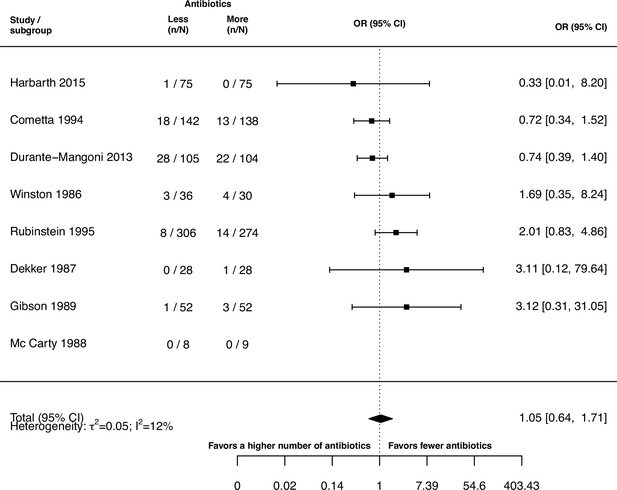
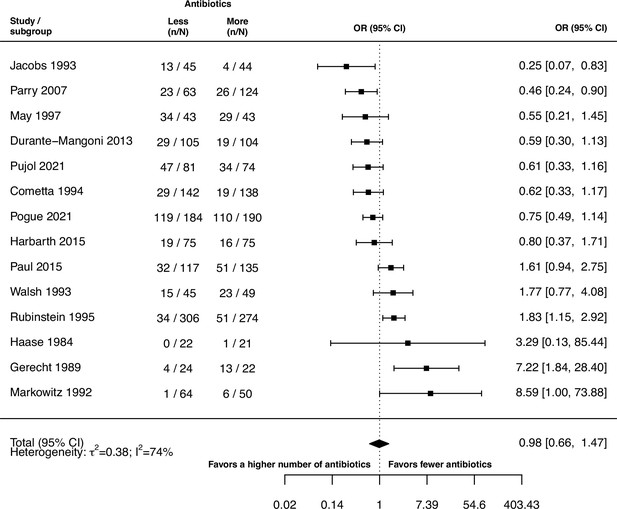
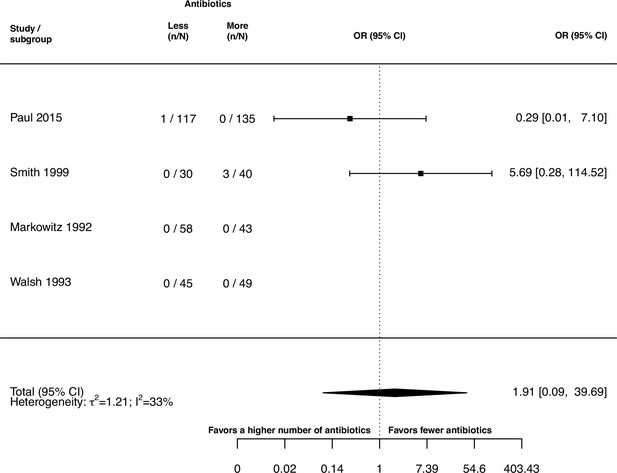
Forest plot of de novo emergence of bacterial resistance stratified by the reason antibiotics were administered.
The colouring indicates the number of antibiotics that were compared in each study. (A) The overall pooled OR of all included studies. (B) The pooled OR of studies with at least one antibiotic in common in the treatment arms. MRSA stands for methicillin-resistant Staphylococcus aureus, MAC for Mycobacterium avium complex, and BSI for blood stream infection.
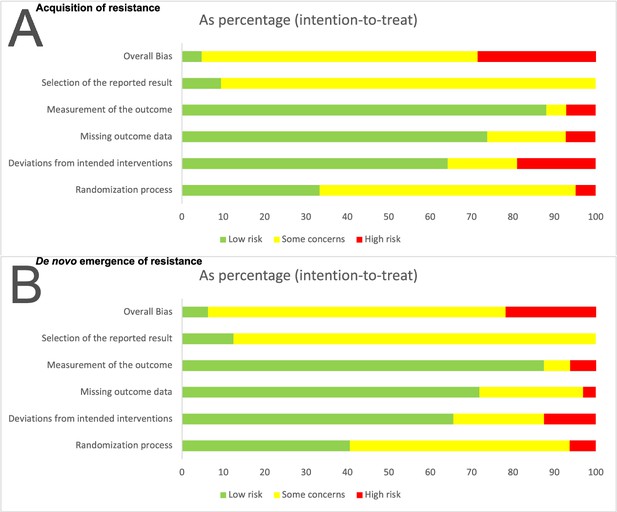
Risk of bias summary for the two main outcomes: (A) Acquisition of resistance, (B) de novo emergence of resistance.
