SPARK regulates AGC kinases central to the Toxoplasma gondii asexual cycle
Figures
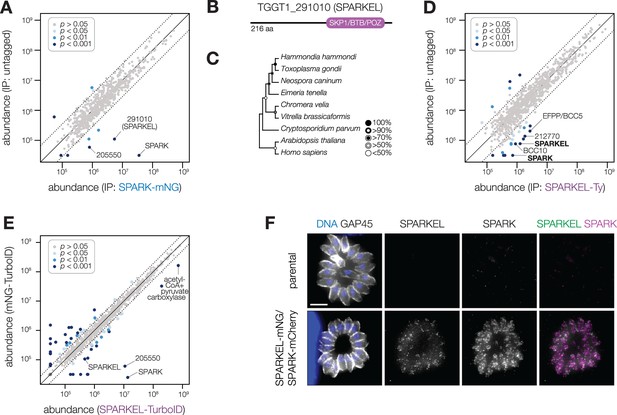
SPARK interacts with an Elongin C-like protein.
(A) Protein abundances from immunopurified SPARK-mNG lysates or an untagged control strain. Dotted lines correspond to one modified z-score. Only proteins quantified by greater than one peptide are shown. Proteins identified in only one IP were assigned a pseudo-abundance of 104.5. (B) TGGT1_291010 SPARK elongin-like protein (SPARKEL) gene model. (C) Neighbor-joining phylogenetic tree of SKP1/BTB/POZ domains of TGGT1_291010 orthologs in apicomplexan species along with human and arabidopsis proteins as outgroups. Bootstrap values determined from 1000 replicates. (D) Protein abundances from immunopurified SPARKEL-Ty lysates or an untagged control strain. Dotted lines correspond to one modified z-score. Only proteins quantified by greater than one peptide are shown. Proteins identified in only one IP were assigned a pseudo-abundance of 104.5. (E) Protein abundances following biotinylation and streptavidin enrichment of samples derived from parasites expressing mNG- or SPARKEL-TurboID fusion constructs. A pseudocount of 104.5 was assigned to proteins identified in only one sample. Point colors correspond to significance thresholds. Dotted lines correspond to one median absolute deviation. (F) Immunofluorescence microscopy of intracellular parasites co-expressing SPARKEL-mNG and SPARK-mCherry. Parasites and nuclei were stained with GAP45 and Hoechst 33342, respectively. Scale bar, 5 µm.
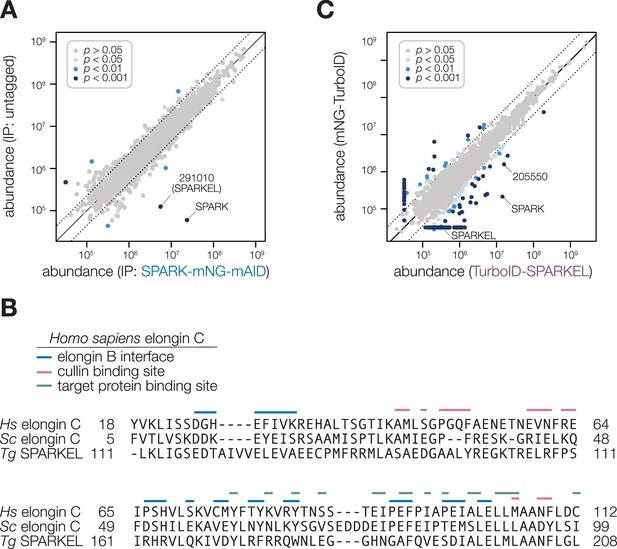
Additional data supporting the interaction between SPARK and SPARKEL.
(A) Protein abundances from immunopurified SPARK-mNG-mAID (Smith et al., 2022) lysates or an untagged control strain from n = 2 biological replicates. Dotted lines correspond to one modified z-score. Only proteins quantified by greater than one peptide are shown. Proteins identified in only one IP were assigned a pseudo-abundance of 104.5. (B) Alignment of the SKP1/BTB/POZ domains of Homo sapiens elongin C, S. cerevisiae elongin C, and SPARKEL. The elongin B interface, cullin binding sites, and target protein binding sites based on the Homo sapiens annotation are shown. (C) Protein abundances following biotinylation and streptavidin enrichment of samples derived from parasites expressing mNG- or TurboID-SPARKEL fusion constructs from n = 2 biological replicates. A pseudocount of 104.5 was assigned to proteins identified in only one sample. Point colors correspond to significance thresholds. Dotted lines correspond to one median absolute deviation.
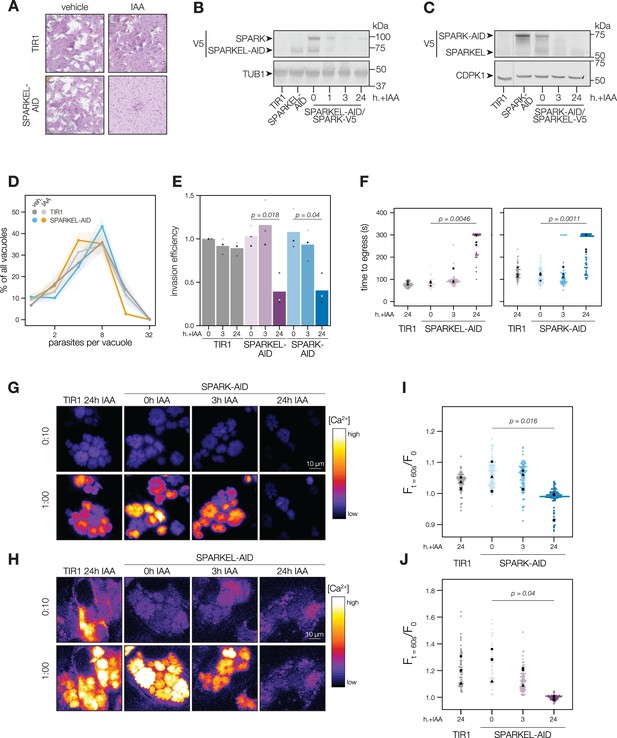
SPARKEL depletion phenocopies the loss of SPARK.
(A) Plaque assays of 500 TIR1 or SPARKEL-AID parasites inoculated onto host cell monolayers and allowed to undergo repeated cycles of invasion, replication, and lysis for 7 days in media with or without 500 µM IAA. (B) Immunoblot of parasites expressing SPARKEL-V5-AID and SPARK-V5 after the indicated h with IAA. TUB1 serves as a loading control. (C) Immunoblot of parasites expressing SPARK-V5-AID and SPARKEL-V5 after the indicated h with IAA. CDPK1 serves as loading control. (D) The number of parasites per vacuole measured for SPARKEL-AID and the parental strain after 24 hr of 500 µM IAA treatment. Mean counts (n=8) are expressed as a percentage of all vacuoles counted. SEM is shown as shaded area. No comparisons yielded significant p-values using ANOVA and Tukey’s test. (E) Invasion assays SPARKEL-AID, SPARK-AID or TIR1 parental strains treated with IAA or vehicle for 3 or 24 hr. Parasites were incubated on host cells for 20 min prior to differential staining of intracellular and extracellular parasites. Parasite numbers were normalized to host cell nuclei for each field. Different shapes correspond to means of n=3 biological replicates. For clarity, only significant comparisons (Welch’s one-sided t-test) are shown. (F) The time to egress of individual intracellular vacuoles following zaprinast treatment. Points correspond to different vacuoles; shapes to different biological replicates (n = 3). Black shapes are the mean for each replicate. p-Values were calculated from a one-tailed t-test. (G, H) Selected frames from live video microscopy of zaprinast-treated SPARK-AID and SPARKEL-AID parasites, respectively, expressing the calcium indicator GCaMP6f, and the corresponding parental strain, 25 hr after infection and with the indicated IAA treatment period. See also Figure 2—video 1 and Figure 2—video 2. (I, J) Relative GCaMP fluorescence of SPARK-AID or SPARKEL-AID vacuoles, respectively, 60 s following zaprinast treatment. Points correspond to different vacuoles; shapes to different biological replicates (n = 3). Black shapes are the mean for each replicate. p-Values were calculated from a one-tailed t-test.
-
Figure 2—source data 1
This file contains source data that was used to generate the blot in Figure 2B.
V5, LICOR.
- https://cdn.elifesciences.org/articles/93877/elife-93877-fig2-data1-v2.zip
-
Figure 2—source data 2
This file contains source data that was used to generate the blot in Figure 2B.
TUB1, LICOR.
- https://cdn.elifesciences.org/articles/93877/elife-93877-fig2-data2-v2.zip
-
Figure 2—source data 3
This file contains source data that was used to generate the blot in Figure 2C.
V5, LICOR.
- https://cdn.elifesciences.org/articles/93877/elife-93877-fig2-data3-v2.zip
-
Figure 2—source data 4
This file contains source data that was used to generate the blot in Figure 2C.
CDPK1, LICOR.
