Combinatorial CRISPR screen reveals FYN and KDM4 as targets for synergistic drug combination for treating triple negative breast cancer
Figures
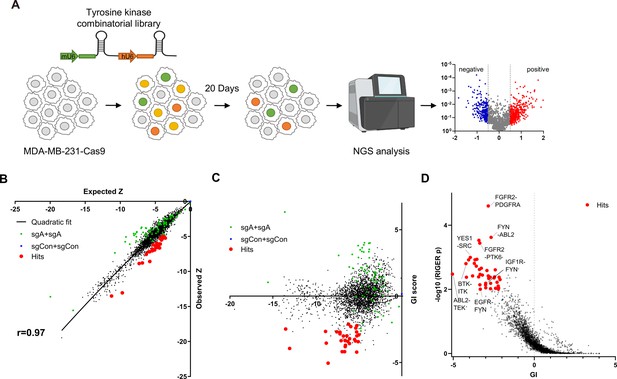
Pairwise CRISPR screen reveals combinations of synthetic lethal tyrosine kinase ablations.
(A) Schematic diagram of combinatorial screens performed in TNBC cell line MDA-MB-231. (B) Scatter plot of expected growth phenotype Z score and observed growth phenotype Z score of each gene combination. Green dots indicate gene combinations where the identical gene is targeted by the two sgRNAs. Red dots indicate candidate synthetic lethal gene pairs listed in Supplementary file 2. (C) Scatter plot of growth phenotype Z score and normalized GI score of each gene combination. (D) Scatter plot of gene level GI score and RIGER p value calculated with GI scores of each sgRNA pairs that target the given gene pair.
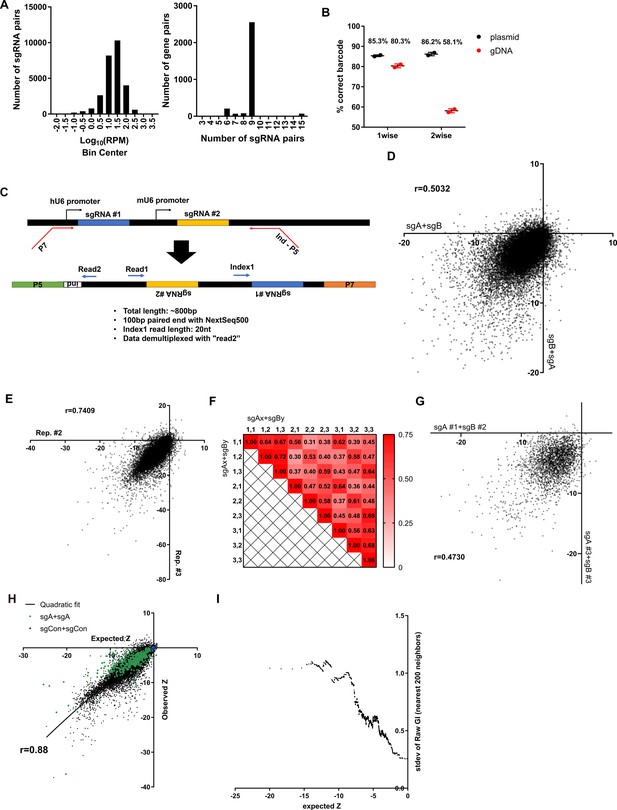
Quality controls for combinatorial sgRNA libraries and screening results.
(A) Summary of representations of each sgRNA pair in the combinatorial library. (Left) Histogram of the frequency of each sgRNA combinations. (Right) Histogram of the number of sgRNA pairs that target a gene pair. (B) Accuracy of sgRNA-barcode pairing in each library (n=2). (C) Sequencing scheme for identifying both sgRNAs in the library by Next Generation Sequencing. (D) Scatter plot of growth phenotype score Z of two different permutations of the same sgRNA pairs. (E) Scatter plot of growth phenotype score Z of two independent biological replicates. (F) Heatmap of the Pearson’s r values between growth phenotype score of each sgRNA pairs that target the same gene pair. (G) Scatter plot of growth phenotype score Z of different sgRNA pairs that target the same gene pair. (H) Expected and Observed growth phenotype score Z of all sgRNA pairs. (I) Standard deviation of the GI score of 200 neighboring gene pairs by expected growth phenotype score. All data are plotted as mean ± s.d. All replicates are biological replicates.