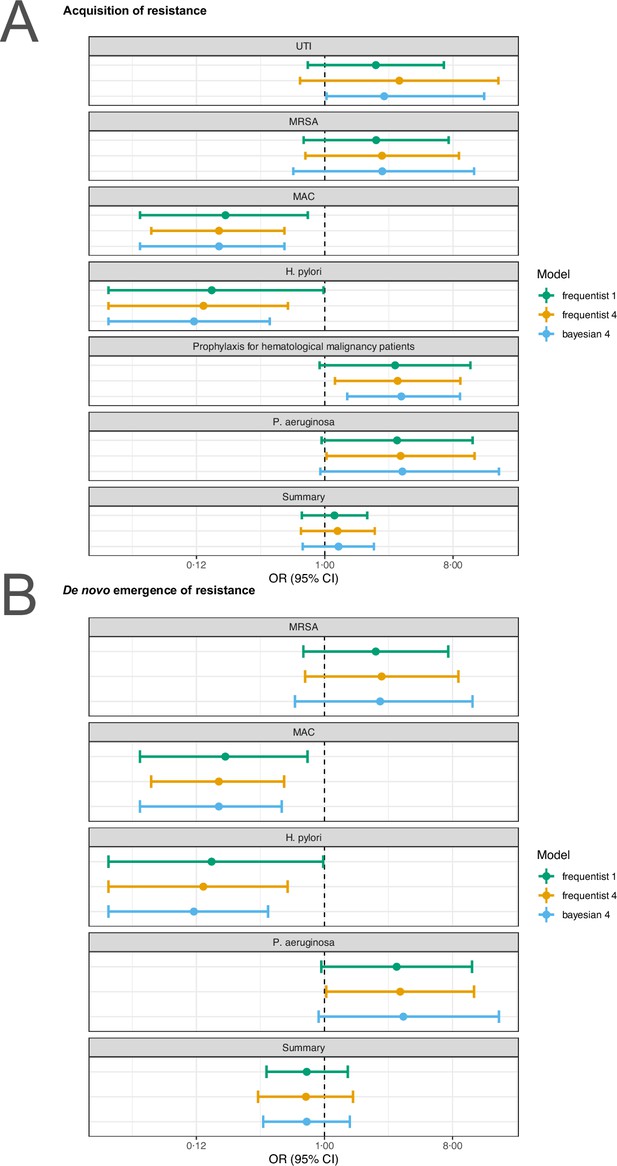
Sensitivity analysis based on model choice for the two main outcomes.
(A) Acquisition of resistance, (B) de novo emergence of resistance. Shown are the frequentist model estimates of model 1, and model 4 presented in Jackson et al., 2018 and a Bayesian estimate of model 4. UTI stands for urinary tract infection, MRSA for methicillin-resistant Staphylococcus aureus, and MAC for Mycobacterium avium complex.
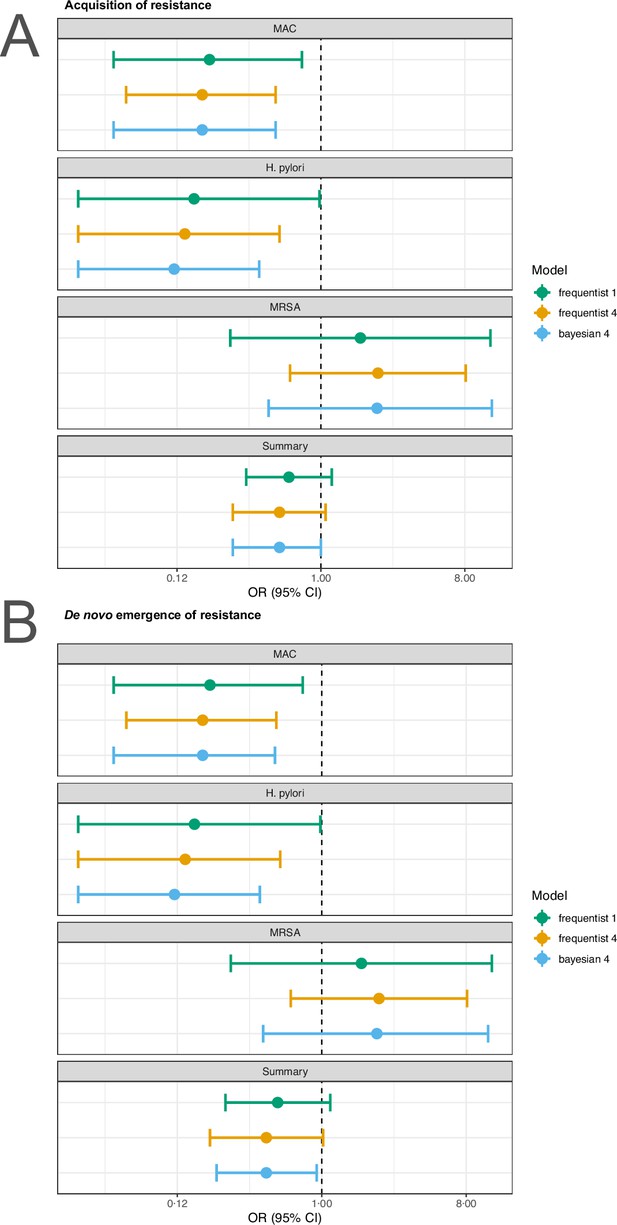
Sensitivity analysis based on model choice for the two main outcomes restricted to studies with at least one common antibiotic in the comparator arms.