- https://cdn.elifesciences.org/articles/93877/elife-93877-fig2-data4-v2.zip
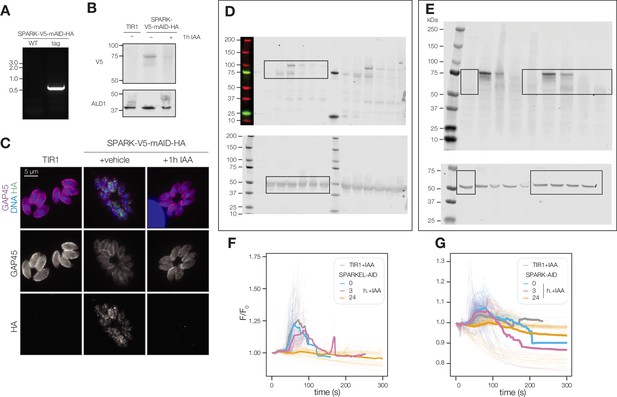
Extended data related to Figure 2.
(A) Primers specific to the 3′ terminus of SPARK and the V5-mAID-HA tagging payload amplified a product in the SPARK-V5-mAID-HA strain but not the untagged parental strain. (B) Confirmation of SPARK-V5-mAID-HA depletion via immunoblot using the V5 epitope. ALD1 was used as a loading control. (C) SPARK-V5-mAID-HA depletion was visualized in formaldehyde-fixed intracellular parasites using the HA epitope and GAP45 staining as a parasite marker. DNA was visualized with Hoechst. HA signal intensity was normalized relative to the TIR1 parental strain. (D) Uncropped immunoblots corresponding to Figure 2B. (E) Uncropped immunoblots corresponding to Figure 2C. (F, G) Normalized GCaMP6f fluorescence of individual SPARK-AID and SPARKEL-AID vacuoles, respectively, after zaprinast treatment and prior to egress (transparent lines) for the indicated period of IAA treatment. The solid line represents the mean normalized fluorescence of all vacuoles across n=3 biological replicates.
-
Figure 2—figure supplement 1—source data 1
This file contains source data that was used to generate the blot in Figure 2—figure supplement 1B.
V5, LICOR.
- https://cdn.elifesciences.org/articles/93877/elife-93877-fig2-figsupp1-data1-v2.zip
-
Figure 2—figure supplement 1—source data 2
This file contains source data that was used to generate the blot in Figure 2—figure supplement 1B.
ALD1, LICOR.
- https://cdn.elifesciences.org/articles/93877/elife-93877-fig2-figsupp1-data2-v2.zip
Representative image series of SPARKEL-AID or TIR1-expresing parental parasites expressing the genetically encoded calcium indicator GcaMP following stimulation with 500 µmM zaprinast after the indicated period of IAA treatment.
Color scale as in Figure 2H.
Representative image series of SPARK-AID or TIR1-expresing parental parasites expressing the genetically encoded calcium indicator GcaMP following stimulation with 500 µmM zaprinast after the indicated period of IAA treatment.
Color scale as in Figure 2G.
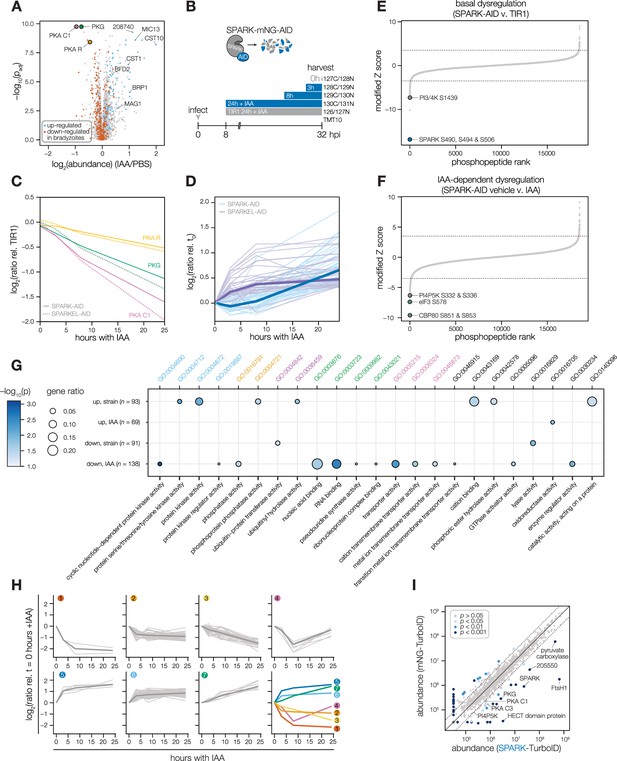
Depletion of SPARK or SPARKEL leads to downregulation of AGC kinases and upregulation of chronic-stage markers.
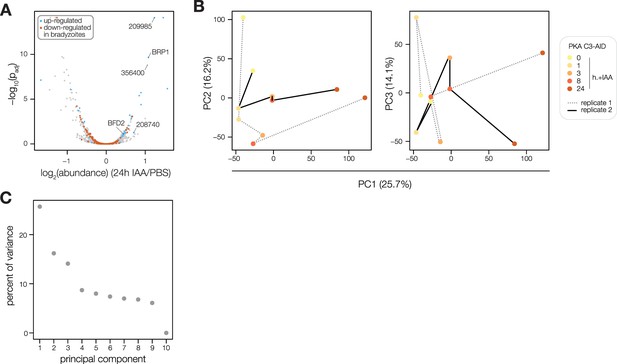
(A) Volcano plot displaying the protein abundance ratios of SPARK-AID parasites treated with IAA or vehicle for 24 hr and adjusted p-values for n = 2 biological replicates. Proteins identified as up- or down-regulated in parasites overexpressing the driver of differentiation (BFD1) (Waldman et al., 2020) are shown in blue and vermilion, respectively. In total, 4474 proteins were quantified, 3847 of which registered more than one peptide. (B) Schematic of the phosphoproteomics experiment following SPARK depletion. Intracellular parasites expressing SPARK-AID were treated with 500 µM IAA for 24, 8, 3, or 0 hr and were harvested at 32 hpi simultaneously with the TIR1 parental strain as a control. Samples were multiplexed with tandem mass tags (TMT). The same experimental design was used for SPARKEL-AID proteomics. Each experiment included two biological replicates. (C) Average protein abundances of PKG, PKA-R, and PKA-C1 relative to the TIR1 parental strain after the indicated period of SPARK (thick lines) or SPARKEL (dotted lines) depletion. (D) Average protein abundances of up-regulated bradyzoite genes relative to the TIR1 parental strain after the indicated period of SPARK (blue lines) or SPARKEL (purple lines) depletion. Up-regulated bradyzoite proteins were defined as those significantly increased in parasites overexpressing BFD1 (Waldman et al., 2020) and two modified Z-scores above the median in the SPARK depletion proteome. Rank-ordered plots of (E) Phosphopeptide basal dysregulation score (peptide abundance in the SPARK-AID strain without IAA treatment relative to the TIR1 parental strain) or (F) IAA-dependent score (summed peptide ratios relative to the SPARK-AID t0 peptide abundance). Dotted lines correspond to 3.5 modified Z scores. Colored points correspond to phosphosites discussed in the main text. (G) Gene ontology (GO) enrichment of phosphoproteins exhibiting SPARK-dependent regulation. Gene ratio is the proportion of proteins with the indicated GO term divided by the total number of proteins. Significance was determined with a hypergeometric test; only GO terms with p<0.05 are shown. Redundant GO terms were removed. Categories discussed in the main text are highlighted with colored text. (H) Gaussian mixture modeling of SPARK-dependent peptides identified by more than one PSM revealed seven kinetically resolved clusters. Individual peptides or the median ratios in each cluster are depicted by light and dark gray lines, respectively. Clusters are numbered according to their discussion in the main text. (I) Protein abundances following biotinylation and streptavidin enrichment of samples derived from parasites expressing mNG- or SPARK-TurboID fusion constructs. A pseudocount of 104.5 was assigned to proteins identified in only one sample. Point colors correspond to significance thresholds for n = 2 biological replicates. Dotted lines correspond to one median absolute deviation.
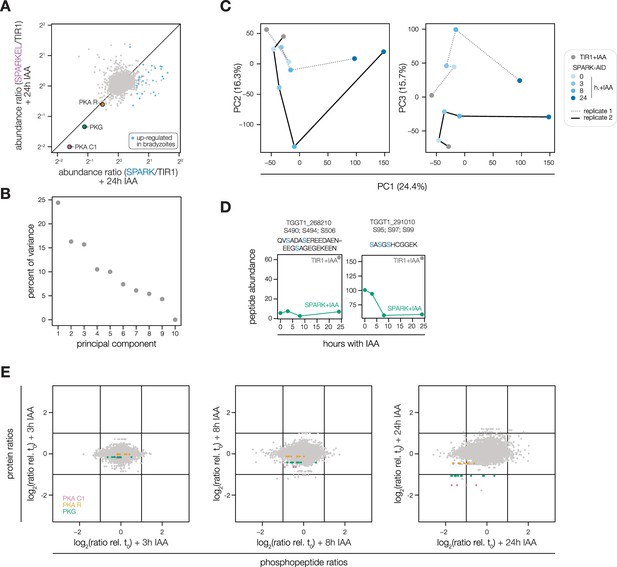
Extended analysis of the SPARK-AID depletion phosphoproteome.
