CyAbrB2 is a nucleoid-associated protein in Synechocystis controlling hydrogenase expression during fermentation
Figures
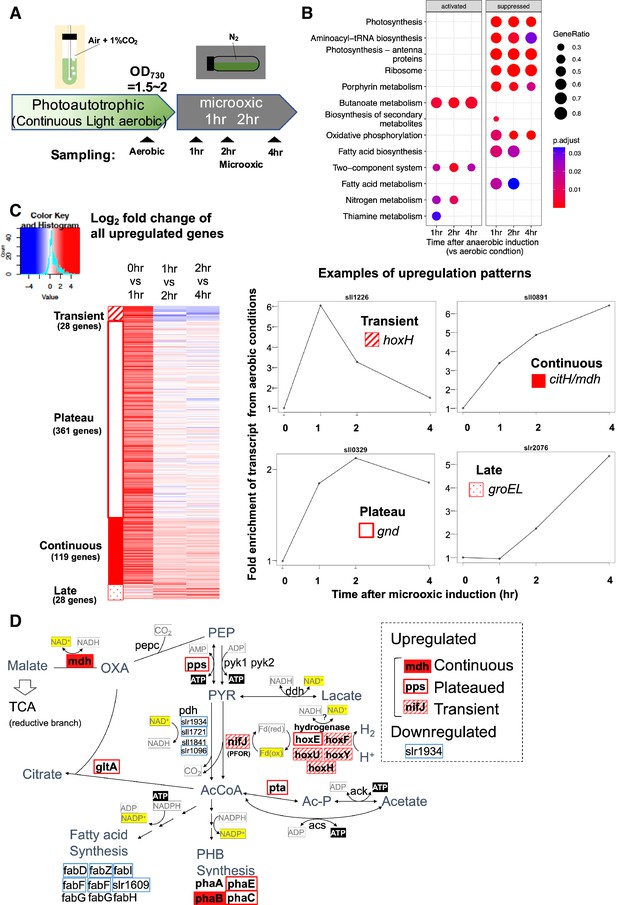
Time-course analysis of the transcriptome of Synechocystis on entry to the microoxic conditions.
(A) Schematic diagram for the sampling of cells under aerobic and microoxic conditions. (B) Gene set enrichment analysis on time-course transcriptome data. KEGG pathways enriched in upregulated or downregulated genes after 1, 2, and 4 hr incubation under microoxic conditions are shown. (C) (Left) Heatmap showing expression change in all upregulated genes over the time course. Genes classified into transient (striped square), plateau (open square), continuous (filled square), and late (dotty square) were clustered into subgroups and sorted by the gene name. (Right) Examples of genes are classified into each expression pattern. (D) The classified genes were mapped to central carbon metabolism, centered on pyruvate. PEP: phosphoenolpyruvate, PYR: pyruvate, AcCoA: acetyl CoA, Ac-P: acetyl phosphate, OXA: oxaloacetate, PHB: polyhydroxy butyrate, TCA: tricarboxylic acid cycle.
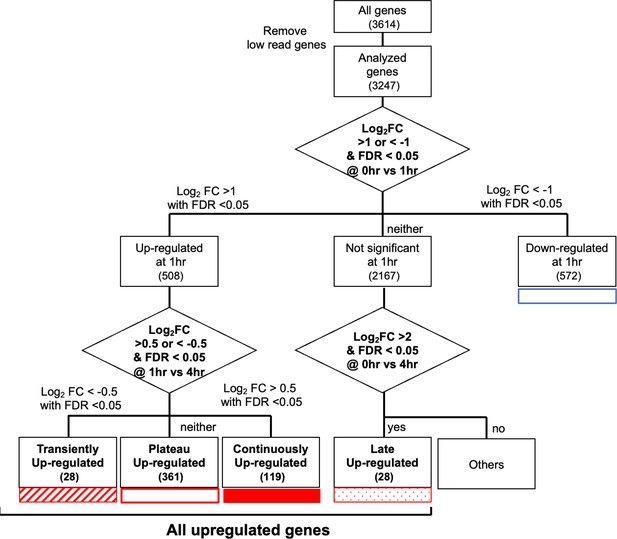
Schematic diagram showing the classification of genes according to the time-course transcriptome.
Transient (striped square), plateau (open square), continuous (filled square), and late (dotty square) are denoted as all upregulated genes.