(A) Acquisition of resistance, (B) de novo emergence of resistance. Shown are the frequentist model estimates of model 1, and model 4 presented in Jackson et al., 2018 and a Bayesian estimate of model 4.
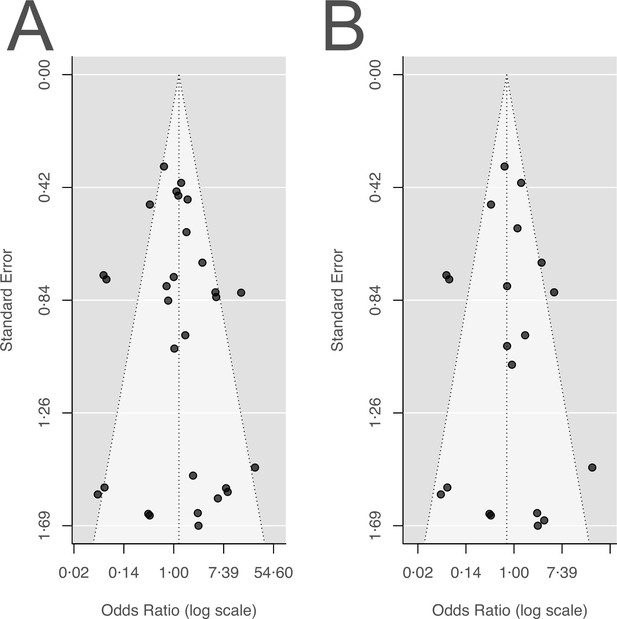
Funnel plots for the two main outcomes.
(A) Acquisition of resistance, (B) de novo emergence of resistance.
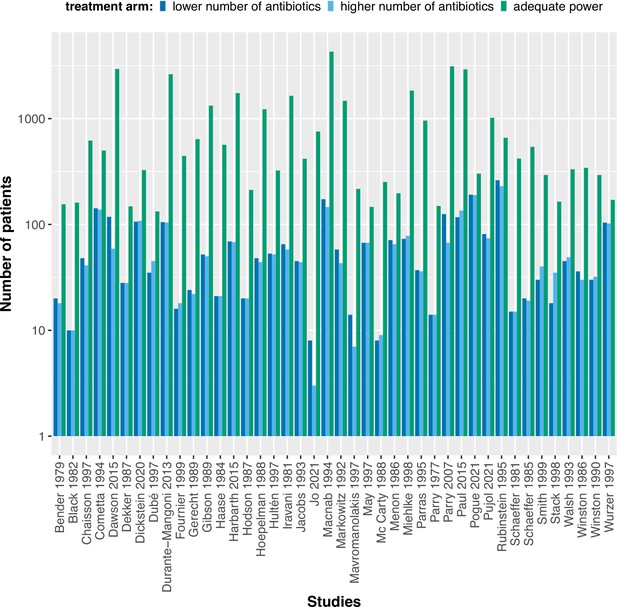
The calculated adequate treatment arm size for each study assuming to detect an odds ratio of 0.5 with 80% power in comparison to the actual treatment arm sizes.
The power calculations were performed using the upper confidence interval for the binomial probability of the treatment arm with fewer antibiotics.
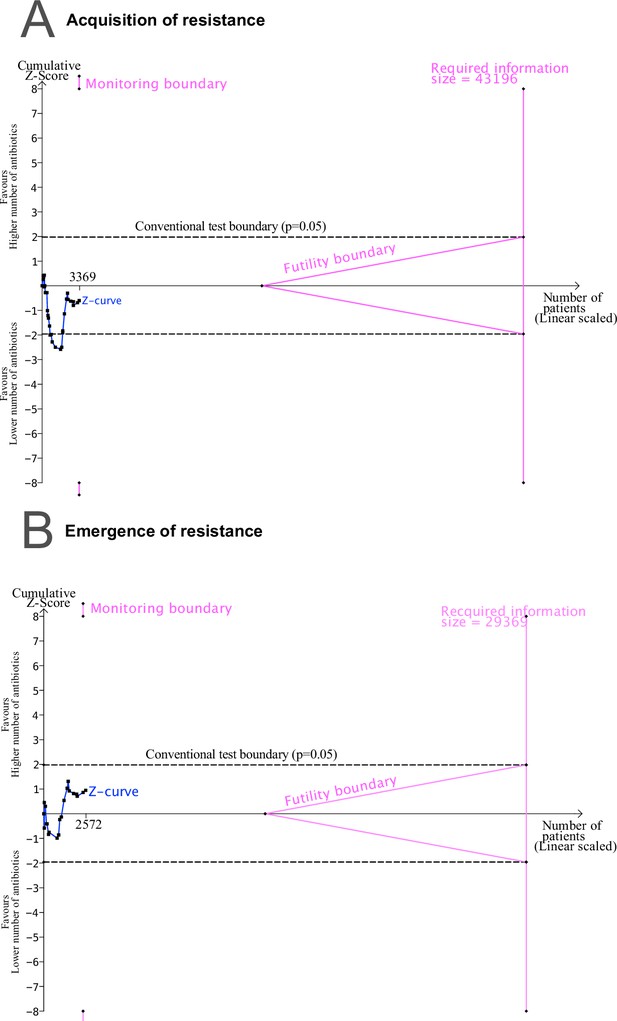
Trial sequential analysis (TSA) output using 80% power, and 5% significance to detect a relative odds reduction of 50%: (A) acquisition of resistance.