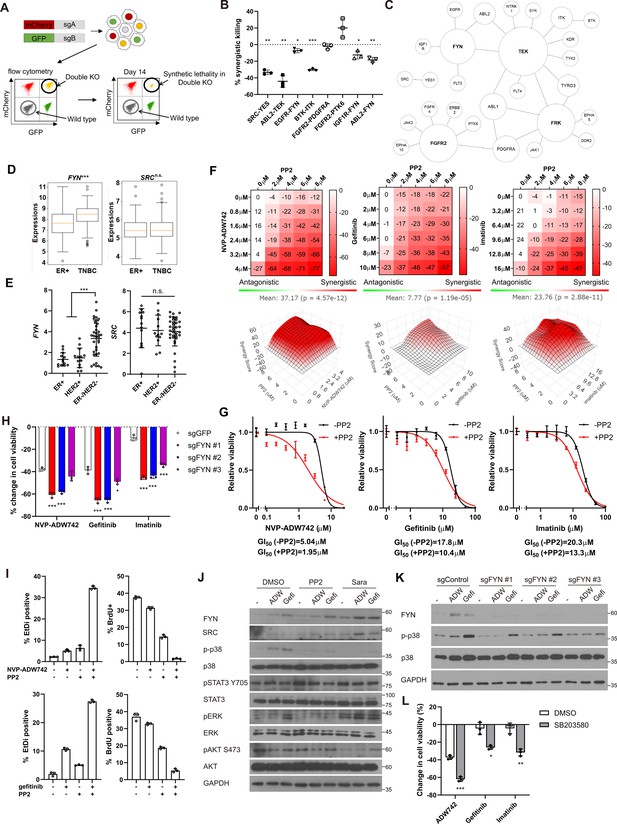
FYN is critical mediator of TKI resistance.
(A) Schematic diagram of in vitro validation of synthetic lethal gene pairs using sgRNAs. (B) Summary of synergistic killing by sgRNAs targeting indicated gene pairs (n=3). (C) Network analysis of the 30 candidate synthetic lethal gene pairs highlighted in Figure 1D. The size of each node is manually drawn to be proportional to the number of connections the gene has. (D, E) FYN and SRC mRNA expressions in (D) microarray data of primary breast cancers of GSE25066 cohort, and (E) in cancer cell line encyclopedia, for indicated subtypes. (F) Summary of MTT assay with MDA-MB-231 cells treated with the TKI combinations at indicated concentrations (n=2). Synergistic killing is calculated using SynergyFinder with Bliss independence model. Raw data for the analysis is available in Figure 2—source data 1. (G) Dose response curve of the indicated TKI in the presence and absence of 5 μM PP2 treated for 72 hr (n=3). (H) MTT assay with MDA-MB-231 Cas9 cells expressing indicated sgRNAs treated with indicated TKIs for 72 hr (n=3). (I) Cell death and cell proliferation in MDA-MB-231 cells treated with NVP-ADW742, gefitinib and PP2 either as single agent or as combination for 72 hr (n=3). (J) Western blot analysis of MDA-MB-231 cells treated with indicated drugs for 48 hr. (K) Western blot analysis of MDA-MB-231 Cas9 cells expressing indicated sgRNA and treated with indicated drugs for 48 hr. (L) MTT assay of MDA-MB-231 cells treated with indicated drugs for 72 hr (SB203580: 10 μM, NVP-ADW742: 4 μM, gefitinib: 10 μM, imatinib: 10 μM) (n=3). PP2, Saracatinib, NVP-ADW742, gefitinib, and imatinib were treated at 5 μM, 5 μM, 4 μM, 12 μM, 12 μM unless otherwise indicated. All data are plotted as mean ± s.d. One sample t-test for B, and unpaired two-sided Student’s t-test in D,E,H, and L. *, p<0.05; **, p<0.01; ***, p<0.001; n.s., p>0.05. All replicates are biological replicates.
