A seven-sex species recognizes self and non-self mating-type via a novel protein complex
Figures

Mating-type recognition in T. thermophila.
(A) Example of self and non-self mating-type recognition. When one cell of mating type I encounters another, costimulation and mating do not occur. When a cell of mating type I encounters a cell of another mating type (II–VII), the cells enter the costimulation stage and go on to form a pair. (B) Two typical phenotypes of the costimulation stage are Tip transformation and concanavalin A (Con-A) receptor appearance. Yellow dashed circle, transformed cell tip (center, single cell) or pairing junction (right, cell pair). Note that Tip transformation may become less obvious after Con-A staining. (C) MTA and MTB gene structure and MTA and MTB protein domain information (Cervantes et al., 2013). MTA and MTB form a head-to-head gene pair. For each gene, the terminal exon is shared by all mating types and the remainder is mating-type-specific (the sequence differs for each mating type). The mating-type-specific region of each protein is predicted to be extracellular.
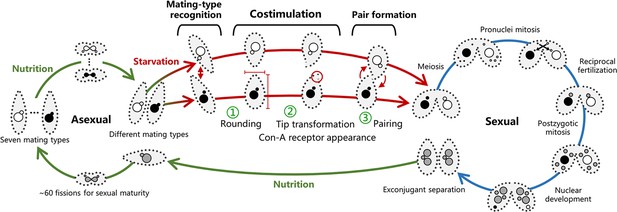
T. thermophila life cycle.
T. thermophila has seven mating types. Cells divide asexually when nutrition is adequate. After starvation, cells of any two different mating types recognize each other and enter a pre-conjugation stage called costimulation. In this stage, cells first become round (Fujishima et al., 1993); subsequently, their cell tips are transformed into a curved shape (Wolfe and Grimes, 1979) and the concanavalin A (Con-A) receptor becomes detectable (Wolfe and Feng, 1988; Wolfe et al., 1986). Costimulated cells then form pairs via a rotating behavior (Videos 1 and 2). T. thermophila possesses two types of nuclei within a single cell: the somatic macronucleus (MAC, ~90N) (Zhou et al., 2022) and the germline micronucleus (MIC, 2N). The mat locus on the MIC chromosome contains several mating-type gene pairs (mtGP, including MTA and MTB) organized in a tandem array. In contrast, the MAC chromosome carries only one mtGP. During the sexual life cycle (conjugation), cells undergo a series of sexual events. MIC undergoes meiosis and reciprocal fertilization (for details, refer to Orias et al., 2011). The new MAC of progeny develops from the fertilization MIC through a series of genome editing events, and increase its ploidy to ~90 via endoreduplication. During this process, mtGP loss occurs, resulting in only one mtGP remaining on the MAC chromosome. The mating-type specificity of mtGPs on each chromosome within one nucleus becomes pure to some extent relatively quickly, a phenomenon termed intranuclear coordination. Exconjugants (separated pairs) finally form parent-like progeny when nutrition becomes available, but remain sexually immature. After ~60 fissions, cells mature and their mating type is determined and becomes fixed (for details of mating-type determination, the interested readers is referred to Orias et al., 2017).
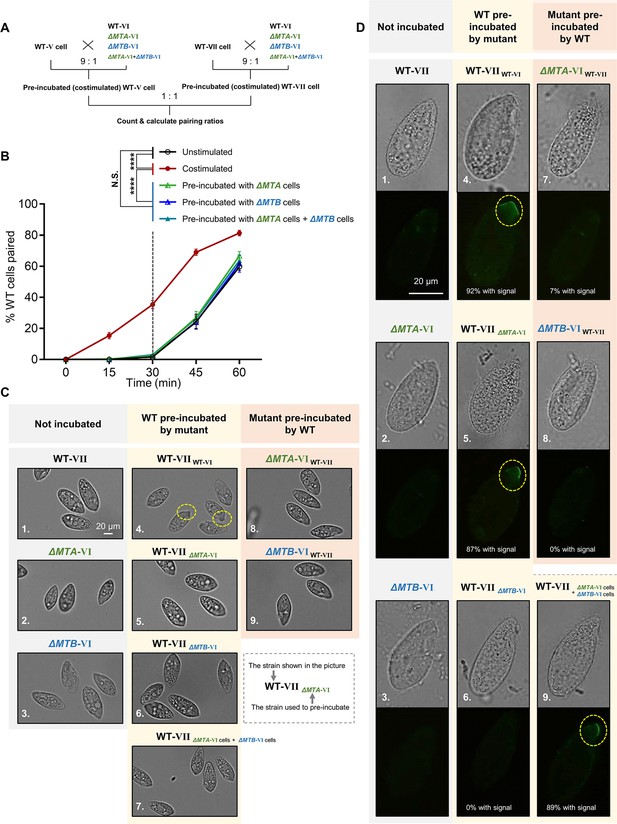
Mating-type proteins are essential for mating-type recognition.