(A) Average protein abundance ratios of SPARK- or SPARKEL-AID parasites treated with IAA for 24 hr relative to the untreated samples. Enriched proteins identified as up-regulated in alkaline-induced bradyzoites (Waldman et al., 2020) are shown in blue. The points corresponding to PKA C1, PKA R, and PKG are highlighted in pink, orange, and green, respectively. (B, C) Principal component analysis of the SPARK-AID depletion phosphoproteome. Plots show the three components accounting for the greatest proportion of the variance (D) Average abundances of SPARK and SPARKEL peptides detected by mass spectrometry. (E) Average protein or phosphopeptide abundance ratios from SPARK-AID parasites treated with IAA for the indicated number of h relative to untreated samples. Values corresponding to PKA C1, PKA R, and PKG are color-coded.
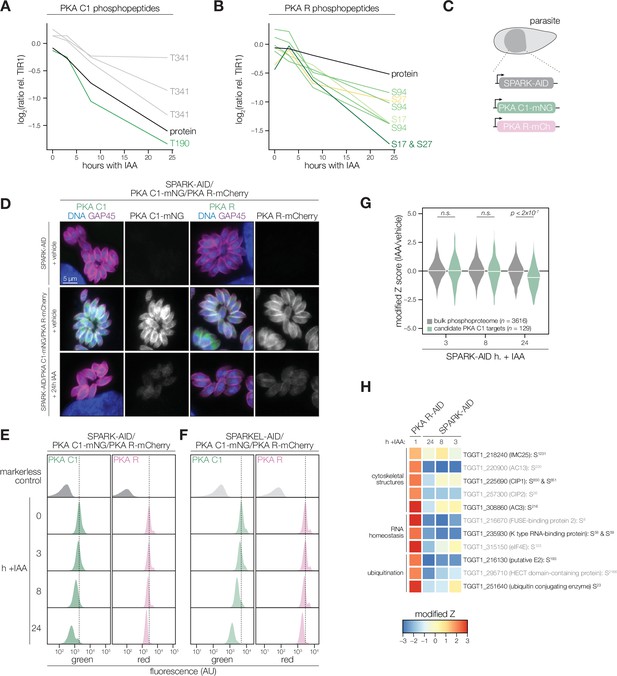
PKA C1 levels are down-regulated upon SPARK depletion.
(A, B) Average protein and phosphopeptide abundances of PKA C1 (A) and PKA R (B) following SPARK depletion. (C) Schematic of the genetic strategy used to monitor PKA C1 and PKA R abundances following SPARK (or SPARKEL) down-regulation with IAA. (D) Immunofluorescence microscopy of parasites expressing SPARK-AID, PKA C1-mNG, and PKA R-mCherry after 0 or 24 hr of IAA treatment to degrade SPARK. Parasites and nuclei were stained with GAP45 and Hoechst 33342, respectively. GAP45 staining and mNG or mCherry staining were normalized to vehicle-treated tagged samples. (E, F) Flow cytometry analysis of parasites expressing PKA C1-mNG, PKA R-mCherry, and SPARK-AID or SPARKEL-AID, respectively, after the indicated period of IAA treatment. The dotted line centers the mode of the vehicle-treated sample. Traces were normalized by unit area. (G) Violin plots displaying the distribution of phosphopeptide abundance values following SPARK depletion. The distributions of candidate PKA C1 targets, as defined in the text and methods, are shown in green. The distributions and p-values (KS test) were derived from the overlapping subset of phosphopeptides identified in each dataset. (H) Heat map of the abundance ratios of candidate PKA C1 targets following SPARK depletion. PKA R depletion results in up-regulation of PKA C1, and candidate PKA C1 targets therefore have positive abundance ratios following PKA R down-regulation.
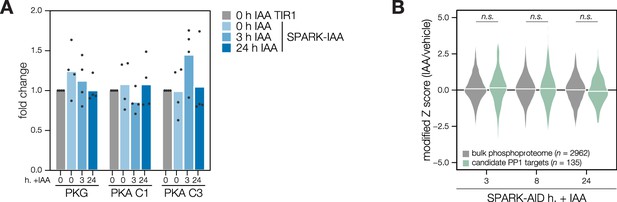
Transcriptional and phosphoproteomic effects of SPARK depletion.
(A) Transcript abundances for PKG, PKA C1, and PKA C3 were compared by qRT-PCR in TIR1 and SPARK-AID, treated with IAA for 0, 3, or 24 hr. Expression levels were normalized to the parental TIR1 strain across n=4 biological replicates. No comparisons yielded significant p-values using a Kruskall-Wallis test. (B) Overlap between the SPARK and PP1 phosphoproteomes. Violin plots displaying the distribution of phosphopeptide abundance values following SPARK depletion. The distributions of candidate PP1 targets, as defined in the text and methods, are shown in green. The distributions and p-values (KS test) were derived from the overlapping subset of phosphopeptides identified in each dataset. PP1 proteome data was obtained from Herneisen et al., 2022.
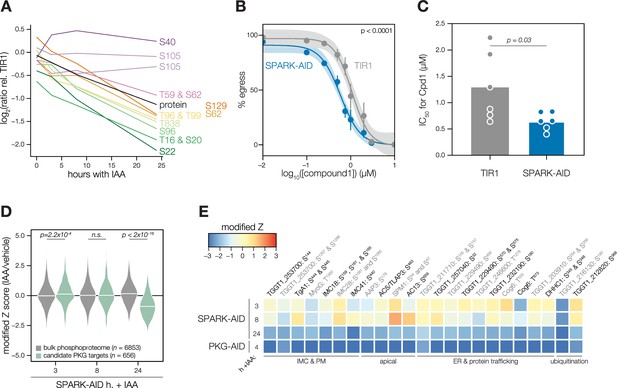
Characterization of PKG function during SPARK depletion.
(A) Protein and phosphopeptide abundances of PKG following SPARK depletion. (B) A23187-stimulated egress assays performed at different concentrations of compound 1 after TIR1 and SPARK-AID parasites had been treated with IAA for 24 hr. Curves were fit to the average values of six replicates and were compared with an extra sum of squares F test. (C) The IC50 values of each strain for compound 1; each point represents a biological replicate (n = 6). Significance was assessed with a two-tailed t-test. (D) Violin plots displaying the distribution of phosphopeptide abundance values following SPARK depletion. The distributions of candidate PKG targets are shown in green. The distributions and p-values (KS test) were derived from the overlapping subset of phosphopeptides identified in each dataset. (E) Heat map of the abundance ratios of candidate PKG targets following SPARK depletion.
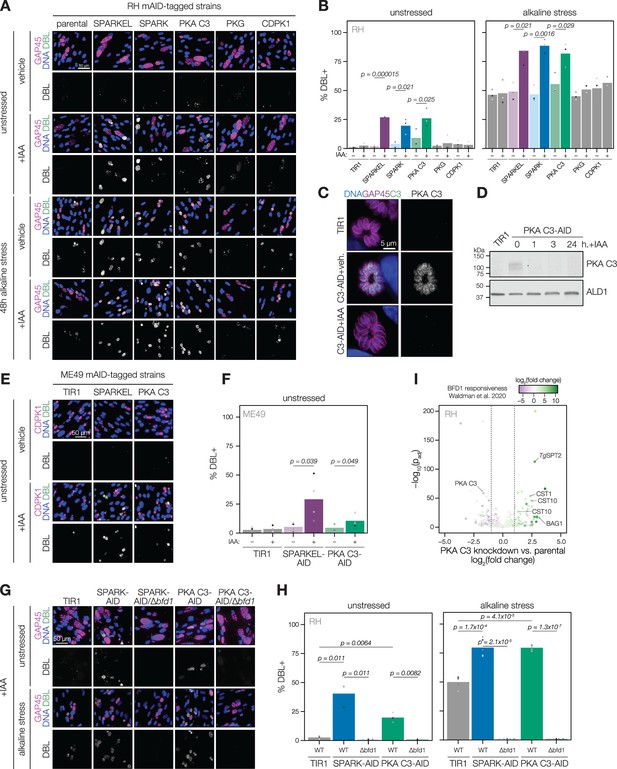
SPARK, SPARKEL, and PKA C3 are negative regulators of differentiation.