-
Figure 2—source data 1
Original files for western blot analysis displayed in Figure 2J and K.
- https://cdn.elifesciences.org/articles/93921/elife-93921-fig2-data1-v1.zip
-
Figure 2—source data 2
Image files containing original western blots for Figure 2J and K, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/93921/elife-93921-fig2-data2-v1.zip
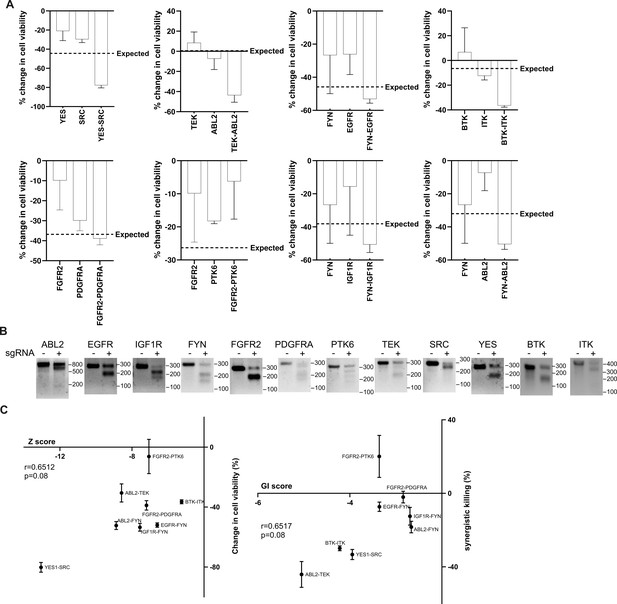
Validation of screening hits.
(A) The changes in cell viability upon each single or double tyrosine kinase knockout in MDA-MB-231 cells analyzed in Figure 2A and B. Expected double knockout phenotype was calculated using Bliss independence model. (B) T7 endonuclease assay of MDA-MB-231 cells infected with lentivirus expressing sgRNA targeting the indicated genes. (C) Scatter plot of (Left) growth phenotype score Z and the change in cell viability and (Right) GI score and synergistic killing by the indicated sgRNA combinations. Correlations were analyzed by Pearson’s r. All data are plotted as mean ± s.d. All replicates are biological replicates.
-
Figure 2—figure supplement 1—source data 1
Original files for gel image displayed in Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/93921/elife-93921-fig2-figsupp1-data1-v1.zip
-
Figure 2—figure supplement 1—source data 2
Image files for gel image displayed in Figure 2—figure supplement 1B, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/93921/elife-93921-fig2-figsupp1-data2-v1.zip
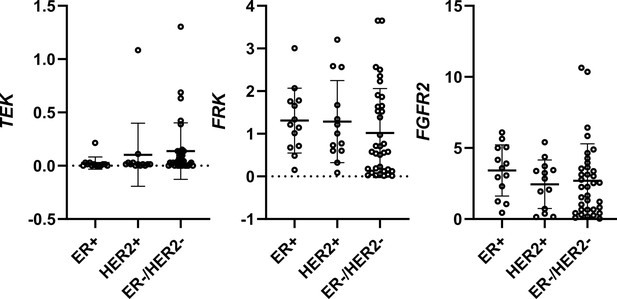
Expression of TEK, FRK, and FGFR2 in breast cancer cell lines of different subtypes.