(B) de novo emergence of resistance. No sufficient evidence on the development of resistance is supported, since the Z-curves do not cross the monitoring nor the futility boundaries, and the required sample size is not reached.
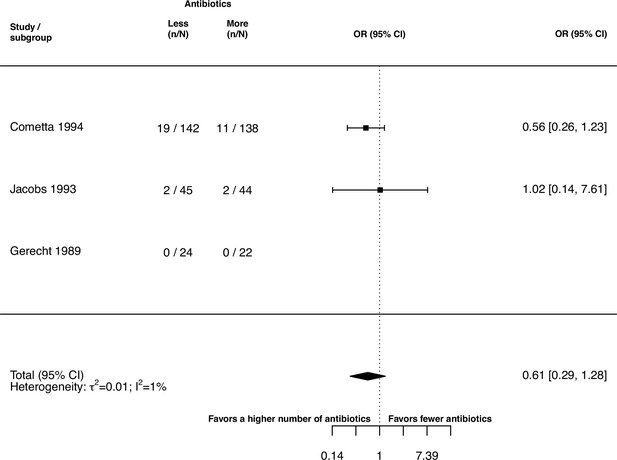
Forest plot of treatment failure due to a change of resistance against the study drugs.
Tables
Justification for extraction of resistance development.
The definitions of resistance development are stated as given by the study authors. In case no explicit definition was given, we state a justification for extraction and indicate it with (*). Note that for data extraction for the publications of Dickstein et al., 2020 and Pogue et al., 2021 additional publications of the same studies were consulted by Paul et al., 2018 and Kaye et al., 2023, respectively. Resistance breakpoints are stated in case numerical values were given in the respective studies. See Supplementary file 1 for which antibiotics the studies tested and reported extractable resistance data.
| Study | Definition of resistance development given by study authors or justification for extraction |
|---|---|
| Bender et al., 1979 | Susceptibility testing for gentamicin of the flora was performed at randomisation and twice weekly after with the Kirby-Bauer disk technique and microtiter minimal inhibitory concentration. (*) |
| Black et al., 1982 | Patients were infected with a known strain and all stool cultures and rectal swabs were plated and tested for trimethoprim resistance. (*) |
| Chaisson et al., 1997 | Testing of isolates to susceptibility for clarithromycin, ethambutol, and clofazimine was performed before the entry of the study and monthly for 6 mo in broth by the method of Heifets.(*) |
| Cometta et al., 1994 | All microorganisms were sensitive to imipenem at randomisation and follow-up cultures were performed. (*) |
| Dawson et al., 2015 | Susceptibility testing at randomisation and for the following cultures by rapid testing. Susceptibilities to isoniazid, rifampicin, and fluoroquinolones were determined by line probe assay. (*) |
| Dekker et al., 1987 | At admission, cultures were performed and surveillance cultures were done twice a week. Gram-negative bacilli were tested for antibiotic susceptibility. The minimal inhibitory concentrations were assessed by agar dilution technique. An MIC of ≥2 µg/mL was considered resistant for ciprofloxacin, an MIC of ≥4 µg/mL for trimethoprim, and an MIC ≥75 µg/mL for sulfamethoxazole. (*) |
| Dickstein et al., 2020 | Development of a new colistin-resistant (ColR) isolates within 28 d from study enrolment. To be considered a new ColR isolate, the ColR isolate had to be detected on day 7 or later in patients for whom the baseline isolate was colistin-susceptible, and for whom no ColR isolate was cultured from the rectal swab taken on day 1. Susceptibility was determined by broth microdilution. Colistin resistance was defined as an MIC >2 mg/L. |
| Dubé et al., 1997 | All available isolates were tested for susceptibility to clarithromycin. Patients were evaluated at the time of enrolment, 2 and 4 wk later, and then every 4 wk. Clarithromycin resistance was defined as detectable growth in a concentration of clarithromycin of 8 µg/mL. (*) |
| Durante-Mangoni et al., 2013 | The identification of a colistin-resistant Acinetobacter baumannii during treatment was defined as resistance emergence. Resistance was determined by the microdilution method and/or E-test. |
| Fournier et al., 1999 | Susceptibility testing was performed at study entry after 2 mo and classification was performed according to Heifets. (*) |
| Gerecht et al., 1989 | Emergence of resistance was defined as one cause of treatment failure. Emergence of resistance was classified as the detection of an infecting microorganism resistant to more than 4 μg/mL of gentamicin sulfate or more than 128 μg/mL of mezlocillin sodium during treatment while the patient shows indications of cholangitis. |
| Gibson et al., 1989 | Microbiological assessment of the blood was performed before treatment and 96 hr after treatment. (*) |
| Haase et al., 1984 | Susceptibility was assessed before therapy, during therapy, and after therapy. Susceptibility testing was performed with disk dilution method, and agar dilution method. Resistance results were reported for reinfections defined as the reappearance of infection with a different organism after completion of therapy. Resistance against norfloxacin and trimethoprim-sulfamethoxazole was defined as a larger inhibition zone diameter of 0.17 and 0.16 mm, respectively, or/and a MIC larger than 16 µg/mL and 3.4–64 µg/mL, respectively. (*) |
| Harbarth et al., 2015 | Susceptibility assessment was performed at baseline and at the end of treatment. Susceptibility was performed with a disc diffusion method phenotypically and genotypically. (*) |