(A) Experimental procedure for the costimulation experiments. Starved wild-type (WT) cells of mating types V (WT-V) and VII (WT-VII) were separately pre-incubated with the indicated mating type VI mutant (9:1 ratio) for 30 min and then the pre-incubated cells were mixed at a 1:1 ratio. Note that before mixing the costimulated cells, any potentially pairing cells were separated by shaking. (B) Effect of pre-incubation with ΔMTA and ΔMTB on the rate of pair formation. Each experiment was repeated three times, with>100 pairs counted at each time point. Matched two-way ANOVA was used for the statistical analysis. N.S., not significant; *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001. Unpaired mutants were excluded when calculating the pairing rate (see Materials and methods). (C) Tip transformation, a hallmark of costimulation. Each strain was pre-incubated with the strain shown in subscript. Yellow dashed circle, transformed cell tip. (D) Appearance of concanavalin A (Con-A) receptors, another hallmark of costimulation. In all, ~90% cells show Con-A receptor fluorescence (panels 4, 5, and 9). The low percentage of cells (7%) with fluorescence in panel 7 were probably WT cells, which comprised 10% of the pre-incubation culture. Each strain was pre-incubated with the strain shown in subscript. Yellow dashed circle, Con-A receptor fluorescence.
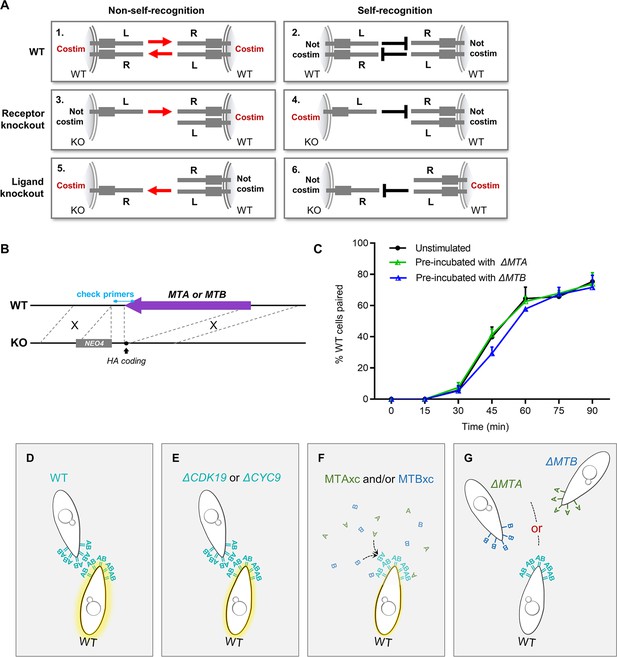
Mating-type gene deletion strains do not costimulate wild-type (WT) cells.
(A) Pre-incubation results predicted by a simple receptor–ligand model of mating-type recognition, in which one mating-type protein is designed ligand (L) and the other receptor (R). The type of cell used to costimulate is shown on the left. The expected result of costimulation if T. thermophila were to use non-self mating-type recognition (1, 3, 5) or self mating-type recognition (2, 4, 6). If this simple model were true, deleting the receptor would have a different effect on costimulation to deleting the ligand. However, the experimental results showed that neither ΔMTA nor ΔMTB can fully transmit/receive mating signal to/from WT cells. Therefore, mating-type recognition in T. thermophila does not occur via the simple receptor–ligand binding mechanism. Costim, costimulation. (B) Construction method of gene deletion strains. (C) Pre-incubation with ΔMTA or ΔMTB does not influence the pairing rate. The experimental method was as described in Figure 2A except that the pre-incubation time was 16 hr. (D–G) Diagram illustrating a theoretical mating-type recognition model between WT cells and ΔCDK19, ΔCYC9 cells, MTAxc, MTBxc proteins and ΔMTA, ΔMTB cells. (D–E) In WT or ΔCDK19, ΔCYC9 cells, where MTA and MTB proteins are expressed on the same cells, they can combine and stimulate another WT cells during attachment. (F) MTAxc and MTBxc proteins, being freely diffusible in solution, can continuously stimulate the WT cell. Moreover, MTAxc and MTBxc proteins have the opportunity to combine, providing a stronger stimulation. (G) In ΔMTA and ΔMTB cells, MTB and MTA proteins integrated into the cytomembrane of different cells and so cannot combine and stimulate the WT cells (i.e. WT cell receives one type of signal at a time, the signal stops when two cells separate).