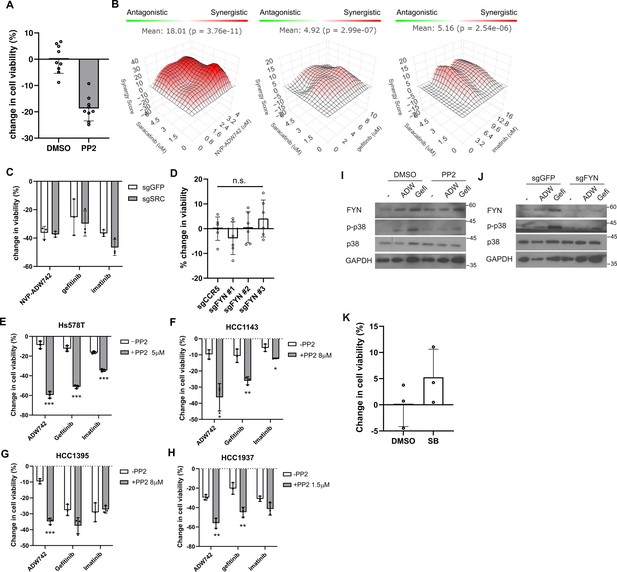
FYN inhibition synergizes with TKIs in multiple TNBC cell lines.
(A) MTT assay with MDA-MB-231 cells treated with 10 μM PP2 for 48 hr. (B) Synergistic killing calculated using SynergyFinder plus by Bliss independence model. (C) MTT assay with MDA-MB-231 Cas9 cells expressing indicated sgRNAs and treated with indicated TKIs (n=3). (D) MTT assay with MDA-MB-231 Cas9 cells expressing indicated sgRNAs. sgCCR5 is used as a control sgRNA that is expected to minimally affect cell viability. (E–H) MTT assay with various TNBC cell lines treated with indicated drug combinations (n=3). Changes in viability are normalized to those treated with DMSO and PP2 at indicated concentrations for cells treated with NVP-ADW742, gefitinib, or imatinib as single agent, and cells treated in combination of PP2, respectively. (I) Western blot analysis of Hs578T cells treated with indicated drugs. (J) Western blot analysis of Hs578T Cas9 cells expressing indicated sgRNA and treated with indicated drug. (K) MTT assay with MDA-MB-231 cells treated with 10 μM SB203580 for 48 hr. All data are plotted as mean ± s.d. Unpaired two-sided Student’s t-test in D-H.*, p<0.05; **, p<0.01; ***, p<0.001; n.s., p>0.05. All replicates are biological replicates.
-
Figure 2—figure supplement 3—source data 1
Original files for western blot analysis displayed in Figure 2—figure supplement 3I and J.
- https://cdn.elifesciences.org/articles/93921/elife-93921-fig2-figsupp3-data1-v1.zip
-
Figure 2—figure supplement 3—source data 2
Image files containing original western blot analysis displayed in Figure 2—figure supplement 3I and J, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/93921/elife-93921-fig2-figsupp3-data2-v1.zip
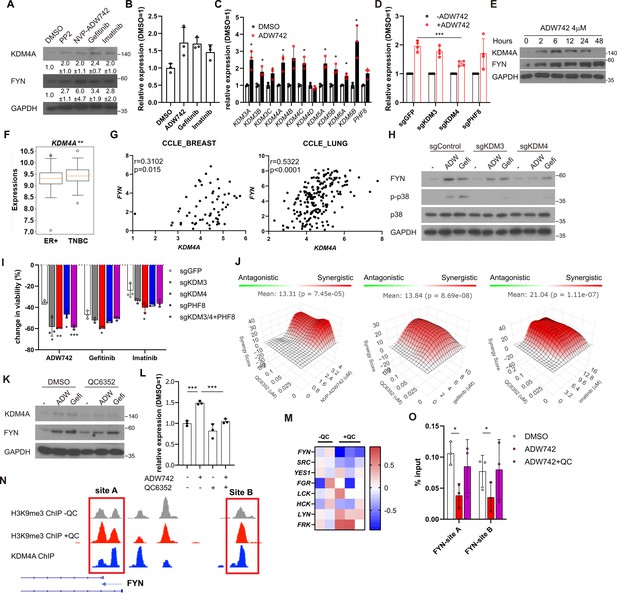
Activation of KDM4 upregulates FYN, conferring drug resistance.