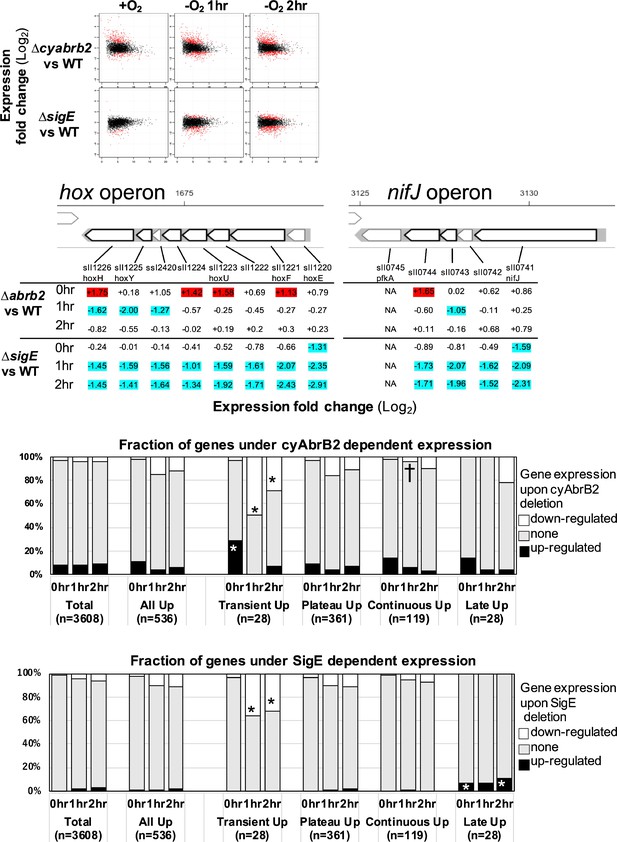
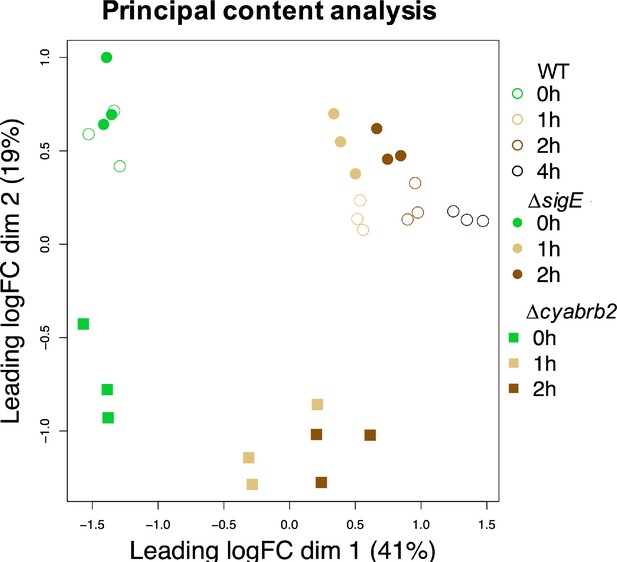
The impacts of ∆sigE and ∆cyabrb2 on the time-course transcriptome.
(A) MA plot showing fold change (y-axis) and average (x-axis) of gene expression between wildtype and mutant strains at each timepoint. Red dots indicate defined differentially expressed genes (DEGs) (|Log2 FC|>1 with false discovery rate [FDR]<0.05). (B) Log2 scaled expression fold change in genes in the hox and nifJ operons upon ∆cyabrb2 and ∆sigE under aerobic conditions (0 hr), 1 hr after microoxic condition (1 hr), and 2 hr after microoxic condition (2 hr). DEGs are marked with sky blue (downregulated upon deletion) or red (upregulated upon deletion). (C and D) Fraction of upregulated and downregulated genes upon the (C) ∆cyabrb2 and (D) ∆sigE at the timepoints of aerobic conditions (0 hr), 1 hr after anoxic condition (1 hr), and 2 hr after anoxic condition (2 hr). Genes are classified according to Figure 1C. Asterisk (*) and dagger (†) denote statistically significant enrichment and anti-enrichment compared with all upregulated genes tested by multiple comparisons of Fisher’s exact test (FDR<0.05).
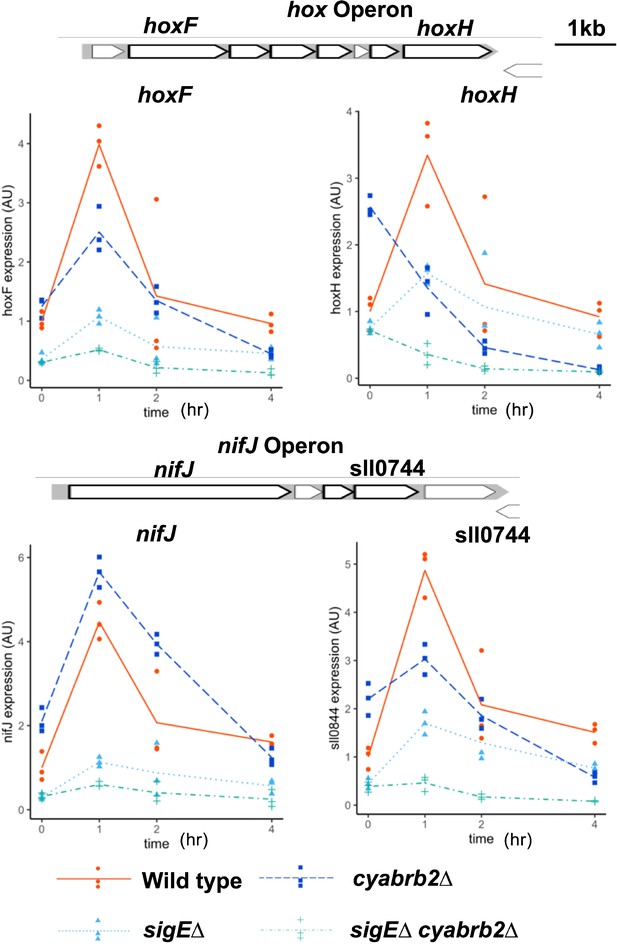
RT-qPCR validated the transiently upregulated genes classified by RNA-seq.
Transcripts extracted from wildtype (solid line), ∆sigE mutant (dotty line), ∆cyabrb2 mutant (dashed line), and ∆sigE ∆cyabrb2 double mutant (dot-dashed line) were assayed in the aerobic condition (0 hr) and 1, 2, 4 hr incubation of microoxic conditions. As the representative of the transiently upregulated genes, expression of hoxF, hoxY, nifJ, and sll0744 were quantified by RT-qPCR. The line represents the mean of n=3, and individual data points are shown as dot plots. Data of each gene is normalized by the mean score of wildtypes in the aerobic condition.