| Hodson et al., 1987 | P. aeruginosa had to be sensitive at inclusion and resistance was measured and reported after 10 d of treatment. Sensitivity was determined by standard disc methods. (*) |
| Hoepelman et al., 1988 | Susceptibility was assessed before, during, and after treatment. Susceptibility testing was performed with the disc diffusion method and minimum inhibitor concentrations were assessed for blood cultures and patients with no response to treatment with the agar dilution technique. Resistance for the agar dilution technique was defined as an MIC of ≥32 µg/mL for ceftriaxone, ≥8 µg/mL for gentamicin, and ≥32 µg/mL for cefuroxime. For the disc diffusion method 30 µg ceftriaxone, 40 µg gentamicin, and 60 µg cefuroxime were used. If the zone of inhibition was ≤18 mm cultures were classified as ceftriaxone resistant and sensitive if the zone was ≥26 mm and intermediate in between. For gentamicin the values were ≤20 mm and ≥28 mm and for cefuroxime ≤20 mm and ≥28 mm, respectively.(*) |
| Hultén et al., 1997 | Susceptibility was assessed by E-test at inclusion and 12 wk after treatment determination. (*) |
| Iravani et al., 1981 | Susceptibility testing at baseline, during treatment and at follow-up. Testing was performed with Bauer’s disc diffusion method using 30 µg nalidixic acid, 1.25 µg trimethoprim, and 23.75 µg sulfamethoxazole. (*) |
| Jacobs et al., 1993 | Emergence of resistance was defined as treatment failure with resistance, i.e., bacteriological failure with the reisolation of original pathogen(s) resistant to the study antibiotic(s) after treatment. |
| Jo et al., 2021 | Susceptibility testing before treatment and after treatment by culture. (*) |
| Macnab et al., 1994 | Susceptibility testing before treatment and after around 90 doses. (*) |
| Markowitz et al., 1992 | Susceptibility was assessed by microdilution method before treatment and for the last continuous positive culture during treatment. Furthermore, susceptibility was assessed for relapse isolates and isolates phenotypically different from the initial one. (*) |
| Mavromanolakis et al., 1997 | Susceptibility was assessed before treatment, after 2 wk, at the end of treatment, and 2 wk after treatment by disk diffusion method. (*) |
| May et al., 1997 | Susceptibility was assessed at treatment start, after 2 mo, and in case of relapse by the Becton Dickinson method. (*) |
| McCarty et al., 1988 | Susceptibility was assessed at admission, every 4 d during treatment, and within 48 hr after treatment by broth microdilution method using the American Microscan Gram Negative-Panel. (*) |
| Menon et al., 1986 | Susceptibility was assessed before therapy, and after 1 and 2 wk after therapy. (*) |
| Miehlke et al., 1998 | Susceptibility was assessed before and after treatment by E-test. An MIC of ≤0.125 mg/L was considered clarithromycin sensitive and an MIC of ≥2 mg/L resistant. An MIC of ≤2 mg/L was considered amoxicillin susceptible and an MIC of ≥4 mg/L resistant. (*) |
| Parras et al., 1995 | Susceptibility was assessed at baseline and at the end of therapy by agar dilution method or automated microdilution methods. (*) |
| Parry et al., 1977 | Susceptibility was assessed before, during, after treatment, after 2 wk, and after 6 mo after treatment by Bauer’s method. (*) |
| Parry et al., 2007 | Susceptibility was assessed before therapy and after treatment by E-test, disk diffusion method. Ofloxacin was tested by disk diffusion method with a 5 µg and organisms were declared susceptible with a breakpoint ≤2 µg/mL and resistant with a breakpoint ≥8 µg/mL. Azithromycin was also tested with the disk diffusion method (15 µg disk), but no clear breakpoints were defined. Instead, azithromycin was determined by E-test according to the manufacture’s guidelines. (*) |
| Paul et al., 2015 | Development of resistance was defined as the acquisition of S. aureus resistant to any of the study drugs or vancomycin-resistant Enterococci. |
| Pogue et al., 2021 | Number of patients, who developed colistin resistance during therapy. Resistance was assessed with broth microdilution and declared as colistin-resistant with an MIC ≥4 mg/L. |
| Pujol et al., 2021 | Emergence of resistance to studying drugs during treatment according to EUCAST. |
| Rubinstein et al., 1995 | Resistance emergence was assessed by measuring MICs before, during, and after treatment. Disk diffusion testing was performed with disks of 30 µg ceftazidime, 30 µg ceftriaxone, and 10 µg tobramycin. An MIC ≤8 mg/L was considered susceptible for ceftazidime and ceftriaxone and a MIC ≥32 mg/L was considered resistant for ceftazidime and an MIC ≥64 mg/L for ceftriaxone. An MIC ≤4 mg/L was classified as susceptible for tobramycin, and an MIC ≥8 mg/L as resistant. |