(A) Western blot analysis of MDA-MB-231 cells treated with indicated drugs. Numbers below blots indicate quantification of average ± s.d. expression level normalized to GAPDH in three independent experiments. (B) RT-qPCR analysis of FYN expression levels in MDA-MB-231 cells treated with indicated drugs for 48 hr (n=3). (C) RT-qPCR analysis of indicated jumonji family histone demethylase expression levels after 48 hr treatment of NVP-ADW742 (n=3). (D) Changes in FYN mRNA levels upon 48 hr of NVP-ADW742 treatment in MDA-MB-231 Cas9 cells expressing indicated sgRNAs (n=4). (E) Western blot analysis of MDA-MB-231 cells treated with NVP-ADW742 for indicated time. (F) KDM4A mRNA levels in primary tumor tissues of indicated subtypes in GSE25066 cohort. (G) Positive correlation of FYN and KDM4A mRNA levels in CCLE database. (H) Western blot analysis of MDA-MB-231 Cas9 cells expressing indicated sgRNA and treated with indicated drugs for 48 hr. (I) MTT assay of MDA-MB-231 Cas9 cells expressing indicated sgRNAs and treated with indicated drugs for 72 hr (n=3). (J) SynergyFinder analysis of MDA-MB-231 cells treated with indicated drug combinations (n=2). (K, L) Western blot analysis (K) and RT-qPCR analysis (L) of MDA-MB-231 cells treated with indicated drugs for 48 hr (n=3). (M) mRNA expression levels of SRC family kinases in breast cancer stem cells treated with QC6352 in RNA sequencing data described in Metzger et al., 2017 (N) H3K9me3 and KDM4A enrichment at genomic locus encoding FYN promoter in ChIP sequencing data described in the same study as (M). (O) H3K9me3 Chromatin immunoprecipitation-qPCR analysis of MDA-MB-231 cells treated with indicated drug for 48 hr at specified genomic loci (n=3). QC6352, PP2, NVP-ADW742, gefitinib and imatinib were treated at 10 μM, 5 μM, 4 μM, 12 μM, 12 μM, respectively, unless otherwise indicated. All data are plotted as mean ± s.d. Unpaired two-sided Student’s t-test in B, C, D, F, I, and L. Paired two-sided Student’s t-test in O. *, p<0.05; **, p<0.01; ***, p<0.001; n.s., p>0.05. All replicates are biological replicates.
-
Figure 3—source data 1
Original files for western blot analysis displayed in Figure 3A, E, H and K.
- https://cdn.elifesciences.org/articles/93921/elife-93921-fig3-data1-v1.zip
-
Figure 3—source data 2
Image files containing original western blots for Figure 3A, E, H and K, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/93921/elife-93921-fig3-data2-v1.zip
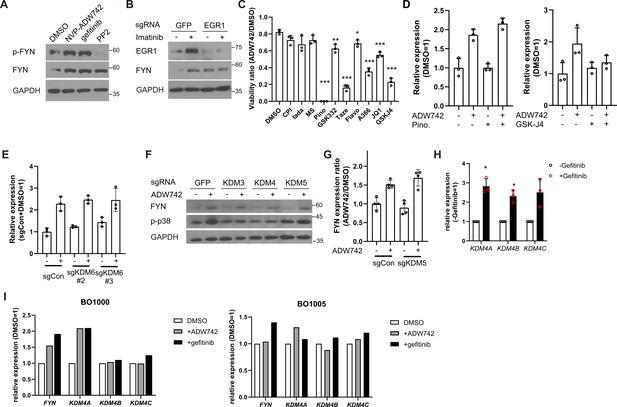
KDM4 regulates FYN expression level, sensitizing MDA-MB-231 cells to TKIs.