(A) Immunofluorescence differentiation assays following knockdown of the indicated AID strains for 48 hr under standard growth conditions (unstressed) or alkaline stress. GAP45 was used to stain parasite vacuoles. Differentiated vacuoles were stained with biotinylated DBA/streptavidin-APC. Nuclei were stained with Hoechst. (B) Quantification of the number of DBL +vacuoles expressed as a percentage of the total stained vacuoles is shown for parasites grown under unstressed or stressed conditions. One-sided t-test, n=3 biological replicates. (C) Fixed, intracellular PKA C3-mNG-AID parasites visualized by immunofluorescence microscopy using the mNG epitope after 1 hr of vehicle or IAA treatment. The mNG signal was internally normalized to the TIR1 parental strain. (D) Immunoblot of PKA C3-AID parasites following the addition of vehicle or 500 µM IAA for 1, 3, or 24 hr. TIR1 was included as an untagged control. PKA C3-AID was detected with V5, and ALD1 was probed as a loading control. (E) Immunofluorescence differentiation assays following knockdown of the indicated ME49/AID-tagged strains under unstressed conditions for 48 hr. Staining was performed as described above, except CDPK1 was used as a parasite vacuole marker. (F) Quantification of the number of DBL +vacuoles expressed as a percentage of the total stained vacuoles in (E). Two-sided t-test, n=5 biological replicates. (G) Immunofluorescence differentiation assays of parasite strains with or without BFD1, depleted of PKA C3 or SPARK with 500 µM IAA and grown under unstressed conditions or alkaline stress for 48 hr. (H) The percentage of DBL +vacuoles corresponding to (G). One-sided t-test. Three biological replicates were quantified under unstressed conditions; five replicates were performed under alkaline stress (I) Effects of 24 hr of PKA C3 knockdown on the transcriptome relative to the untagged strain. Dotted lines correspond to an absolute log2 change of 1. Genes significantly affected by BFD1 overexpression p-value <0.001 as previously defined (Waldman et al., 2020) are colored according to log2 change in the chronic-stage transcriptome. Highlighted points are discussed in the text. P-values were determined from a Wald test implemented in DESeq2.
-
Figure 6—source data 1
This file contains source data that was used to generate the blot in Figure 6D.
V5, LICOR.
- https://cdn.elifesciences.org/articles/93877/elife-93877-fig6-data1-v2.zip
-
Figure 6—source data 2
This file contains source data that was used to generate the blot in Figure 6D.
ALD1, LICOR.
- https://cdn.elifesciences.org/articles/93877/elife-93877-fig6-data2-v2.zip
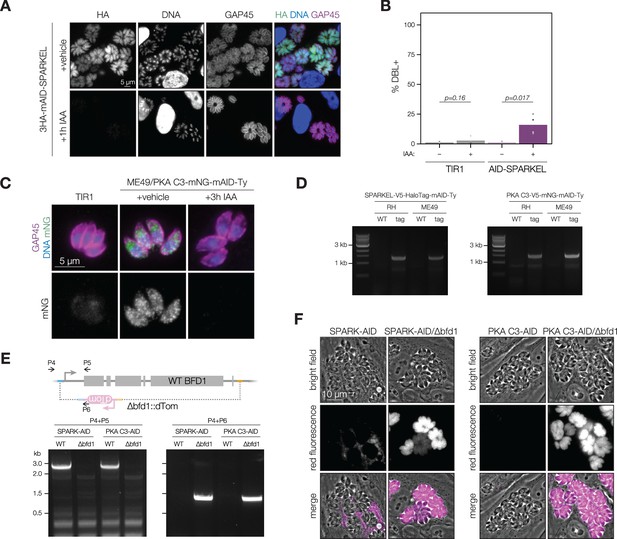
Extended data related to Figure 6.
(A) Fixed, intracellular RH/3HA-mAID-SPARKEL parasites visualized by immunofluorescence microscopy using the HA epitope after 1 hr of vehicle or IAA treatment. The HA signal was normalized to the vehicle-treated sample. (B) Quantification of the number of DBL +vacuoles expressed as a percentage of the total stained vacuoles following 48 hr of AID-SPARKEL knockdown. Two-sided t-test from n = 4 biological replicates. (C) Fixed, intracellular ME49/PKA C3-mNG-AID parasites visualized by immunofluorescence microscopy using the mNG epitope after 3 hr of vehicle or IAA treatment. The mNG signal was normalized to the vehicle-treated sample. (D) Amplification of the SPARKEL-AID and PKA C3-AID genomic loci using tag-specific primers to confirm correct integration of the tagging payload. The integration was confirmed with Sanger sequencing between the 3′ gene junction and CDPK3 3′UTR from the tag (Smith et al., 2022). (E) The strategy used to knock out and replace BFD1 with a dTomato cassette containing homology to sequenced flanking the BFD1 locus (Waldman et al., 2020). Amplification of sequences specific to the intact locus or dTom knockout for the indicated strains are shown below the schematic. Oligonucleotide sequences are listed in Supplementary file 5. (F) Live microscopy images of intact or ∆bfd1::dTom parasites showing red fluorescence arising from the knockout cassette.
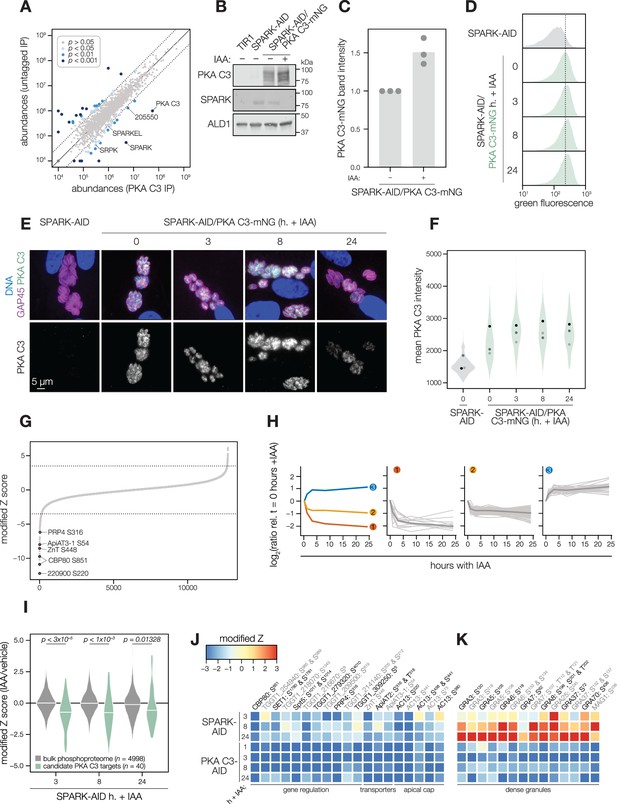
SPARK and PKA C3 physically and genetically interact.
(A) Protein abundances from immunopurified PKA C3-mNG-AID lysates or an untagged control strain from n = 2 biological replicates. Dotted lines correspond to one modified z-score. Only proteins quantified by greater than one peptide are shown. Proteins identified in only one IP were assigned a pseudo-abundance of 104.5. Point colors correspond to significance thresholds. (B) Immunoblot of parasites expressing SPARK-V5-AID-HA and PKA C3-mNG after 24 hr of IAA treatment. ALD1 serves as a loading control. Band intensity normalized to the dual-tagged strain is shown in (C) from three replicates. (D) Flow cytometry analysis of SPARK-AID parasites expressing PKA C3-mNG treated with IAA for the indicated number of h. Traces are representative of two biological replicates. The dotted line centers the mode of the vehicle-treated sample. Traces were normalized by unit area. (E, F) Fixed, intracellular SPARK-AID/PKA C3-mNG parasites visualized by immunofluorescence microscopy using the mNG epitope after the indicated period of IAA treatment (E). The mNG signal was internally normalized to the SPARK-AID parental strain. Quantification of the PKA C3 signal intensity of three replicates is shown in (F). (G) Phosphopeptide IAA-dependent score (summed peptide ratios relative to the PKA C3-AID t0 peptide abundance). Dotted lines correspond to 3.5 modified Z scores. Colored points correspond to phosphosites discussed in the main text. (H) Gaussian mixture modeling of PKA C3-dependent peptides identified by more than one PSM revealed three kinetically resolved clusters. Individual peptides or the median ratios in each cluster are depicted by light and dark gray lines, respectively. Clusters are numbered according to their discussion in the main text. (I) Violin plots displaying the distribution of phosphopeptide abundance values following SPARK depletion. The distributions of candidate PKA C3 targets are shown in green. The distributions and p-values (KS test) were derived from the overlapping subset of phosphopeptides identified in each dataset. (J) Heat map of the abundance ratios of candidate PKA C3 targets following SPARK depletion. (K) Heat map of the abundance ratios of dense granule proteins following SPARK depletion, as discussed in the text.
-
Figure 7—source data 1
This file contains source data that was used to generate the blot in Figure 7D.
mNG, LICOR.
- https://cdn.elifesciences.org/articles/93877/elife-93877-fig7-data1-v2.zip
-
Figure 7—source data 2
This file contains source data that was used to generate the blot in Figure 7D.
V5, LICOR.
- https://cdn.elifesciences.org/articles/93877/elife-93877-fig7-data2-v2.zip
-
Figure 7—source data 3
This file contains source data that was used to generate the blot in Figure 7D.
ALD1, LICOR.
- https://cdn.elifesciences.org/articles/93877/elife-93877-fig7-data3-v2.zip
Extended analysis of the PKA C3 depletion proteome.
(A) Volcano plot displaying the protein abundance ratios of SPARK-AID parasites treated with IAA or vehicle for 24 hr and adjusted p-values. Proteins identified as up- or down-regulated in parasites overexpressing the driver of differentiation (BFD1) (Waldman et al., 2020) are shown in blue and vermilion, respectively. (B, C) Principal component analysis of the PKA C3-AID depletion phosphoproteome. Plots show the three components accounting for the greatest proportion of the variance.