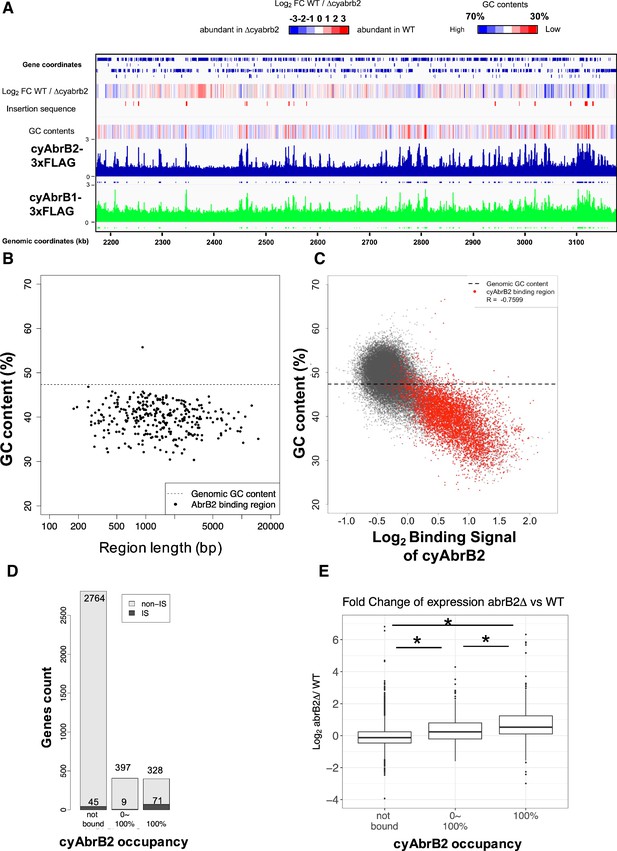
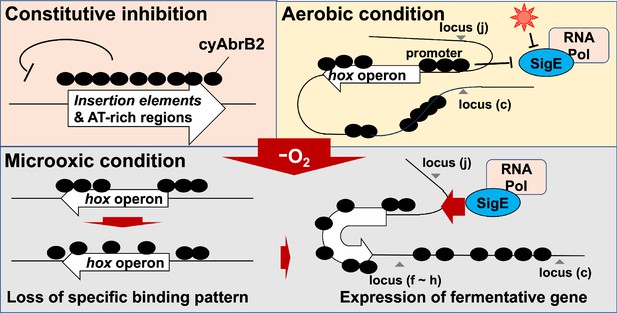
The long-tract distribution of cyAbrB2 on the Synechocystis genome and the repressive effect of cyAbrB2 on the gene expression.
(A) Snapshot of ChIP-seq data for cyAbrB2 and cyAbrB1 under aerobic conditions. The heatmap in the second column indicates expression fold change upon ∆cyabrb2 under aerobic conditions. Positive values (colored in red) indicate that the gene expression is higher in wildtype than in ∆cyabrb2, and negative values (colored in blue) indicate the opposite. The positions for the insertion elements are marked with red in the third column. The heatmap in the fourth column indicates GC contents. High GC contents are colored in blue and low GC contents are colored in blue. (B) GC contents and region length of cyAbrB2 binding regions (black dots). The horizontal dotted line indicates the genomic average of GC content. (C) Scatter plot of GC content and binding signal of cyAbrB2. The x-axis is the binding signal of cyAbrB2 in each 100 bp region, and the y-axis indicates GC contents within 500 bp windows sliding every 100 base pairs. CyAbrB2 binding regions are marked with red dots. (D) Histogram of genes showing the extent of occupancy (not bound, partially overlapped, or entirely overlapped) by the cyAbrB2 binding region. The gray bars indicate non-IS genes, and the count numbers of the non-IS genes are displayed on the gray bars. The black bars indicate the IS genes, and the count numbers of the IS genes are displayed above the black bars. (E) Boxplot showing fold change in gene expression by ∆cyabrb2 under aerobic conditions. Genes are classified according to the extent of occupancy by the cyAbrB2 binding region. Asterisk (*) denotes statistical significance tested by multiple comparisons of the Wilcoxon-rank test. Actual FDRs of "not bound vs 0~100%", "not bound vs 100%", and "0~100% vs 100%" are <2e-16, <2e-16, and 5e-06, respectively.
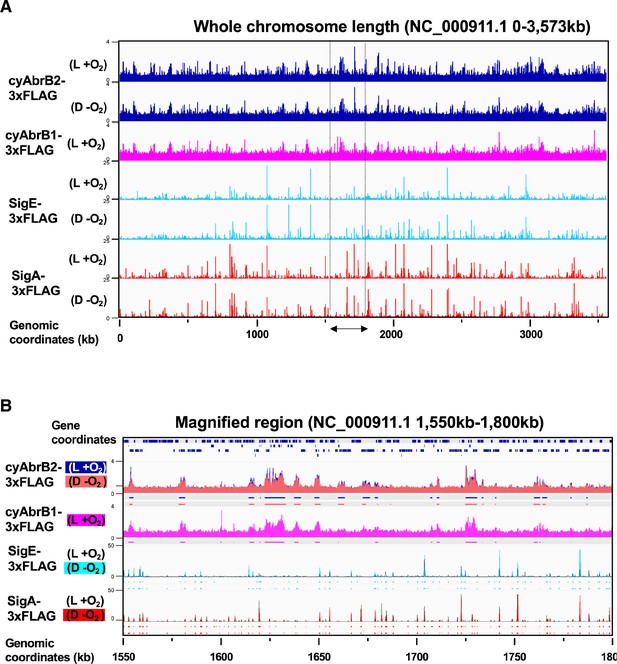
Overview of genome occupancy of cyAbrB2, cyAbrB1, SigE and SigA under the aerobic and microoxic conditions.
(A and B) Overview for ChIP-seq of FLAG-tagged cyAbrB2, cyAbrB1, SigE, and SigA. Y-axis indicates [normalized IP read count/normalized input read count at each 25 bp window], and x-axis indicates chromosome position. (A) Distribution of cyAbrB2, cyAbrB1, SigE, and SigA across the whole genome of Synechocystis. Aerobic (L +O2) and dark microoxic (D – O2) data are displayed. (B) Magnified image for chromosome position of 1550–1800 kb. ChIP-seq data of cyAbrB2, cyAbrB1, SigE, and SigA in aerobic and dark microoxic conditions are overlayed. The dots below the graph indicate the binding region of each protein calculated by peak caller.