| Schaeffer et al., 1981 | Susceptibility was assessed before therapy, after 7 d, and after 5 to 9 d after therapy by plating. Susceptibility testing was performed by plating 0.1 mL of culture on Mac Conkey agar containing 100 µg/mL cinoxacin or 1–24 µg/mL trimethoprim-sulfamethoxazole. Any growing culture was considered resistant and resistance tests were confirmed with standard agar sensitivity testing to a maximum concentration of 100 µg cinoxacin or 80–400 µg trimethoprim-sulfamethoxazole. (*) |
| Schaeffer and Sisney, 1985 | Susceptibility testing was performed before therapy, during therapy, and after 5 to 7 d after therapy by plating. 0.1 mL of cultures were plated on either Mueller-Hinton agar containing 10 µg/mL agar of norfloxacin or 1–24 µg/mL agar trimethoprim-sulfamethoxazole with 5% lysed red blood cells from the horse. Any growing culture was considered resistant and resistance tests were confirmed with tube dilution sensitivity testing to a maximum concentration of 100 µg/mL norfloxacin or 32–608 µg/mL trimethoprim-sulfamethoxazole. (*) |
| Smith et al., 1999 | Susceptibility was assessed at inclusion, and at the end of treatment by disk-susceptibility testing. An MIC of ≥100 µg/mL was considered resistant for azlocillin and resistant to tobramycin if the MIC was ≥8 µg/mL.(*) |
| Stack et al., 1998 | Susceptibility was assessed at baseline, and at 4 or 8 wk after treatment by E-test. Resistance was considered with bacterial growth at a drug concentration of >2 µg/mL for clarithromycin. (*) |
| Walsh et al., 1993 | Susceptibility was assessed at baseline and for organisms culturable after the end of therapy and a 2 wk follow-up period by a microtiter tube dilution technique. Organisms were declared resistant if the MIC was greater than 2 µg/mL for rifampicin, greater than 8 µg/mL for novobiocin, and greater than 2 µg/mL and 38 µg/mL for trimethoprim and sulfamethoxazole. |
| Winston et al., 1986 | Susceptibility of surveillance cultures was assessed at baseline, twice weekly during the study period and after study completion. Acquired organisms were defined as new organisms isolated during the study period, that were not present at baseline. An MIC ≤16 µg/mL was considered as sensitive for norfloxacin, polymyxin. For disc sensitivity testing cultures were considered sensitive to norfloxacin if a zone of ≥17 mm was present in a 10 µg norfloxacin disk. (*) |
| Winston et al., 1990 | New organisms that were isolated during the study period but had not been present before the study were defined as acquired organisms. Susceptibility tests were done by agar dilution method, or by antibiotic disks. An MIC of ≤4, 16, or 4 µg/mL for ofloxacin, polymyxin, or vancomycin was considered susceptible to the antibiotics, respectively. For ofloxacin additional disk sensitivity testing was performed. Susceptibility was declared if a zone of 16 mm or greater was present around a 5 µg disk of ofloxacin. (*) |
| Wurzer et al., 1997 | Susceptibility was assessed pre-treatment and between 4 and 6 wk of follow-up by agar dilution, and micro broth dilution. An MIC concentration of ≤2 µg/mL indicated susceptibility for clarithromycin, and an MIC above 2 µg/mL resistance. An MIC lower or equal to 0.125 µg/mL for amoxycilin was considered susceptible and classified resistant if above 0.125 µg/mL. (*) |
Summary of the results of the sub-group analyses stratifying according to the overall risk of bias for the two main outcomes.
Note that the listing of eligible studies also includes studies reporting zero cases in both treatment arms and were, therefore, not included in the statistical analysis.
Summary of the results of the predefined sub-group analyses for the outcome acquisition of resistance.
Note that the listing of eligible studies also includes studies reporting zero cases in both treatment arms, which are not included in the statistical analysis.
Summary of the results of the predefined sub-group analyses for the outcome de novo emergence of resistance.
Note that the listing of eligible studies also includes studies reporting zero cases of resistance in both treatment arms, which were, therefore, not included in the statistical analysis.
Summary of the results of the post-hoc sub-group analyses for the outcome acquisition of resistance.
Note that the listing of eligible studies also includes studies reporting zero cases in both treatment arms and were, therefore, not included in the statistical analysis.
Summary of the results of the post-hoc sub-group analyses for the outcome de novo emergence of resistance.
Note that the listing of eligible studies also includes studies reporting zero cases in both treatment arms and were, therefore, not included in the statistical analysis.
Overview of the model-averaged coefficients obtained by the multi-model inference for the main outcome acquisition of resistance.