Tables
Proteins discussed in the text.
| Gene ID | Description | Context in text | Modifications | Reference | Dataset |
|---|---|---|---|---|---|
| TGGT1_268210 | AGC kinase | SPARK | Smith et al., 2022 | ||
| TGGT1_291010 | hypothetical protein | SPARKEL | SPARK IP-MS | ||
| TGGT1_205550 | AGC kinase | AGC kinase | SPARK TurboID, SPARKEL TurboID | ||
| TGGT1_310220 | hypothetical protein | BCC10 | Engelberg et al., 2022 | SPARKEL IP-MS | |
| TGGT1_269460 | Ser/Thr phosphatase family protein | EFPP/BCC5 | S550 | Engelberg et al., 2022; Liang et al., 2021; Roumégous et al., 2022 | SPARKEL IP-MS, SPARK depletion timecourse phosphoproteome |
| TGGT1_311360 | protein kinase G AGC kinase family member PKG | PKG | Brown et al., 2017 | 24 hr SPARK depletion proteome, SPARK depletion timecourse phosphoproteome | |
| TGGT1_226030 | AGC kinase | PKA C1 | Jia et al., 2017; Uboldi et al., 2018 | 24 hr SPARK depletion proteome, SPARK depletion timecourse phosphoproteome | |
| TGGT1_242070 | cAMP-dependent protein kinase regulatory subunit | PKA R | S27 | Jia et al., 2017; Uboldi et al., 2018 | 24 hr SPARK depletion proteome, SPARK depletion timecourse phosphoproteome, PKA C3 depletion proteome |
| TGGT1_270240 | MAG1 protein | MAG1 | S238 | Parmley et al., 1994 | 24 hr SPARK depletion proteome, PKA C3 depletion phosphoproteome |
| TGGT1_314250 | bradyzoite rhoptry protein | BRP1 | Schwarz et al., 2005 | 24 hr SPARK depletion proteome, 24 hr PKA C3 depletion proteome | |
| TGGT1_208740 | putative microneme protein | Waldman et al., 2020 | 24 hr SPARK depletion proteome, 24 hr PKA C3 depletion proteome | ||
| TGGT1_264660 | SAG-related sequence SRS44 | CST1 | Tomita et al., 2013 | 24 hr SPARK depletion proteome, PKA C3 depletion transcriptome | |
| TGGT1_312330 | hypothetical protein | CST10 | Tu et al., 2020; Waldman et al., 2020 | 24 hr SPARK depletion proteome, PKA C3 depletion transcriptome | |
| TGGT1_260190 | microneme protein MIC13 | MIC13 | Fritz et al., 2012 | 24 hr SPARK depletion proteome, PKA C3 depletion transcriptome | |
| TGGT1_311100 | zinc finger (CCCH type) motif-containing protein | BFD2 | S183 | Licon et al., 2023 | 24 hr SPARK depletion proteome, 24 hr PKA C3 depletion proteome, PKA C3 depletion phosphoproteome |
| TGGT1_266010 | phosphatidylinositol 3- and 4-kinase | PI3/4 K | S1439 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_202550 | NLI interacting factor family phosphatase | CTD3/BCC6 | T384 and T386 | Engelberg et al., 2022 | SPARK depletion timecourse phosphoproteome |
| TGGT1_224240 | protein phosphatase 2 C domain-containing protein | PPM1 | S557 | Yang and Arrizabalaga, 2017 | SPARK depletion timecourse phosphoproteome |
| TGGT1_268770 | dual specificity phosphatase, catalytic domain-containing protein | phosphosite down-regulated with SPARK depletion | S11 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_219682 | putative pyruvate dehydrogenase kinase | S633 | Ferrarini et al., 2021 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_225960 | STE kinase | S2905 | SPARK depletion timecourse phosphoproteome | ||
| TGGT1_305860 | calcium-dependent protein kinase 3 | CDPK3 | T40 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_267580 | cyclin2-related protein | S491 | SPARK depletion timecourse phosphoproteome | ||
| TGGT1_204280 | cell-cycle-associated protein kinase DYRK | T642, S645, and S649 | Smith et al., 2022 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_239885 | hypothetical protein | S933 and S936 | Smith et al., 2022 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_267100 | protein phosphatase 2 C domain-containing protein | PPM2B | S/T778-788 | Yang et al., 2019 | SPARK depletion timecourse phosphoproteome |
| TGGT1_311310 | protein phosphatase 2B catalytic subunit, calcineurin family phosphatse superfamily protein | CnA | S104 | Paul et al., 2015 | SPARK depletion timecourse phosphoproteome |
| TGGT1_277895 | ubiquitin carboxyl-terminal hydrolase | UBP1 | S44, S52, S1691, and S1695 | Koreny et al., 2023; Wan et al., 2023 | SPARK depletion timecourse phosphoproteome |
| TGGT1_294360 | putative ubiquitin specific protease 39 isoform 2 | USP39 | S20 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_226050 | hypothetical protein | RBR E3 ligase | S319 and S322 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_239410 | hypothetical protein | putative CCR4-NOT complex subunit | S80 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_295658 | zinc finger in N-recognin protein | UBR box E3 ligase | S3335 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_295710 | HECT-domain (ubiquitin-transferase) domain-containing protein | HECT E3 | S2166, S4313 | SPARK depletion timecourse phosphoproteome, PKA R depletion timecourse phosphoproteome | |
| TGGT1_216130 | putative ubiquitin conjugating enzyme E2 | putative E2 enzyme | S193 | SPARK depletion timecourse phosphoproteome, PKA R depletion timecourse phosphoproteome | |
| TGGT1_293670 | transcription elongation factor A TFIIS | TFIIS | S356 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_226810 | histone lysine methyltransferase SET1 | SET1 | S1780 and S1781 | PKA C3 and SPARK depletion timecourse phosphoproteome | |
| TGGT1_218070 | hypothetical protein | NOC3p | S1343 | PKA C3 and SPARK depletion timecourse phosphoproteome | |
| TGGT1_280800 | SWI2/SNF2 SRCAP/Ino80 | TgSRCAP | S635 and S638 | Sullivan et al., 2003 | SPARK depletion timecourse phosphoproteome |
| TGGT1_306660 | RNA pseudouridine synthase superfamily protein | PUS1 | S506 | Anderson et al., 2009 | SPARK depletion timecourse phosphoproteome |
| TGGT1_312370 | RNA pseudouridine synthase superfamily protein | PUS protein | S951 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_231970 | pre-mRNA processing splicing factor PRP8 | PRP8 | T820 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_310820 | putative SLU7 splicing factor | SLU7 | S[516-519] | SPARK depletion timecourse phosphoproteome | |
| TGGT1_267600 | FHA domain-containing protein | a FHA protein | S406 and S413 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_216670 | FUSE-binding protein 2/KH type splicing regulatory protein | KH protein | S8 | PKA R, PKA C3, and SPARK depletion phosphoproteome | |
| TGGT1_241170 | hypothetical protein | KH protein | S362, S368, and S665 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_235930 | domain K- type RNA binding proteins family protein | KH protein | S58 and S59 | Farhat et al., 2021 | SPARK depletion timecourse phosphoproteome |
| TGGT1_227450 | hydrolase, NUDIX family protein | DCP2 homolog | T1982 and S1985 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_260600 | Pumilio-family RNA binding repeat-containing protein | PUF1 | S225 | Joyce et al., 2013; Liu et al., 2014 | SPARK depletion timecourse phosphoproteome |
| TGGT1_246040 | MIF4G domain-containing protein | MIF4G domain protein | S341 and S383 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_249610 | hypothetical protein | CBP80 | S851 and S853 | Gissot et al., 2013 | PKA C3 and SPARK depletion timecourse phosphoproteome |
| TGGT1_317720 | putative eukaryotic translation initiation factor 3 subunit 7 | eIF3 | S587 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_231480 | putative GCN1 | GCN1 | S1100 | SPARK depletion timecourse phosphoproteome | |