Proteins that interact with MTA and MTB.
(A) Construction of HA-tagged strains. All of the tagged strains mated like wild-type (WT) cells. (B) Statistical analysis of immunoprecipitation-coupled mass spectrometry (IP-MS) data. A total of 13 experiments were carried out. WT samples (untagged) were run in parallel for each sample. All 13 WT controls were combined as the background control. Red dot, high-confidence interaction; dark gray dot, low-confidence interaction. Gene identifiers are summarized in Figure 3—source data 2. Note that the wash buffer contained 1% Triton X-100 and 600 mM NaCl. (C) Interaction network based on IP-MS data. Orange oval, bait; blue oval, high-confidence prey; light gray dot, low-confidence prey; black line, high-confidence interaction; dark gray dashed line, low-confidence interaction; light gray dotted line, interaction supported by a few peptides (these proteins were shown because their coding genes are coexpressed with MTA and MTB and deleting them affects mating behavior). (D) Diagram of functional domain annotation of mating-type recognition complex (MTRC) components. GFR, growth factor receptor domain; PLF, pectin lyase fold; Poly-E, poly-glutamic acid region; NTH, P-loop-containing nucleotide triphosphate hydrolase. (E) Expression profiles of genes whose protein products were identified by IP-MS as potentially components of the MTRC. Expression data is derived from TetraFGD (Xiong et al., 2011).
-
Figure 3—source data 1
Immunoprecipitation-coupled mass spectrometry (IP-MS) results.
- https://cdn.elifesciences.org/articles/93770/elife-93770-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Gene identifiers.
- https://cdn.elifesciences.org/articles/93770/elife-93770-fig3-data2-v1.xlsx

Mating-type proteins are cell surface proteins but do not localize to cilia.
(A) Fractionation of MTA7-HA cells (please see Figure 4—figure supplement 1A for the experimental process). Red arrowhead, MTA7-HA; F, flow through; P, pellet; R, resin; S, supernatant; W, wash. The MTA signal is undetectable until S3 (enriched membrane proteins), and only appears after affinity chromatography (R). (B) Western blotting (WB) analysis of cell surface proteins. Red arrowhead, MTA7-HA; M, marker; C, negative control (unbiotinylated). (C) Cilia isolation and purification. (D) WB analysis of IP products of membrane and ciliary proteins. Mem, membrane; R, resin; S, supernatant. The same amount of MTA7-HA cells was used for the membrane and ciliary protein IPs. The full blot is shown in Figure 4—figure supplement 1B. (E) Construction scheme for eGFP-tagged MTB2 strains. (F) Costimulated MTB2-eGFP cell. (G) Paired MTB2-eGFP × WT cell. To induce MTB2-eGFP overexpression, cells were treated with 10 ng/ml Cd2+ for 5 hr. Green, eGFP signal; red, tubulin signal; yellow dashed line, cell outline. The focal plane of these images is the cell surface.
-
Figure 4—source data 1
MS analysis of MTA7-HA cilia protein.
- https://cdn.elifesciences.org/articles/93770/elife-93770-fig4-data1-v1.xlsx
-
Figure 4—source data 2
MS analysis of MTB2-eGFP cilia protein.
- https://cdn.elifesciences.org/articles/93770/elife-93770-fig4-data2-v1.xlsx
-
Figure 4—source data 3
TIF containing Figure 4A and original scan of the relevant western blot analysis (anti-HA) with highlighted bands and sample labels.