Significant model estimates are displayed in a bold font.
| Model-averaged coefficients (full-average) | Estimated | Standard error | z value | Pr(>|z|) |
|---|---|---|---|---|
| Intercept | 0.73 | 1.69 | 0.60 | 0.54 |
| Length of follow-up | 1.00 | 1.00 | 0.46 | 0.65 |
| Treatment length | 0.99 | 0.01 | 0.77 | 0.43 |
| 1 vs 3 antibiotics | 0.78 | 2.19 | 0.31 | 0.75 |
| 2 vs 3 antibiotics | 1.16 | 2.16 | 0.19 | 0.85 |
| Antibiotics in common: no | 5.67 | 1.89 | 2.73 | 0.01 |
| Comorbidity: yes | 1.35 | 1.77 | 0.53 | 0.60 |
| Gram-positive and negative bacteria | 1.52 | 1.91 | 0.65 | 0.52 |
| Gram-positive bacteria | 1.08 | 2.07 | 0.11 | 0.91 |
| Year difference of youngest antibiotics | 1.00 | 1.01 | 0.15 | 0.88 |
Model output for a meta-regression for acquisition of resistance including as a covariable, whether at least one antibiotic was in common in the treatment arms.
Significant model estimates are displayed in a bold font.
| OR (95% CI) | z value | Pr(>|z|) | Study heterogeneity (I2; τ2) | |
|---|---|---|---|---|
| Intercept | 0.63 (0.33–1.21) | –1.39 | 0.17 | 59%; 0.90 |
| Antibiotics common: no | 5.86 (2.05–16.76) | 3.30 | <0.01 |
Overview of the model-averaged coefficients obtained by the multi-model inference for the main outcome de novo emergence of resistance.
| Model-averaged coefficients (full-average) | Estimated | Standard error | z value | Pr(>|z|) |
|---|---|---|---|---|
| Intercept | 2.22 | 2.42 | 0.90 | 0.37 |
| Length of follow-up | 0.99 | 1.01 | 0.73 | 0.46 |
| Treatment length | 1.00 | 1.01 | 0.41 | 0.68 |
| 2 vs 3 antibiotics | 1.29 | 2.51 | 0.28 | 0.78 |
| Antibiotics in common: yes | 0.32 | 2.66 | 1.16 | 0.25 |
| Comorbidity: yes | 1.40 | 2.03 | 0.47 | 0.64 |
| Gram-positive and negative bacteria | 1.45 | 2.23 | 0.47 | 0.64 |
| Gram-positive bacteria | 1.16 | 1.98 | 0.21 | 0.83 |
| Year difference of youngest antibiotics | 0.99 | 1.02 | 0.30 | 0.72 |
Oerview of different treatment failure definitions.
| Study | Definition of treatment failure given by the study authors |
|---|---|
| Cometta et al., 1994 | Lack of improvement of primary infection, development of a sepsis syndrome or septic shock during treatment, superinfection |
| Durante-Mangoni et al., 2013 | No improvement of clinical conditions by day 21 or worsening of the condition at any time, given persistently positive Acinetobacter baumannii cultures |
| Gerecht et al., 1989 | Continued presence of infecting organism(s) in bile cultures, with persistent indications of cholangitis, or superinfection, or the presence of new infecting organism(s) during or at the end of antibiotic treatment, with indications of cholangitis, or the emergence of an infecting organism(s) resistant to gentamicin or mezlocillin during treatment, with indications of cholangitis, or the emergence of an infecting organism(s) resistant to gentamicin or mezlocillin during treatment, with indications of cholangitis, or relapse, or recurrence of indications of cholangitis, with the original infecting organism(s) present in cultures of bile or blood within 8 wk after treatment, or death due to uncontrolled infection. |
| Haase et al., 1984 | The persisting presence of the pretherapy infecting organism, with or without pyuria, during treatment. |
| Harbarth et al., 2015 | No improvement or worsening in the clinical condition, or a change of the assigned therapy at any time, or death. |
| Jacobs et al., 1993 | No apparent response to therapy and no definitive identification of an alternative etiology that would explain this lack of response. |
| Markowitz et al., 1992 | Persistence of septic pulmonary emboli, persistence of positive blood or deep tissue cultures, or relapse after the end of presumably adequate treatment. |
| May et al., 1997 | Treatment failure was defined as all other situations than success, whereas the primary determinants of success were as follows: patient living, either not fever or a reduction of ≥1 °C in initial body temperature, and a blood culture negative for M. avium |
| Parry et al., 2007 | Continuing fever with at least one other typhoid-related symptom for more than 7 d after the start of treatment, or a required change in therapy due to the development of severe complications during treatment (severe gastrointestinal bleeding, intestinal perforation, visible jaundice, myocarditis, pneumonia, renal failure, shock, or an altered conscious level) |
| Paul et al., 2015 | Treatment failure at 7 d was defined as a composition of death, persistence of fever, persistence of hypotension, non-improving Sequential Organ Failure Assessment score, or persistent bacteraemia on day 7. |
| Pogue et al., 2021 | Clinical failure was defined by meeting any of the following criteria: death either during therapy or within 7 d after; receipt of rescue therapy for the trial pathogen within 7 d after treatment, exclusion from the trial due to an adverse event considered related to trial treatment; bacteremia more than 5 d after the begin of therapy for patients with blood stream infections; or failure to improve or worsening of oxygenation by the end of trial treatment in patients with pneumonia. |
| Pujol et al., 2021 | No clinical improvement after 3 d of therapy, persistent MRSA bacteraemia at day 7 or later, early discontinuation of therapy due to adverse events or based on clinical judgment, recurrent MRSA bacteraemia before or at the test of cure, missing blood cultures at the test of cure, and/or death due to any cause before the test of cure. |
| Rubinstein et al., 1995 | Use of a new antibiotic due to a worsening in clinical condition, isolation of resistant organism, or superinfection at the initial site during treatment, no clinical response or death attributed to infection. |
List of studies for which study authors or institutions were contacted.