(A) Western blot analysis of MDA-MB-231 cells treated with 4 μM NVP-ADW742, 10 μM gefitinib, or 10 μM PP2 for 48 hr. (B) Western blot analysis of MDA-MB-231 Cas9 cells expressing indicated sgRNAs and treated with imatinib. (C) MTT assay (n=3) with MDA-MB-231 cells treated with indicated epigenetic drug in combination of 4 μM NVP-ADW742 for 72 hr. (D) RT-qPCR analysis of FYN mRNA level in MDA-MB-231 cells treated with 4 μM NVP-ADW742 and (left) 20 μM pinometostat, or (right) 10 μM GSK-J4 for 48 hr (n=3). (E) RT-qPCR analysis of MDA-MB-231 Cas9 cells expressing indicated sgRNAs and treated with 4 μM NVP-ADW742 for 48 hr. (F) Western blot analysis of MDA-MB-231 Cas9 cells expressing indicated sgRNAs and treated with 4 μM NVP-ADW742 for 48 hr. (G) RT-qPCR analysis of FYN mRNA level in MDA-MB-231 Cas9 cells expressing indicated sgRNAs treated with NVP-ADW742 (n=4). (H) RT-qPCR analysis of KDM4 mRNA levels in MDA-MB-231 cells treated with gefitinib (n=3). (I) RT-qPCR analysis of indicated genes in TNBC primary tumor derived organoids treated with indicated TKIs (n=1). All data are plotted as mean ± s.d. Unpaired two-sided Student’s t-test in C and H.*, p<0.05; **, p<0.01; ***, p<0.001; n.s., p>0.05. All replicates are biological replicates.
-
Figure 3—figure supplement 1—source data 1
Original files for western blot analysis displayed in Figure 3—figure supplement 1A, B and F.
- https://cdn.elifesciences.org/articles/93921/elife-93921-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Image files containing original western blot analysis displayed in Figure 3—figure supplement 1A, B and F, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/93921/elife-93921-fig3-figsupp1-data2-v1.zip
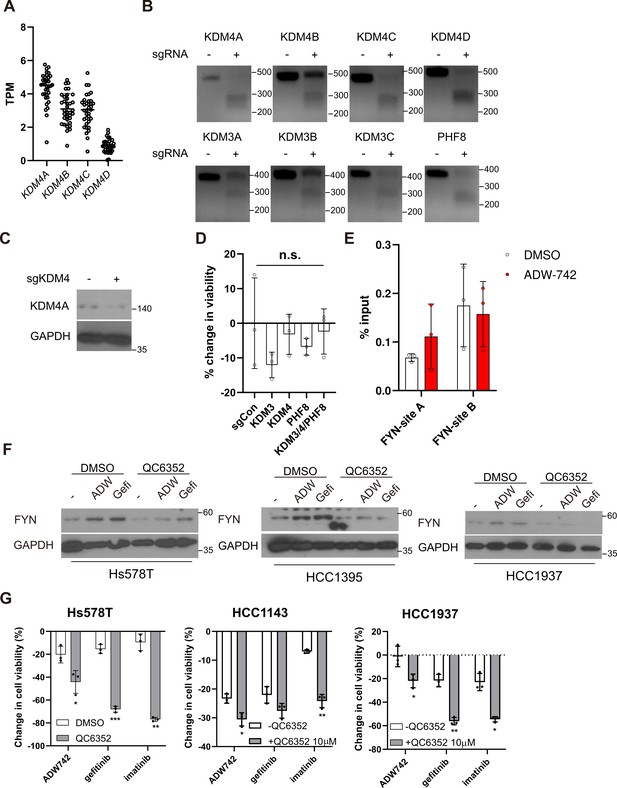
KDM4 inhibition synergizes with TKIs in multiple TNBC cell lines.