- https://cdn.elifesciences.org/articles/93770/elife-93770-fig4-data3-v1.zip
-
Figure 4—source data 4
TIF containing Figure 4B and original scan of the relevant western blot analysis (anti-HA) with highlighted bands and sample labels.
- https://cdn.elifesciences.org/articles/93770/elife-93770-fig4-data4-v1.zip
-
Figure 4—source data 5
TIF containing Figure 4D, Figure 4—figure supplement 1B, and original scan of the relevant western blot analysis (anti-HA) with highlighted bands and sample labels.
- https://cdn.elifesciences.org/articles/93770/elife-93770-fig4-data5-v1.zip
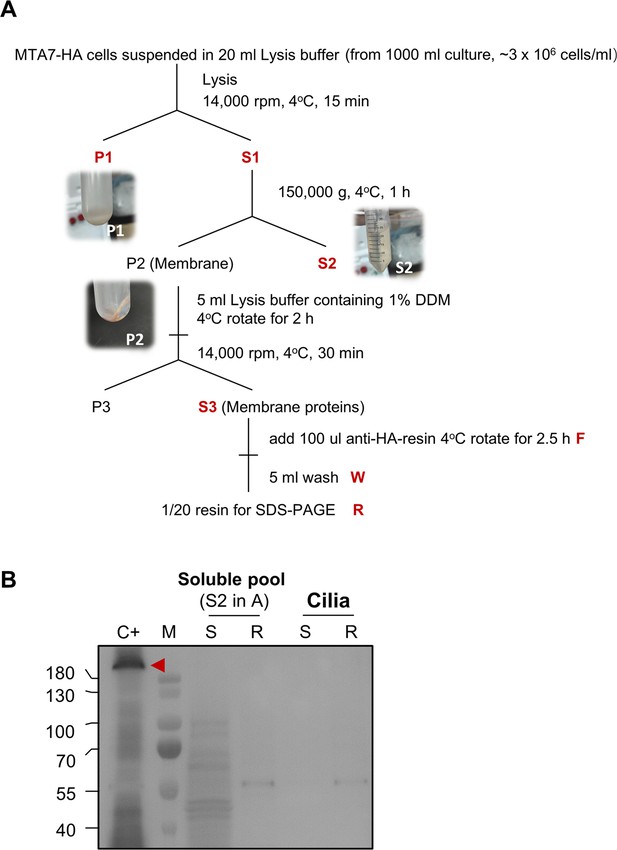
Fractionation of MTA7-HA cells.
(A) Experimental process for cell fractionation. (B) Affinity purification results of the soluble pool (S2, shown in panel A) and ciliary protein. C+, positive control (i.e. IP product of MTA7-HA cell membrane); R, resin; S, supernatant. No MTA7-HA signal was detected in the soluble pool or in ciliary protein samples, even after affinity purification.
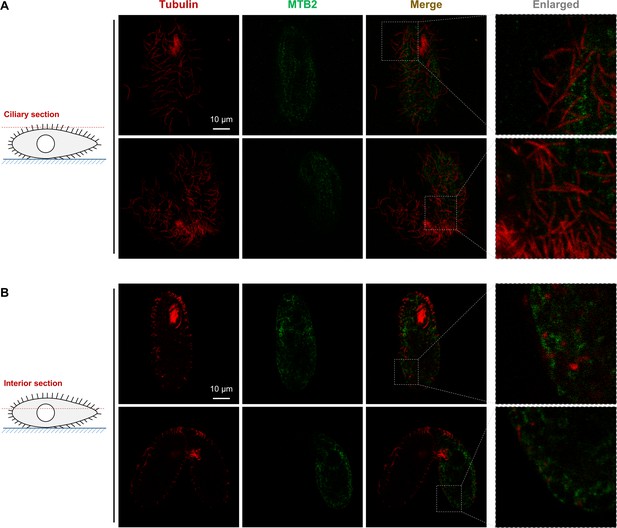
Confocal images of ciliary sections and cell interior sections of MTB2-eGFP cells.
(A) No MTB2-eGFP signal was associated with cilia. (B) In the cell interior section, MTB2-eGFP protein was detected on the plasma membrane and in intracellular structures, probably the endoplasmic reticulum (ER) and Golgi.