| TGGT1_251630 | slc30a2 protein | TgZnT | S448 | Chasen et al., 2019 | PKA C3 and SPARK depletion timecourse phosphoproteome |
| TGGT1_288540 | nucleoside transporter protein | nucleoside transporter | SPARK depletion timecourse phosphoproteome | ||
| TGGT1_260310 | ATP-binding cassette transporter ABC.B1 | ABC transporter | SPARK depletion timecourse phosphoproteome | ||
| TGGT1_318710 | ATP-binding cassette sub-family F member 1 | ABC transporter | SPARK depletion timecourse phosphoproteome | ||
| TGGT1_280660 | HECT-domain (ubiquitin-transferase) domain-containing protein | putative HECT-domain E3 ubiquitin ligase | S5275 and S5279 | SPARK TurboID, SPARK depletion timecourse phosphoproteome | |
| TGGT1_286470 | AGC kinase | PKA C3 | Sugi et al., 2016 | SPARK TurboID | |
| TGGT1_218240 | hypothetical protein | IMC25 | S1231 | Wang et al., 2016 | PKA R and SPARK depletion phosphoproteome |
| TGGT1_220900 | hypothetical protein | AC13 | S101, S220, S659 | Back et al., 2023 | PKA R, PKG, PKA C3, and SPARK depletion phosphoproteome |
| TGGT1_225690 | hypothetical protein | CIP1 | S850 and S851 | Long et al., 2017 | PKA R and SPARK depletion phosphoproteome |
| TGGT1_257300 | hypothetical protein | CIP2 | S26 | Long et al., 2017 | PKA R and SPARK depletion phosphoproteome |
| TGGT1_308860 | hypothetical protein | AC3 | S216 | Chen et al., 2015 | PKA R and SPARK depletion phosphoproteome |
| TGGT1_315150 | putative eukaryotic initiation factor-4E | eIF4E | S1231 | PKA R and SPARK depletion phosphoproteome | |
| TGGT1_235930 | domain K- type RNA binding proteins family protein | KH protein | S58 and S59 | Farhat et al., 2021 | PKA R and SPARK depletion phosphoproteome |
| TGGT1_251640 | ubiquitin-conjugating enzyme subfamily protein | E2 protein | S23 | PKA R and SPARK depletion phosphoproteome | |
| TGGT1_253700 | transporter, major facilitator family protein | MFS transporter | S144, S1287, S1288 | PKG and SPARK depletion phosphoproteome | |
| TGGT1_312100 | plasma membrane-type Ca(2+)-ATPase A1 PMCAA1 | TgA1 | S444 and S445 | PKG and SPARK depletion phosphoproteome | |
| TGGT1_314780 | myosin G | MyoG | T1300 | Frénal et al., 2017 | PKG and SPARK depletion phosphoproteome |
| TGGT1_295360 | hypothetical protein | IMC18 | S159, S161, and S165 | Chen et al., 2015 | PKG and SPARK depletion phosphoproteome |
| TGGT1_239400 | hypothetical protein | IMC28 | S1081 and S1082 | Chen et al., 2017 | PKG and SPARK depletion phosphoproteome |
| TGGT1_225560 | hypothetical protein | IMC41 | S440 | Back et al., 2023 | PKG and SPARK depletion phosphoproteome |
| TGGT1_313480 | hypothetical protein | AAP3 | S178 | Engelberg et al., 2020 | PKG and SPARK depletion phosphoproteome |
| TGGT1_235380 | hypothetical protein | AC5/TLAP3 | S463 | Chen et al., 2015; Liu et al., 2013 | PKG and SPARK depletion phosphoproteome |
| TGGT1_263520 | microtubule associated protein SPM1 | SPM1 | S54 and S57 | Tran et al., 2012 | PKG and SPARK depletion phosphoproteome |
| TGGT1_211710 | TB2/DP1, HVA22 family protein | TB2/DP1, HVA22 family proteins | S155 and S177 | PKG and SPARK depletion phosphoproteome | |
| TGGT1_257040 | TB2/DP1, HVA22 family protein | TB2/DP1, HVA22 family proteins | S32 | PKG and SPARK depletion phosphoproteome | |
| TGGT1_229490 | tetratricopeptide repeat-containing protein | TPR protein TGGT1_229490 | S456, S570 and S575 | PKG and SPARK depletion phosphoproteome | |
| TGGT1_246600 | ABC1 family protein | ER ABC transporter | T1278 | PKG and SPARK depletion phosphoproteome | |
| TGGT1_232190 | Sec7 domain-containing protein | Sec7 protein | S180 | PKG and SPARK depletion phosphoproteome | |
| TGGT1_224150 | hypothetical protein | TgCOG6 | T533 and T673 | Marsilia et al., 2023 | PKG and SPARK depletion phosphoproteome |
| TGGT1_203910 | TBC domain-containing protein | TgTBC10 | T254 and S256 | Quan et al., 2023 | PKG and SPARK depletion phosphoproteome |
| TGGT1_250870 | DHHC zinc finger domain-containing protein | TgDHHC1 | S345 and S348 | Frénal et al., 2013 | PKG and SPARK depletion phosphoproteome |
| TGGT1_212820 | ubiquitin family protein | ubiquitin family protein with a C-terminal extension | S368 | PKG and SPARK depletion phosphoproteome | |
| TGGT1_290970 | 8-amino-7-oxononanoate synthase | TgSPT2 | Nyonda et al., 2022 | PKA C3 depletion transcriptome | |
| TGGT1_259020 | bradyzoite antigen BAG1 | BAG1 | Behnke et al., 2008 | PKA C3 depletion transcriptome | |
| TGGT1_253440 | cell-cycle-associated protein kinase SRPK, putative | SRPK | Talevich et al., 2011 | PKA C3 IP-MS | |
| TGGT1_268960 | putative 5'-AMP-activated protein kinase subunit beta-1 family protein | AMPK subunit beta | Yang et al., 2022 | PKA C3 IP-MS | |
| TGGT1_209985 | cAMP-dependent protein kinase | secreted cAMP-dependent protein kinases | PKA C3 depletion proteome | ||
| TGGT1_356400 | cAMP-dependent protein kinase | secreted cAMP-dependent protein kinases | PKA C3 depletion proteome | ||
| TGGT1_277790 | hypothetical protein | DEP domain protein | S586-600 | PKA C3 depletion phosphoproteome | |
| TGGT1_318150 | transporter, major facilitator family protein | TgApiAT3-1 | S54 | PKA C3 and SPARK depletion phosphoproteomes | |
| TGGT1_259260 | membrane protein FtsH1 | FtsH1 | S553 | PKA C3 depletion phosphoproteome | |
| TGGT1_227280 | dense granule protein GRA3 | GRA3 | S120, T145, S197 | PKA C3 depletion phosphoproteome | |
| TGGT1_312420 | hypothetical protein | GRA38 | S457 | Nadipuram et al., 2016 | PKA C3 depletion phosphoproteome |
| TGGT1_204340 | hypothetical protein | CST8 | T601 | Tu et al., 2020 | PKA C3 depletion phosphoproteome |
| TGGT1_230180 | hypothetical protein | GRA24 | T144 | Braun et al., 2013 | PKA C3 depletion phosphoproteome |
| TGGT1_269180 | MIF4G domain-containing protein | eIF4G1 | S1312 | Holmes et al., 2023 | PKA C3 depletion phosphoproteome |
| TGGT1_218300 | zinc finger (CCCH type) motif-containing protein | zinc finger protein | S663 | PKA C3 depletion phosphoproteome | |
| TGGT1_209500 | hypothetical protein | DNA repair protein | S619 | PKA C3 depletion phosphoproteome | |
| TGGT1_310450 | putative myosin heavy chain | IAP2 | T1269 | PKA C3 depletion phosphoproteome | |
| TGGT1_273050 | hypothetical protein | BCC8 | Liang et al., 2021 | PKA C3 depletion phosphoproteome | |
| TGGT1_287240 | hypothetical protein | condensin 2 | PKA C3 depletion phosphoproteome | ||
| TGGT1_278660 | putative P-type ATPase4 | ATP4 | S83 and T166 | PKA C3 depletion phosphoproteome | |
| TGGT1_233130 | nucleoside transporter protein | nucleoside transporter | S6 | PKA C3 depletion phosphoproteome | |
| TGGT1_292020 | GCC2 and GCC3 domain-containing protein | CRMPb | S7300, S7303, S7363, and S7365 | Singer et al., 2023; Sparvoli et al., 2022 | PKA C3 depletion phosphoproteome |
| TGGT1_221180 | hypothetical protein | microneme protein | S788 | PKA C3 depletion phosphoproteome | |
| TGGT1_304490 | hypothetical protein | microneme protein | S10 | PKA C3 depletion phosphoproteome | |
| TGGT1_309590 | rhoptry protein ROP1 | ROP1 | T248 | PKA C3 depletion phosphoproteome | |
| TGGT1_211290 | rhoptry protein ROP15 | ROP13 | S312 | PKA C3 depletion phosphoproteome | |
| TGGT1_258580 | rhoptry protein ROP17 | ROP17 | T51 | PKA C3 depletion phosphoproteome | |
| TGGT1_291960 | rhoptry kinase family protein ROP40 (incomplete catalytic triad) | ROP40 | S98 | PKA C3 depletion phosphoproteome | |