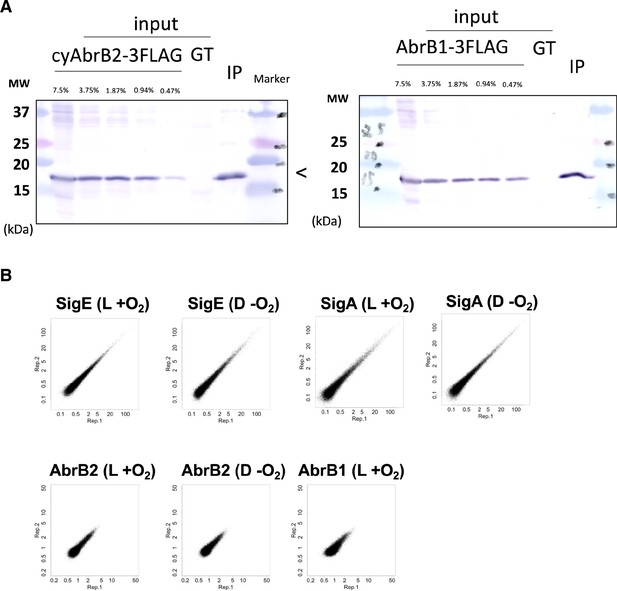
Validation of procedure for ChIP-seq of FLAG-tagged cyAbrB2, SigE, and SigA.
(A) The immunoblot for inputs and immunoprecipitants (IP) of ChIP for cyAbrB2-FLAG and cyAbrB1-FLAG. Input lysate of untagged control (GT) is also loaded. Inputs equivalent to the indicated portion of IP were loaded. (B) Scatter plots showing the reproducibility of two replicates for ChIP-seq assay. ChIP-seq data of SigE, SigA, and cyAbrB2 in aerobic and microoxic conditions and ChIP-seq data of cyAbrB1 in the aerobic condition are shown. Dots indicate normalized IP read count/normalized input read count in each 100 bp window. X-axis is the value of replicate1, and y-axis is the value of replicate 2.
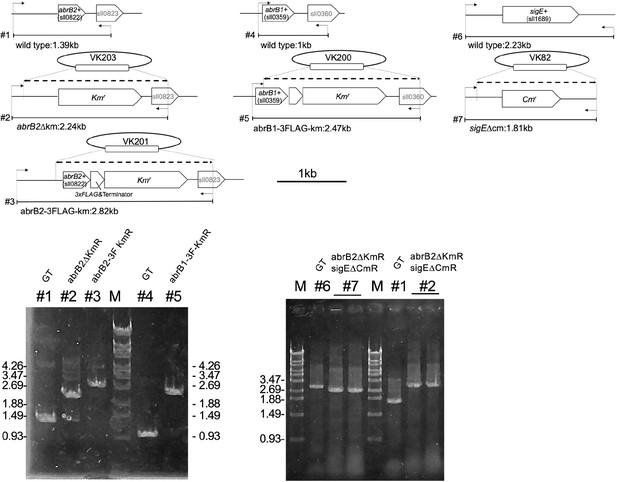
Confirmation of genomic deletion and the epitope tagging of abrB2 (#1-#3), the epitope tagging of abrB1 (#4 and #5), and deletion of sigE (#6 and #7).
Dotty lines are homologous regions between plasmids and the genome. Arrows indicate the position of check primers.
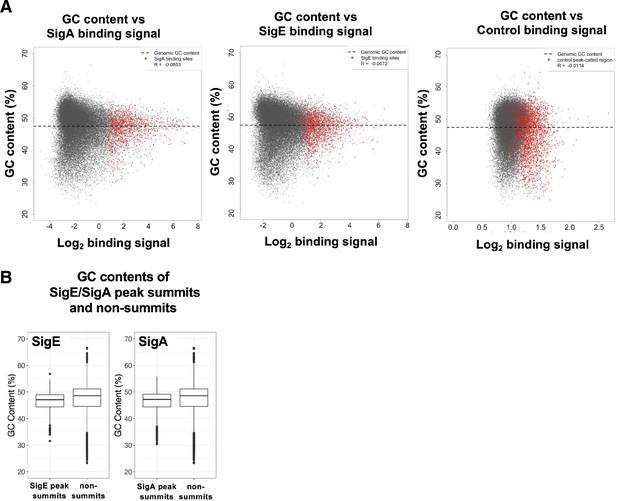
Relationships between GC content and binding patterns for SigE and SigA.
GC content vs ChIP enrichment score of SigA and SigE. (A) Scatter plot showing GC contents in each 100 bp vs. binding signal of SigA, SigE, and control IP. Data are displayed as in Figure 3C. (B) GC content in each 100 bp of (left) SigE peaks and non-SigE peaks, and (right) SigA peaks and non-SigA peaks.