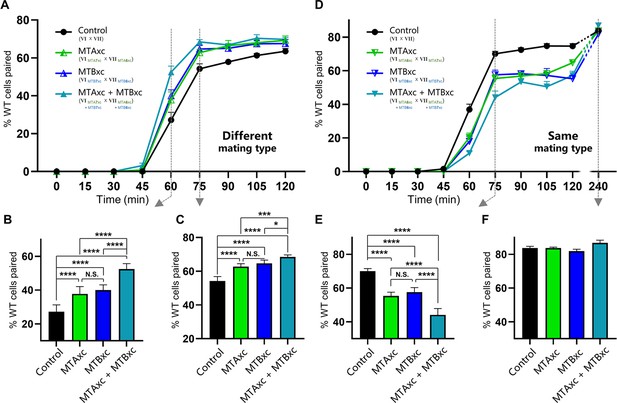
Stimulation experiments using MTAxc and/or MTBxc.
(A) Wild-type (WT) cells were treated with MTAxc and/or MTBxc proteins (30 pg/ml, 1 hr) of different mating-type specificities. WT-VI cells were treated with MTA/B7xc protein, and WT-VII cells were treated with MTA/B6xc protein. Treated cells were washed twice before mixing to remove residual proteins from the starvation medium. Note that the starvation medium used for washing should contain mating-essential factors secreted by T. thermophila cells during starvation (Adair et al., 1978). The mating types used in each experiment is shown in the figure. Each strain was pre-incubated with the strain shown in subscript. Each experiment was repeated five times, with >100 pairs counted at each time point. Matched two-way ANOVA was used for the statistical analysis. N.S., not significant; *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001. Error bars, SEM. (B, C) The percentages of cells paired at 60 min (B) and 75 min (C) were used for the statistical analysis (method described in Figure 2B). (D) WT cells (mating types VI and VII) were treated with MTAxc and/or MTBxc proteins of the same mating-type specificity (as described in A). (E, F) The percentages of cells paired at 75 min (E) and 240 min (F) were used for the statistical analysis (method described in Figure 2B).
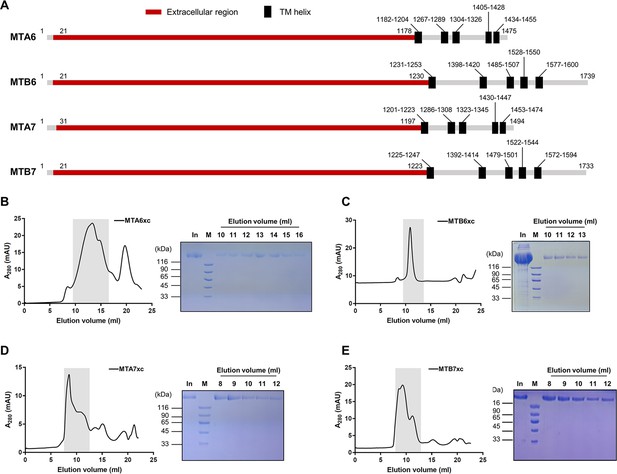
Expression and purification of extracellular regions of mating-type proteins.
(A) Schematic diagram showing truncated and full-sized proteins. (B–E) Size-exclusion chromatography for MTA6xc (B), MTB6xc (C), MTA7xc (D), and MTB7xc (E). Left, elution profile; right, Coomassie blue staining. In, input; M, marker.
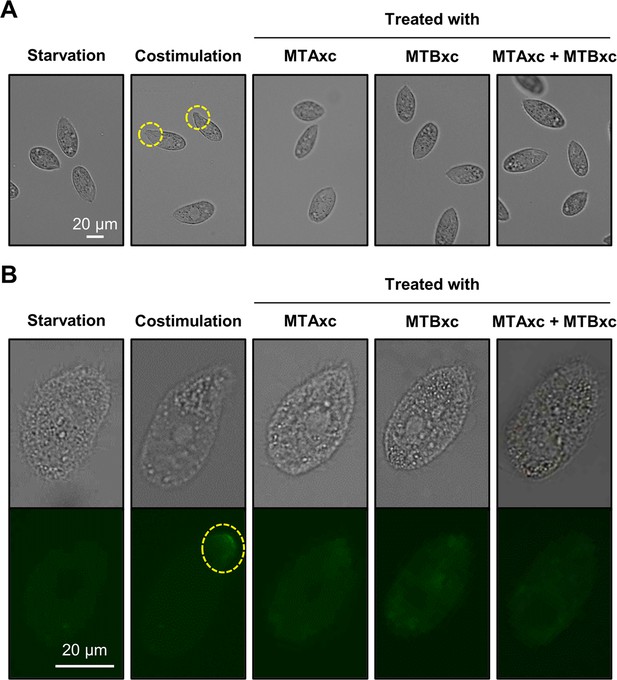
Treatment with MTA6xc and/or MTB6xc proteins fails to induce costimulation.