| TGGT1_308810 | rhoptry neck protein RON9 | RON9 | S190, S239, S327, and S375 | PKA C3 depletion phosphoproteome | |
| TGGT1_310780 | dense granule protein GRA4 | GRA4 | S248 | PKA C3 depletion phosphoproteome | |
| TGGT1_275440 | dense granule protein GRA6 | GRA6 | S133 and S134 | PKA C3 depletion phosphoproteome | |
| TGGT1_203310 | dense granule protein GRA7 | GRA7 | S62, S72, S77, S135, S153 | PKA C3 depletion phosphoproteome | |
| TGGT1_254720 | dense granule protein GRA8 | GRA8 | S198, T202, T262 | PKA C3 depletion phosphoproteome | |
| TGGT1_220240 | hypothetical protein | GRA31 | S420 | Nadipuram et al., 2020; Young et al., 2020 | PKA C3 depletion phosphoproteome |
| TGGT1_217680 | hypothetical protein | GRA57 | S806 | Krishnamurthy et al., 2023; Nadipuram et al., 2020; Young et al., 2020 | PKA C3 depletion phosphoproteome |
| TGGT1_215360 | hypothetical protein | GRA62 | S319 | Cygan et al., 2021 | PKA C3 depletion phosphoproteome |
| TGGT1_249990 | hypothetical protein | GRA70 | S436 | Krishnamurthy et al., 2023; Lockyer et al., 2023 | PKA C3 depletion phosphoproteome |
| TGGT1_289540 | hypothetical protein | SFP1 | S897 | Young et al., 2020 | PKA C3 depletion phosphoproteome |
| TGGT1_251740 | AP2 domain transcription factor AP2XII-9 | AP2XII-9 | S1697 | PKA C3 depletion phosphoproteome | |
| TGGT1_273870 | SWI2/SNF2 ISWI-like (AT hook) | ISWI protein | T765 | PKA C3 depletion phosphoproteome | |
| TGGT1_260240 | hypothetical protein | CAF1 | S272 | PKA C3 depletion phosphoproteome | |
| TGGT1_253750 | PLU-1 family protein | PLU-1 | S3874 and S3877 | Wang et al., 2014 | PKA C3 depletion phosphoproteome |
| TGGT1_300330 | hypothetical protein | GCFC | S27 | PKA C3 depletion phosphoproteome | |
| TGGT1_223880 | zinc finger, C3HC4 type (RING finger) domain-containing protein | zinc finger protein | S3701 | PKA C3 depletion phosphoproteome | |
| TGGT1_288380 | heat shock protein HSP90 | HSP90 | S600 | PKA C3 depletion phosphoproteome | |
| TGGT1_321520 | hypothetical protein | p23 | S109 | PKA C3 depletion phosphoproteome | |
| TGGT1_225450 | hypothetical protein | CSN3 | S509 and S517 | PKA C3 depletion phosphoproteome | |
| TGGT1_242890 | hypothetical protein | PSME4 | S3127 | PKA C3 depletion phosphoproteome | |
| TGGT1_218960 | AP2 domain transcription factor AP2XII-1 | AP2XII-1 | T1697, S1702, T1667, T1703 | Antunes et al., 2024 | PKA C3 depletion phosphoproteome |
| TGGT1_262150 | kelch repeat and K+channel tetramerisation domain containing protein | Kelch13 | S139 | Harding et al., 2020; Koreny et al., 2023; Wan et al., 2023 | PKA C3 depletion phosphoproteome |
| TGGT1_254940 | MIF4G domain-containing protein | eIF4G2 | S985 and S989 | Holmes et al., 2023 | PKA C3 and SPARK depletion phosphoproteomes |
| TGGT1_320020 | transporter, major facilitator family protein | TgApiAT2 | S316 and T318 | PKA C3 and SPARK depletion phosphoproteomes | |
| TGGT1_233000 | KOW motif domain-containing protein | Spt5 | S1011 and S1014 | PKA C3 and SPARK depletion phosphoproteomes | |
| TGGT1_279320 | hypothetical protein | nucleotidyltransferase | S4010 | PKA C3 and SPARK depletion phosphoproteomes | |
| TGGT1_313180 | putative cell-cycle-associated protein kinase PRP4 | PRP4 | S316 | Swale et al., 2022 | |
| TGGT1_214140 | hypothetical protein | associates with AP2IX4/MORC complex | S205 and S212 | ||
| TGGT1_309250 | hypothetical protein | associates with GCN5b complex | PMID: 31381949 and 24391497 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background RH (T. gondii) | RH/TIR1 | PMID: 28465425 | RH/TIR1/∆KU80/∆HXGPRT. Mycoplasma negative. | |
| Strain, strain background RH (T. gondii) | ME49/TIR1 | PMID: 37081202 | ME49/TIR1/∆KU80/∆HXGPRT. Mycoplasma negative. | |
| Strain, strain background RH (T. gondii) | RH/TIR1/GCaMP6f | PMID: 35484233 | RH/TIR1/pTUB1-GCaMP6f/∆KU80/∆HXGPRT. Mycoplasma negative. | |
| Strain, strain background RH (T. gondii) | RH/TIR1/GCaMP6f | PMID: 35976251 | RH/TIR1/pMIC2-MIC2-GLuc-myc-P2A-GCaMP6f/∆KU80/∆HXGPRT. Mycoplasma negative. | |
| Strain, strain background RH (T. gondii) | RH/TIR1/mNG-TurboID | PMID: 36712004 | RH/TIR1/mNG-TurboID-Ty/∆KU80/∆HXGPRT. Mycoplasma negative. | |
| Strain, strain background RH (T. gondii) | RH/TIR1/PKG-AID | PMID: 28465425 | TGGT1_ 311360 | RH/TIR1/∆KU80/∆HXGPRT/PKG-mAID-HA/HXGPRT. Mycoplasma negative. |
| Strain, strain background RH (T. gondii) | RH/TIR1/CDPK1-AID | PMID: 37610220 | TGGT1_ 301440 | RH/TIR1/∆KU80/∆HXGPRT/CDPK1-V5-mNeonGreen-mAID-Ty. Mycoplasma negative. |
| Strain, strain background RH (T. gondii) | SPARK-mNG-AID | PMID: 35484233 | TGGT1_ 268210 | RH/TIR1/∆KU80/∆HXGPRT/SPARK-V5-mNG-mAID-Ty. Mycoplasma negative. |
| Strain, strain background RH (T. gondii) | SPARK-mNG | This paper | TGGT1_ 268210 | RH/∆KU80/∆HXGPRT/SPARK-V5-mNG-Ty. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | SPARKEL-TurboID | This paper | TGGT1_ 291010 | RH/TIR1/∆KU80/∆HXGPRT/SPARKEL-TurboID-Ty. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | TurboID-SPARKEL | This paper | TGGT1_ 291010 | RH/TIR1/∆KU80/∆HXGPRT/3xHA-TurboID-SPARKEL. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | SPARKEL-Ty | This paper | TGGT1_ 291010 | RH/TIR1/∆KU80/∆HXGPRT/SPARKEL-V5-mNG-Ty. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | SPARKEL-mNG/SPARK-mCherry | This paper | TGGT1_ 268210, TGGT1_ 291010 | RH/TIR1/∆KU80/∆HXGPRT/SPARKEL-V5-mNG-Ty/SPARK-V5-mCherry-HA/HXGPRT. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | SPARKEL-AID | This paper | TGGT1_ 291010 | RH/TIR1/∆KU80/∆HXGPRT/SPARKEL-V5-HaloTag-mAID-Ty. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | SPARKEL-AID/ME49 | This paper | TGGT1_ 291010 | ME49/TIR1/∆KU80/∆HXGPRT/SPARKEL-V5-HaloTag-mAID-Ty. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | SPARKEL-AID/GCaMP | This paper | TGGT1_ 291010 | RH/TIR1/pMIC2-MIC2-GLuc-myc-P2A-GCaMP6f/∆KU80/∆HXGPRT/SPARKEL-V5-HaloTag-mAID-Ty. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | SPARKEL-AID/SPARK-mCherry | This paper | TGGT1_ 268210, TGGT1_ 291010 | RH/TIR1/∆KU80/∆HXGPRT/SPARKEL-V5-HaloTag-mAID-Ty/SPARK-V5-mCherry-HA/HXGPRT. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | SPARK-AID | This paper | TGGT1_ 268210 | RH/TIR1/∆KU80/∆HXGPRT/SPARK-V5-mAID-HA/HXGPRT. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | SPARK-AID/SPARKEL-mNG | This paper | TGGT1_ 268210, TGGT1_ 291010 | RH/TIR1/∆KU80/∆HXGPRT/SPARKEL-V5-mNG-Ty/SPARK-V5-mAID-HA/HXGPRT. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | PKA R-AID | This paper | TGGT1_ 242070 | RH/TIR1/∆KU80/∆HXGPRT/PKA R-V5-mAID-HA/HXGPRT. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | AID-SPARKEL | This paper | TGGT1_ 291010 | RH/TIR1/∆KU80/∆HXGPRT/3xHA-mAID-SPARKEL. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | SPARK-TurboID | This paper | TGGT1_ 268210 | RH/TIR1/∆KU80/∆HXGPRT/SPARK-TurboID-Ty. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | PKA C1-mNG/PKA R-mCherry | This paper | TGGT1_ 226030, TGGT1_ 242070 | RH/TIR1/∆KU80/∆HXGPRT/PKA C1-mNG-3myc/PKA R-mCherry-Ty. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | SPARKEL-AID/PKA C1-mNG/PKA R-mCherry | This paper | TGGT1_ 291010, TGGT1_ 226030, TGGT1_ 242070 | RH/TIR1/∆KU80/∆HXGPRT/PKA C1-mNG-3myc/PKA R-mCherry-Ty/SPARKEL-V5-HaloTag-mAID-Ty. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | SPARK-AID/PKA C1-mNG | This paper | TGGT1_ 268210, TGGT1_ 226030 | RH/TIR1/∆KU80/∆HXGPRT/SPARK-V5-mAID-HA/HXGPRT/PKA C1-mNG-3myc. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | SPARK-AID/PKA C1-mNG/PKA R-mCherry | This paper | TGGT1_ 268210, TGGT1_ 226030, TGGT1_ 242070 | RH/TIR1/∆KU80/∆HXGPRT/SPARK-V5-mAID-HA/HXGPRT/PKA C1-mNG-3myc/PKA R-mCherry-P2A-DHFR. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | PKA C3-AID | This paper | TGGT1_ 286470 | RH/TIR1/∆KU80/∆HXGPRT/PKA C3-V5-mNG-mAID-Ty. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | PKA C3-AID/ME49 | This paper | TGGT1_286470 | ME49/TIR1/∆KU80/∆HXGPRT/PKA C3-V5-mNG-mAID-Ty. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | SPARK-AID/∆BFD1 | This paper | TGGT1_ 268210, TGGT1_ 200385 | RH/TIR1/∆KU80/∆HXGPRT/∆BFD1::dTomato/SPARK-V5-mAID-HA/HXGPRT. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | PKA C3-AID/∆BFD1 | This paper | TGGT1_ 286470, TGGT1_ 200385 | RH/TIR1/∆KU80/∆HXGPRT/∆BFD1::dTomato/PKA C3-V5-mNG-mAID-Ty. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Strain, strain background RH (T. gondii) | PKA C3-mNG/SPARK-AID | This paper | TGGT1_ 286470, TGGT1_ 268210 | RH/TIR1/∆KU80/∆HXGPRT/PKA C3-mNG-P2A-DHFR/SPARK-V5-mAID-HA/HXGPRT. Mycoplasma negative. Strain construction and validation described in the Materials and Methods section. Strain available upon request. |
| Cell line (Homo sapiens) | Human Foreskin Fibroblasts (HFFs) | ATCC | SCRC- 1041 | Mycoplasma negative |
| Antibody | Rat monoclonal (16D7) anti-mCherry | Thermo Fisher | IFA (1/1,000) | |
| Antibody | Mouse monoclonal (32F6) anti-mNG | ChromoTek | IFA (1/1,000) and WB (1/500) | |
| Antibody | Guinea pig monoclonal anti-CDPK1 | Covance | Custom antibody | WB (1/50,000) |
| Antibody | Rabbit polyclonal anti-GAP45 | Lampire Biological Laboratory | Provided by R. Drew Etheridge Lab. IFA (1/1,000). Used for differentiation assays. | |
| Antibody | Rabbit polyclonal anti-GAP45 | PMID: 18312842 | Provided by Dominique Soldati-Favre Lab. IFA (1/10,000) | |
| Antibody | Mouse polyclonal anti-SAG1 | PMID: 3183382 | Provided by L. David Sibley Lab. IFA (1/1,000). Used for invasion assays. | |
| Antibody | Mouse monoclonal anti-V5 | Invitrogen | Invitrogen: R960-25 | WB (1/1000) |
| Antibody | Mouse monoclonal anti-TUB1 (clone 12G10) | Developmental Studies Hybridoma Bank at the University of Iowa | RRID: AB_1157911 | WB (1/5000) |
| Antibody | Rat monoclonal (3F10) anti-HA | Roche | RRID: AB_390919 | IFA (1/1000) |
| Antibody | Mouse monoclonal (BB2) anti-Ty1 | PMID: 8813669 | Protein G crosslinking (60 µg/1 mg beads) | |
| Antibody | Rabbit polyclonal clone WU1614 anti-ALD | PMID: 16923803 | Provided by L. David Sibley Lab. WB (1/10,000) | |
| Antibody | biotinylated dolichos | Vector labs B-1035 | IFA (1/1000) | |
| Chemical compound, drug | Hoechst 33342 | Invitrogen | Invitrogen: H3570 | IFA (1/20,000) |
| Chemical compound, drug | DAPI (4',6-Diamidino-2-Phenylindole, Dihydrochloride) | Invitrogen | Invitrogen: D1306 | |
| Chemical compound, drug | Prolong Diamond | Thermo Fisher | Thermo Fisher: P36965 | |
| Chemical compound, drug | Zaprinast | Calbiochem | Calbiochem: 684500 | Egress assay (500 µM) |
| Chemical compound, drug | A23187 | Calbiochem | Calbiochem: 100105 | Egress assay (2 µM) |
| Chemical compound, drug | Compound 1 | PMID: 12455981 | Egress assay (as indicated) | |
| Chemical compound, drug | Biotin | Sigma Aldrich | Sigma Aldrich: B4501-1G | TurboID (500 µM) |
| Chemical compound, drug | TRIzol reagent | Thermo Fisher | Thermo Fisher Scientific: 15596018 | |
| Commercial assay or kit | S-trap micro | Protifi | Protifi: C02-micro-80 | |
| Commercial assay or kit | Pierce Quantitative Fluorometric Peptide Assay | Thermo Fisher Scientific | Thermo Fisher Scientific: 23290 | |
| Commercial assay or kit | TMT10plex Label Reagent Set | Thermo Fisher Scientific | Thermo Fisher Scientific: 90111 | |
| Commercial assay or kit | TMTpro 16plex Label Reagent Set | Thermo Fisher Scientific | Thermo Fisher Scientific: A44522 | |
| Commercial assay or kit | EasyPep MS Sample Prep Kits - Maxi | Thermo Fisher Scientific | Thermo Fisher Scientific: A45734 | |
| Commercial assay or kit | High-Select TiO2 Phosphopeptide Enrichment Kit | Thermo Fisher Scientific | Thermo Fisher Scientific: A32993 | |
| Commercial assay or kit | High-Select Fe-NTA Phosphopeptide Enrichment Kit | Thermo Fisher Scientific | Thermo Fisher Scientific: A32992 | |
| Commercial assay or kit | Pierce High pH Reversed-Phase Peptide Fractionation Kit | Thermo Fisher Scientific | Thermo Fisher Scientific: 84868 | |
| Commercial assay or kit | Pierce Streptavidin Magnetic Beads | Thermo Scientific | Thermo Scientific: 88817 | |
| Commercial assay or kit | Sep-Pak C18 Plus Short Cartridge, 360 mg Sorbent per Cartridge, 55–105 µm | Waters | Waters: WAT020515 | |
| Commercial assay or kit | Pierce Protein G Magnetic Beads | Thermo Fisher | Thermo-Fisher: 88847 | |
| Commercial assay or kit | mNeonGreen-Trap Magnetic Agarose | ChromoTek | ChromoTek:ntma-20 | |
| Commercial assay or kit | NEXTFLEX Poly(A) Beads 2.0 Kit | Perkin Elmer | NOVA-512993 | |
| Commercial assay or kit | SuperScriptIII Reverse Transcriptase | Invitrogen | Invitrogen: 18080044 | |
| Commercial assay or kit | PowerUp SYBR Green master mix for qPCR | Applied Biosystems | A25779 | |
| Software, algorithm | Proteome Discoverer 4.2 | Thermo Fisher | ||
| Software, algorithm | R version 4.0 | R Foundation for Statistical Computing | ||
| Software, algorithm | Prism 8 | GraphPad | ||
| Software, algorithm | HHPRED | PMID: 29258817 | ||
| Software, algorithm | MEGA | PMID: 33892491 | ||
| Software, algorithm | ClustalW | PMID: 17846036 | ||
| Software, algorithm | TrimGalore version 0.6.7 | doi:10.5281/zenodo. 7598955 | ||
| Software, algorithm | STAR version 2.7.1 a | PMID: 23104886 | ||
| Software, algorithm | featureCounts version 1.6.2 | PMID: 24227677 | ||
| Software, algorithm | DESeq2 version 1.30.1 | PMID: 25516281 |
Additional files
-
Supplementary file 1
Protein quantification and statistical tests for immunoprecipitation-mass spectrometry experiments (SPARK, SPARKEL, and PKA C3-tagged strains), exported from the Proteome Discoverer 2.4 software.
- https://cdn.elifesciences.org/articles/93877/elife-93877-supp1-v2.xlsx
-
Supplementary file 2
Protein quantification and statistical tests for TurboID mass spectrometry experiments (SPARKEL and SPARK TurboID-tagged strains), exported from the Proteome Discoverer 2.4 software.
- https://cdn.elifesciences.org/articles/93877/elife-93877-supp2-v2.xlsx
-
Supplementary file 3
Protein and peptide quantification and statistical tests for SPARK, SPARKEL, and PKA C3 depletion proteomes, exported from the Proteome Discoverer 2.4 software.
- https://cdn.elifesciences.org/articles/93877/elife-93877-supp3-v2.xlsx
-
Supplementary file 4
Peptide ratios quantified in the PKA R-AID, and PKG-AID, and SPARK-AID depletion phosphoproteomes.
Ratios are reported relative to the vehicle-treated sample within each experiment. Modified Z-scores were calculated by standardizing each value with respect to the median and median absolute deviation within each ratio.
- https://cdn.elifesciences.org/articles/93877/elife-93877-supp4-v2.xlsx
-
Supplementary file 5
Oligonucleotides and DNA sequences used in this study.
- https://cdn.elifesciences.org/articles/93877/elife-93877-supp5-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/93877/elife-93877-mdarchecklist1-v2.docx