(A) Expression levels of KDM4 family genes in TNBC cell lines in CCLE. (B) T7 endonuclease assay validation of sgRNAs against indicated genes used in Figure 3I. (C) Validation of KDM4 knockout by western blot in MDA-MB-231 cells expressing indicated sgRNA. (D) MTT assay with MDA-MB-231 Cas9 cells expressing indicated sgRNAs. sgCCR5 is used as a control sgRNA that is expected to minimally affect cell viability. (E) H3K27me3 Chromatin immunoprecipitation-qPCR analysis of MDA-MB-231 cells treated with 4 μM NVP-ADW742 for 48 hr at specified genomic loci (n=3). (F) Western blot analysis of FYN expression level in indicated TNBC cell lines treated with QC6352 in combination with TKIs. (G) Summary of MTT assay with cancer cell lines treated with indicated drugs for 72 hr (n=3). TKIs are used at concentration of 4 μM, 10 μM, 10 μM for NVP-ADW742, gefitinib, and imatinib, respectively, unless otherwise indicated. All data are plotted as mean ± s.d. Unpaired two-sided Student’s t-test in C.*, p<0.05; **, p<0.01; ***, p<0.001; n.s., p>0.05. All replicates are biological replicates.
-
Figure 3—figure supplement 2—source data 1
Original files for gel image displayed in Figure 3—figure supplement 2B, and western blot analysis displayed in Figure 3—figure supplement 2C and F.
- https://cdn.elifesciences.org/articles/93921/elife-93921-fig3-figsupp2-data1-v1.zip
-
Figure 3—figure supplement 2—source data 2
Images files for gel image displayed in Figure 3—figure supplement 2B, and western blot analysis displayed in Figure 3—figure supplement 2C and F, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/93921/elife-93921-fig3-figsupp2-data2-v1.zip

Combination therapy targeting FYN +IGF1 R and KDM4 +EGFR synergistically eliminates tumor in vivo.
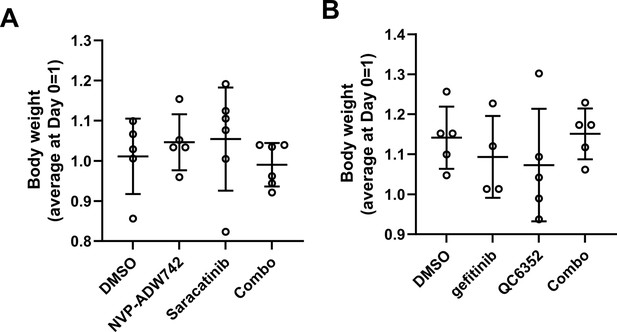
(A, B) Tumor volume for MDA-MB-231 xenografts treated with indicated drugs. Additive effects were calculated by Bliss independence model (n=5). (C) Distant relapse free survival of GSE25066 patient cohort classified by FYN (left) and KDM4A (right) mRNA expression. (D) Schematics diagram of the mechanism of KDM4-FYN conferring TKI resistance. All data are plotted as mean ± s.d. *, p<0.05; **, p<0.01; ***, p<0.001; n.s., p>0.05. All replicates are biological replicates.
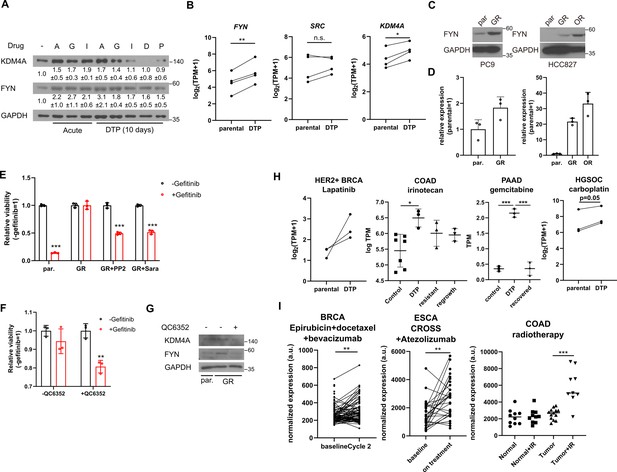
FYN and KDM4 are associated with drug tolerance.