(A) Tip transformation. Yellow dashed circle, transformed cell tip. (B) Concanavalin A (Con-A) receptor appearance. Yellow dashed circle, Con-A signal. Cells of mating type VII were used for these experiments.

Results of treatment with either MTAxc or MTBxc proteins.
(A–F) Dose–response effect of treatment with MTA6xc or MTB6xc protein. (A–C) MTA6xc results. (D–F) MTB6xc results. Cells of mating types I and VII were used for these experiments. Experimental and statistical methods were as described for Figure 5, except for protein concentrations. (G–J) MTA6xc or MTB6xc proteins affect the mating of various combinations of other WT mating types. (G) MTA6xc results. Both mating partners were treated. (H) MTB6xc results. Both mating partners were treated. (I) MTA6xc results. Cells of only one mating type were treated. Note that mating type VI cells were used in these experiments. (J) MTB6xc results. Note that mating type VI cells were used in these experiments. Each experiment was repeated five times, with >100 pairs counted at each time point. Matched two-way ANOVA was used for the statistical analysis. N.S., not significant; *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001. Error bars, SEM. Experimental and statistical methods were as described for Figure 5. The mating types used in each experiment are shown in the figure. Red subscript ‘A6’ or ‘B6’ in (G–J) indicates the strain treated with MTA6xc or MTB6xc, respectively.
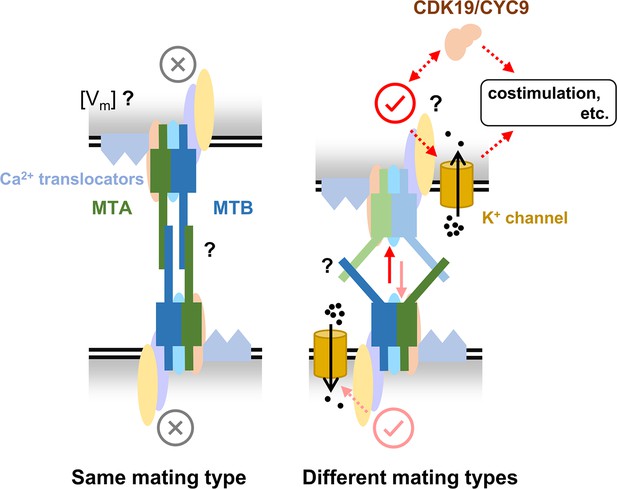
A hypothesized mating-type recognition model.
MTA and MTB function by forming mating-type recognition complex (MTRC) with several other proteins. When cells of the same mating type contact each other, the interaction between MTRCs inhibits mating. Conversely, when cells of different mating types contact each other, the interaction between MTRCs initiates mating. Many details remain unknown, such as (i) during self-recognition, whether the MTRC is only blocked or generates an inhibitory signal (e.g. membrane potential by Ca2+), (ii) how two MTRCs interact during cell–cell recognition, and the differences between self- and non-self-recognition, and (iii) the downstream pathway when MTRC is activated. We hope future studies will help refine and advance this model, contributing to a comprehensive understanding of how mating types are recognized in multiple mating systems.
Videos
Mating behavior of T. thermophila.
Mating behavior of T. thermophila.
To distinguish cells of different mating types, smaller mating type VI cells and larger mating-type VII cells were used in this experiment.
Cells of different mating types form a pair, whereas cells of the same mating type become separated after a short contact.
To distinguish cells of different mating types, smaller mating type VI cells and larger mating-type VII cells were used in this experiment.
Additional files
-
Supplementary file 1
Strains used in this study.
- https://cdn.elifesciences.org/articles/93770/elife-93770-supp1-v1.xlsx
-
Supplementary file 2
Primers used in this study.
- https://cdn.elifesciences.org/articles/93770/elife-93770-supp2-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/93770/elife-93770-mdarchecklist1-v1.pdf