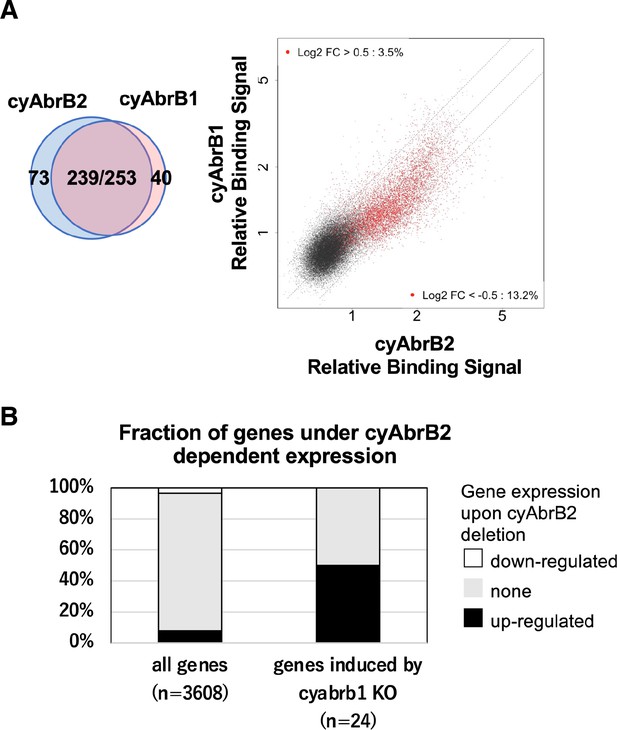
cyAbrB2 and cyAbrB1 show similar binding pattern and overlapping gene regulation.
(A) Venn diagram showing overlap of the binding region of cyAbrB1 and cyAbrB2 (left), and scatter plot showing ChIP binding signal of cyAbrB2 (y-axis) and cyAbrB1(x-axis) in the aerobic condition. Data is plotted as in Figure 5A. (B) Fractions of upregulated and downregulated genes upon the ∆cyabrb2 mutant in the aerobic conditions. Fractions of all genes (left n=3608) and genes induced by cyAbrB1 knockout (right n=24) are shown. Genes induced by cyAbrB1 knockout are from the previous study (Hishida et al., 2024).
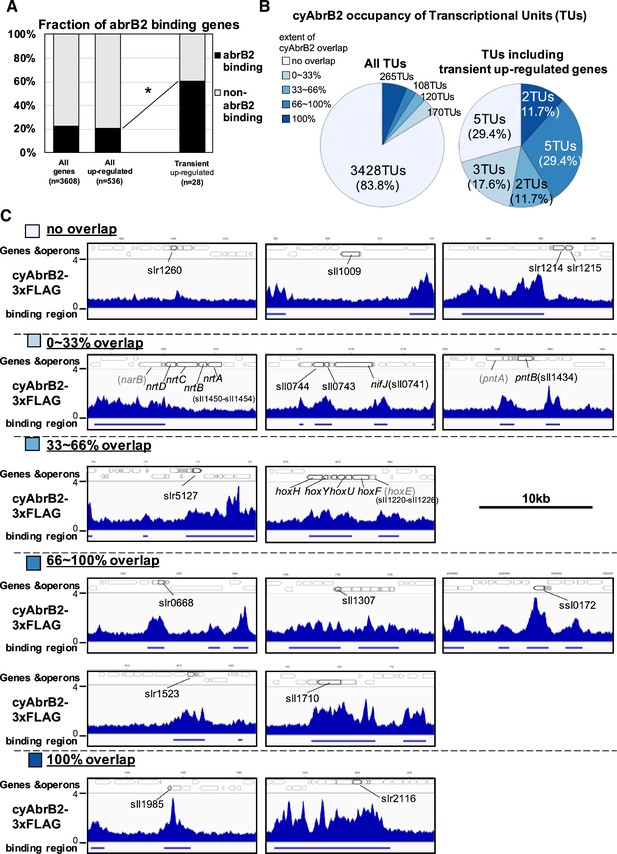
Transient up-regulated genes are enriched in cyAbrB2 binding regions.
(A) Fraction of genes overlapped or non-overlapped with cyAbrB2 binding regions at the timepoints of aerobic conditions. Genes are classified according to Figure 1—figure supplement 1. Asterisk (*) denotes statistically significant enrichment compared with all upregulated genes tested by multiple comparisons of Fisher’s exact test. (B) Pie charts of transcriptional units (TUs) classified by extent of overlapping with cyAbrB2 binding region. The left pie represents all TUs, and the right pie represents only TUs containing the transient upregulated genes. (C) Distribution of cyAbrB2 in the aerobic condition around transiently upregulated genes. Arrows with bold lines indicate transiently upregulated genes. Shaded arrows indicate operons whose data were obtained from a previous study. The bars below the graph indicate the binding regions of each protein. The black bar at the top of the figure indicates a length of 10 kbp.
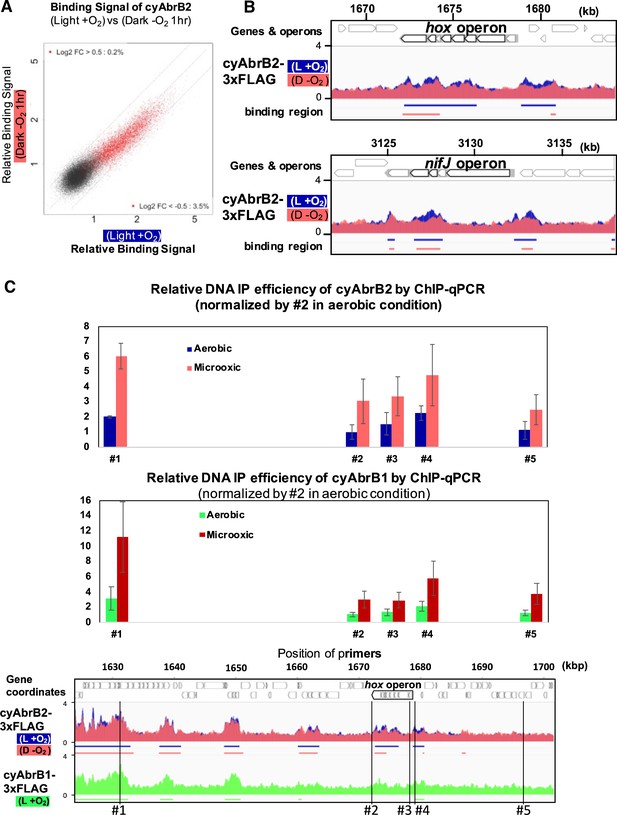
Changes of cyAbrB2 binding pattern on entry to the microoxic condition.