(A) MDA-MB-231 cells treated with indicated drugs for short (acute: 2 days) and long (DTP: 10 days) time periods. A: 1 μM NVP-ADW742, G: 5 μM gefitinib, I: 5 μM imatinib, D: 100 nM doxorubicin, P: 5 nM paclitaxel. Numbers below blots indicate quantification of average ± s.d. expression level normalized to GAPDH in three independent experiments. (B) Summary of mRNA expressions of indicated genes in EGFR mutant lung cancer cells (parental) and their derivative osimertinib tolerant persisters (DTP). (C, D) Western blots (C) and RT-qPCR (D) analyses of indicated parental and EGFR inhibitor resistant lung cancer cells. (E, F) MTT assay with PC9 parental (par.) and gefitinib-resistant (GR) cells treated with indicated drug combinations (gefi: 2 μM gefitinib, PP2: 5 μM PP2, Sara: 5 μM saracatinib, QC6352: 10 μM QC6352) for 72 hr. (G) Western blot analysis with PC9 cells treated with 10 μM QC6352 for 48 hr. (H) FYN mRNA expression levels of parental and DTP populations in various cancers treated with indicated drugs (I) FYN mRNA expression levels of residual disease after indicated treatments. All data are plotted as mean ± s.d. Paired two-sided Student’s t-test in B, H (HER2 +BRCA set and HGSOC carboplatin set), and I (BRCA set and ESCA set), and unpaired two-sided Student’s t-test in H (COAD and PAAD sets) and I (COAD set). The expression data in B, H, and I are obtained from NCBI gene expression omnibus. The accession numbers of the expression data analyzed are listed in Supplementary file 4. *, p<0.05; **, p<0.01; ***, p<0.001; n.s., p>0.05. All replicates are biological replicates.
-
Figure 5—source data 1
Original files for western blot analysis displayed in Figure 5A, C and G.
- https://cdn.elifesciences.org/articles/93921/elife-93921-fig5-data1-v1.zip
-
Figure 5—source data 2
Image files containing original western blots for Figure 5A, C and G, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/93921/elife-93921-fig5-data2-v1.zip
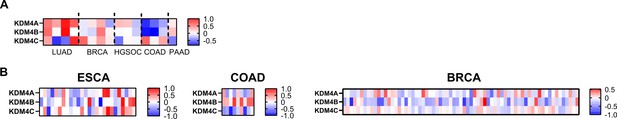
KDM4 expression levels in drug tolerant cancers and residual tumor after therapy.
(A) Heatmap of log transformed ratio of KDM4 mRNA expression of drug tolerant persisters and parental cells the datasets used are identical to those used in Figure 5B and H. (B) Heatmap of log transformed ratio of KDM4 mRNA expression of residual tumor after therapy and primary tumor before therapy. The datasets used are identical to those used in Figure 5I.
Additional files
-
Supplementary file 1
Tyrosine kinases subject to CRISPR screens in this study.
- https://cdn.elifesciences.org/articles/93921/elife-93921-supp1-v1.xlsx
-
Supplementary file 2
Growth phenotype scores Z and gene interaction scores GI calculated with combinatorial screens in Figure 1.
- https://cdn.elifesciences.org/articles/93921/elife-93921-supp2-v1.xlsx
-
Supplementary file 3
List of sgRNA used in the study.
- https://cdn.elifesciences.org/articles/93921/elife-93921-supp3-v1.xlsx
-
Supplementary file 4
List of GEO data used for analysis.
- https://cdn.elifesciences.org/articles/93921/elife-93921-supp4-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/93921/elife-93921-mdarchecklist1-v1.docx
-
Source data 1
Source data for Figure 2F, Figure 3J and Figure 2—figure supplement 3B.
- https://cdn.elifesciences.org/articles/93921/elife-93921-data1-v1.xlsx