An indication is given as to whether clarifying information was obtained.
| Study | Person/Institution contacted | Information sufficient for paper inclusion obtained (yes/no) |
|---|---|---|
| African and Councils, 1972 | Research office of the Royal Brompton & Harefield hospitals | no |
| Bazzoli et al., 1998 | Franco Bazolli | no |
| Benson et al., 2000 | Constance Benson | no |
| Bochenek et al., 2003 | David Yates Graham; Wieslaw Bochenek | no |
| Bosso and Black, 1988 | John Bosso | no |
| Bow et al., 1987 | Eric Bow | no |
| Cruciani et al., 1989 | Mario Cruciani | no |
| Dalgic et al., 2014 | Nazan Dalgic | no |
| de Pauw et al., 1985 | Ben de Pauw | no |
| de Pauw, 1987 | Ben de Pauw | no |
| DiNubile et al., 2005 | Mark Dinubile | no |
| Frank et al., 2002 | Elliot Frank | no |
| Gold et al., 1985 | Ronald Gold | no |
| Grossman et al., 1994 | Ronald Grossman | no |
| Grabe et al., 1986 | Magnus Grabe | no |
| Guerrant et al., 1981 | Richard Guerrant | no |
| Heyland et al., 2008 | Daren Heyland | no |
| Hodson et al., 1987 | Margaret Hodson | no |
| Hoepelman et al., 1988 | Andy I.M. Hoepelman | no |
| Jackson et al., 1986 | Mary Anne Jackson | no |
| Liang et al., 1990 | Raymond Hin Suen Liang | no |
| McLaughlin et al., 1983 | John McLaughlin | no |
| Muder et al., 1994 | Robert Muder | no |
| Padoan et al., 1987 | Rita Padoan | no |
| Paul et al., 2015 | Mical Paul | yes |
| Parry et al., 2007 | Christopher Parry | yes |
| Pujol et al., 2021 | Miquel Pujol | yes |
| Schaad et al., 1997 | Urs Schaad | no |
| Shawky et al., 2022 | Sherief Bad-Elsalam | no |
| Sun et al., 2022 | Jia Fan | no |
Table of studies, which were included in previous meta-analyses, but excluded in our study.
The reason for exclusion is indicated. *In our protocol we stated, that we would include articles in the Russian language. However, since VNK, the only Russian-speaking author, did not screen all the papers from our systematic search for inclusion, we excluded studies in the Russian language.
| Study | Inclusion in previous meta-analyses | Reason for exclusion | Identified with our search strategy |
|---|---|---|---|
| Carbon et al., 1987 | Paul et al., 2014 | Not accessible via ETH Zurich library services | no |
| Cone et al., 1985 | Bliziotis et al., 2005 | No data on resistance emergence, due to no clear statement of how many resistances are measured in the treatment arm with more antibiotics | no |
| Croce et al., 1993 | Bliziotis et al., 2005 | No proper randomisation of treatment strategies, i.e., the trial was conducted in different phases | yes |
| German and Austrian Imipenem/Cilastatin Study Group, 1992 | Bliziotis et al., 2005, Paul et al., 2014 | No fixed treatment, as an additional antibiotic was allowed to be administered only in the treatment arm with more antibiotics | no |
| Gribble et al., 1983 | Bliziotis et al., 2005 | No fixed treatment, since antibiotics could be substituted during treatment | yes |
| Iakovlev et al., 1998 | Paul et al., 2014 | Russian language* | no |
| Klastersky et al., 1973 | Paul et al., 2014 | Not clearly extractable how many patients developed resistance | no |
| Mandell et al., 1987 | Bliziotis et al., 2005, Paul et al., 2014 | Treatment is not fixed due to alterations of treatment based on the infecting organism | no |
| Sculier et al., 1982 | Paul et al., 2014 | No proper comparison, since the study does not compare per se a different number of antibiotics but adds an additional way of administration of the same antibiotic | no |
Additional files
-
Supplementary file 1
Overview of the 42 randomised controlled trials (RCTs) or quasi-RCTs included in the systematic review and meta-analysis.
The underlined antibiotics indicate that resistance measurements were made for this antibiotic, reported and extractable from the studies. Justification for resistance outcome extraction is given in Appendix 3—table 1.
- https://cdn.elifesciences.org/articles/93740/elife-93740-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/93740/elife-93740-mdarchecklist1-v1.pdf