(A) Scatter plot showing changes of the binding signal by 1 hr cultivation in the microoxic condition. The binding signal of each 100 bp window is plotted. Red dots are cyAbrB2 binding regions in either aerobic or microoxic conditions. The dotty lines indicate Log2 fold enrichment of 0.5, 0, and –0.5 between aerobic and microoxic conditions. (B) Distribution of cyAbrB2 around hox operon and nifJ operon. ChIP-seq data in aerobic (L + O2) and dark microoxic (D − O2) conditions are overlayed. The bars below the graph indicate the binding regions of each protein. (C) Quantification for IP efficiency of cyAbrB2 (top) and cyAbrB1 (middle) by qPCR in the aerobic and microoxic conditions. The position of primers and ChIP-seq data of cyAbrB2 are shown at the bottom. Scores are normalized by the IP% at position #2 in the aerobic condition. Error bars represent standard deviation (n=3).
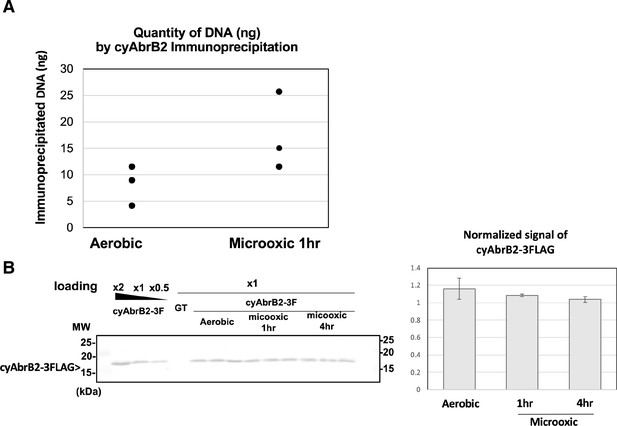
Alteration of cyAbrB2 binding to genome under the microoxic condition.
(A) Amount of precipitated DNA by cyAbrB2 ChIP. Three experiments were performed in the aerobic and microoxic conditions. (B) Western blot images of cyAbrB2-3FLAG. Proteins were extracted in the aerobic condition and 1 and 4 hr incubation under microoxic conditions. The total protein concentration of each sample was adjusted to 4 mg/mL, measured by the BCA method. Quantification of cyAbrB2 from western blot image was performed by ImageJ (ver. 2.0.0-rc-65) and plotted in the right graph.
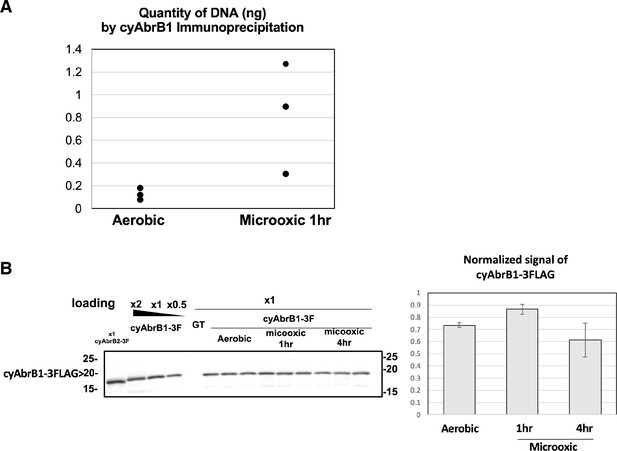
Alteration of cyAbrB1 binding to genome under the microoxic condition.
(A) Amount of precipitated DNA by cyAbrB1 ChIP. Three experiments were performed in the aerobic and microoxic conditions. (B) Western blot images of cyAbrB1-3FLAG. The experiment and data analysis were performed as in Figure 1.
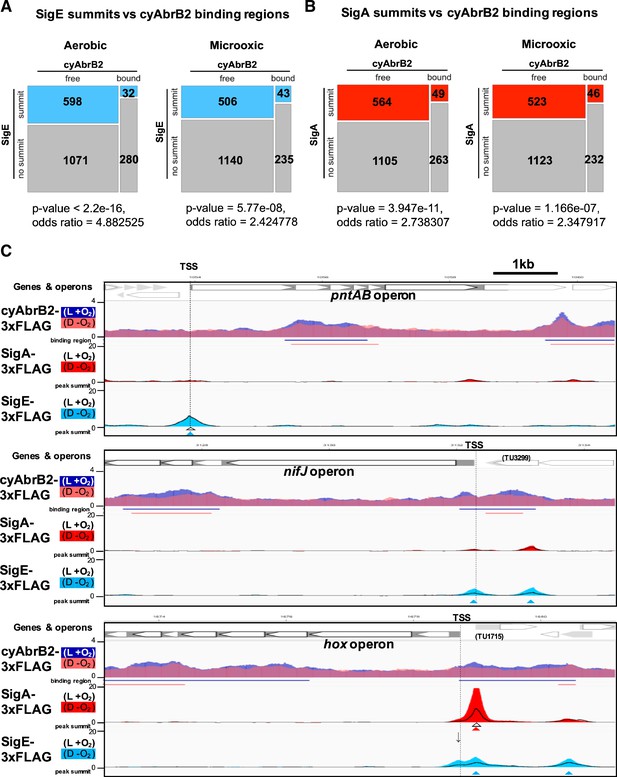
Sigma factors are excluded from cyAbrB2 binding regions.
(A and B) Anti-co-occurrence of cyAbrB2 binding regions and sigma factors. Mosaic plots of cyAbrB2 binding regions and SigE peaks (A) or SigA binding peaks (B) are shown. Odds and p-values were calculated by Fisher’s exact test. (C) Snapshots of ChIP-seq data for CyAabrB2, SigE, and SigA at the nifJ region (top) and hox region (bottom). ChIP-seq data for cyAbrB2, SigE, and SigA under aerobic and dark microoxic conditions are overlayed. ChIP-seq data of cyAbrB2 under aerobic and microoxic conditions are colored blue and pink, respectively. ChIP-seq data for SigE and SigA are shown in solid lines (aerobic conditions) and the area charts (microoxic conditions). The positions of transcription start sites (TSSs) were obtained from a previous study (Kopf et al., 2014) and indicated by vertical dotted lines. Open triangles indicate peak summits under aerobic conditions, and solid triangles indicate peak summits under microoxic conditions.
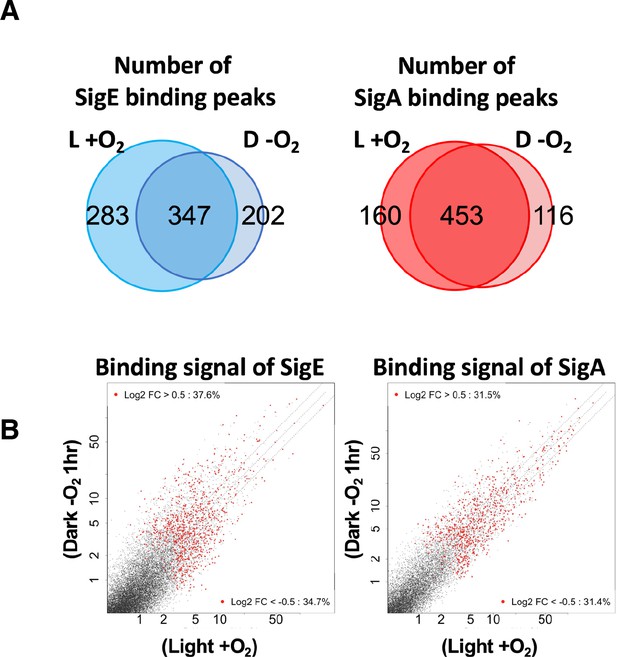
Changes of SigE and SigA distribution on the entry to the microoxic condition.
(A) Venn diagram showing the number of peaks of SigE (left) and SigA (right) in aerobic (L + O2) and dark microoxic (D − O2) conditions. (B) Scatter plot showing changes in the binding signal of SigE and SigA by 1 hr cultivation under microoxic conditions. The binding signal of each 100 bp window is plotted.
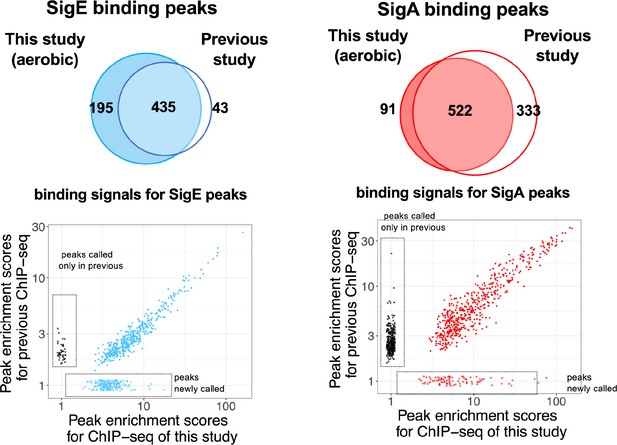
Reproducibility of ChIP-seq data of SigA and SigE, compared with the previous study (Kariyazono and Osanai, 2022).
(Top) Venn diagrams show the overlapping of peaks called in this study and the previous study. (Bottom) Scatter plot comparing ChIP binding signals of SigA and SigE peaks commonly called in present and previous studies. Plots boxed by dashed lines are peaks called only in the present or previous study.
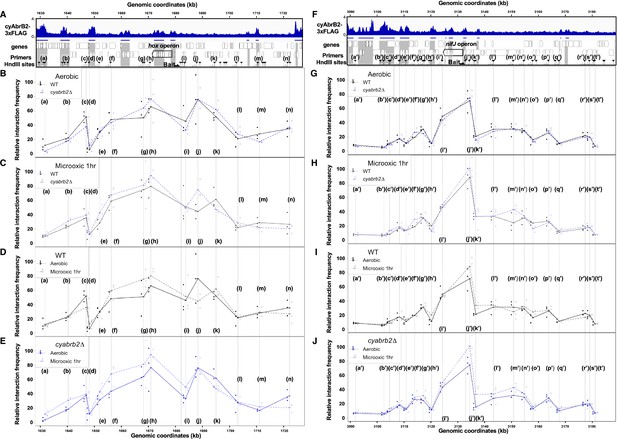
3C analysis showed changes of DNA conformation around hox and nifJ operon on entry to microoxic condition and the impact of cyabrb2 deletion on DNA conformation.
(A and F) Schematic diagram of 3C analysis around hox operon (A) and nifJ operon (F). In the panels (A) and (F), the black horizontal arrow shows the location of the bait primer, and white horizontal arrows ((a) to (n) in hox operon (A) and (a’) to (t’) in nifJ operon (F)) indicate loci where the interaction frequency with bait were assayed. Vertical black arrowheads indicate the position of HindIII sites. ChIP-seq data of cyAbrB2 in the aerobic condition is displayed in the bottom, and cyAbrB2 binding regions are marked with shade. (B–E) The line plot showing the interaction frequency of each locus with hox fragment. Two of data sets are presented; (B) wildtype vs ∆cyabrb2 in aerobic condition, (C) wildtype vs ∆cyabrb2 in 1 hr of microoxic condition, (E) wildtype in aerobic vs 1 hr of microoxic condition, and (E) ∆cyabrb2 in aerobic vs 1 hr of microoxic condition are compared. (G–J) The line plot showing the interaction frequency of each locus with nifJ fragment. Two data sets are selected and presented; (G) wildtype vs ∆cyabrb2 in aerobic condition, (H) wildtype vs ∆cyabrb2 in 1 hr of microoxic condition, (I) wildtype in aerobic vs 1 hr of microoxic condition, and (J) ∆cyabrb2 in aerobic vs 1 hr of microoxic conditions are compared. The line plots indicate the average interaction frequency over the random ligation (n=3). Individual data are plotted as dots.
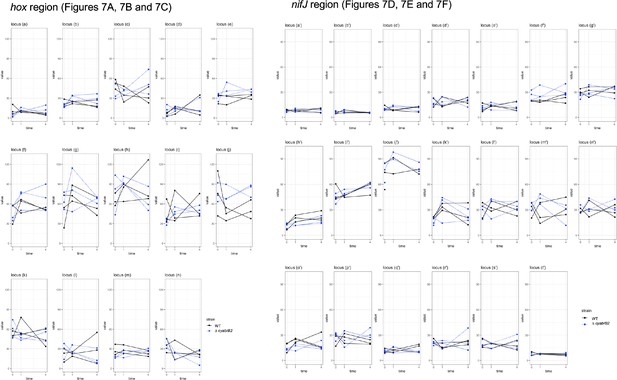
Dynamics of individual 3C scores.
Re-plotting of Figure 7 with the x-axis showing time (0, 1, 4 hr in microoxic conditions) and the y-axis showing the interaction frequency. Plots from the individual samples are connected by solid (wildtype) or dotty (∆cyabrb2) lines.
The validation of unidirectional primer sets for 3C assay is shown in Figure 7.
The 3C sample in this assay is the mixture of all 3C samples assayed in Figure 7.
Tables
List of transiently upregulated genes.
| Operon | ||
|---|---|---|
| Oxidoreductase | ||
| sll0741 | nifJ/‘pyruvate-ferredoxin/flavodoxin oxidoreductase’ | TU3296 |
| sll0743 | Hypothetical protein | |
| sll0744 | Dihydroorotate dehydrogenase (fumarate) | |
| sll1221 | hoxF/‘bidirectional [NiFe] hydrogenase diaphorase subunit’ | TU1714 |
| sll1222 | Unknown protein | |
| sll1223 | hoxU/‘bidirectional [NiFe] hydrogenase diaphorase subunit’ | |
| sll1224 | hoxY/‘NAD-reducing hydrogenase small subunit’ | |
| sll1225 | Unknown protein | |
| sll1226 | hoxH/‘NAD-reducing hydrogenase large subunit’ | |
| slr1434 | pntB/‘H+-translocating NAD(P) transhydrogenase subunit beta’ | TU1089 |
| Transporter | ||
| sll1450 | nrtA/‘nitrate/nitrite transport system substrate binding protein’ | TU1023 |
| sll1451 | nrtB/‘nitrate/nitrite transport system permease protein’ | |
| sll1452 | nrtC/‘nitrate/nitrite transport system ATP binding protein’ | |
| sll1453 | nrtD/‘nitrate/nitrite transport system ATP binding protein’ | |
| Two-component system | ||
| slr1214 | Twitching motility two-component system response regulator PilG | TU905 |
| slr1215 | Unknown protein | TU907 |
| Glycosyl transferase | ||
| slr2116 | spsA/‘spore coat polysaccharide biosynthesis protein; SpsA’ | TU1673 |
| Protease | ||
| sll1009 | frpC/‘iron-regulated protein’ | TU491 |
| Insertion sequence (transposase) | ||
| slr1523 | Transposase | TU1659 |
| sll1985 | Transposase | TU1589 |
| sll7001 | Transposase | NA |
| sll7003 | Toxin FitB | TU7001 |
| ssl0172 | Transposase | TU3163 |
| Other | ||
| slr1260 | Hypothetical protein | TU1446 |
| slr0668 | Unknown protein | TU3532 |
| slr5127 | Unknown protein | TU5127 |
| sll0710 | Unknown protein | TU97 |
| sll1307 | Unknown protein | TU1224 |
-
The list of transiently upregulated genes was merged by transcriptional units and sorted by function. The transcriptional unit information was obtained from a previous study (Kopf et al., 2014).
Fold changes of transcripts from sigA, sigB, sigC, sigD, and sigE.
| 0 hr vs 1 hr | 1 hr vs 4 hr | ||||
|---|---|---|---|---|---|
| Sigma factor | Locus | Log2FC | FDR | Log2FC | FDR |
| SigA | slr0653 | –0.873248 | 0.00766486 | –0.0013514 | 0.99797563 |
| SigB | sll0306 | 1.38098826 | 8.42E-06 | 0.77453605 | 0.04057775 |
| SigC | sll0184 | 2.97101055 | 1.75E-16 | 1.30743549 | 0.00067892 |
| SigD | sll2012 | 0.4701823 | 0.1498473 | –0.4522181 | 0.32402556 |
| SigE | sll1689 | –1.9111759 | 1.96E-11 | –1.1223298 | 0.00633142 |
-
Data is extracted from Supplementary file 1d.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Synechocystis sp. PCC6803) | Wildtype | https://doi.org/10.1016/0076-6879(88)67088-1 | GT | |
| Strain, strain background (Synechocystis sp. PCC6803) | ∆sigE::KmR | https://doi.org/10.1074/jbc.M505043200 | G50 | |
| Strain, strain background (Synechocystis sp. PCC6803) | SigA-8His-KmR | https://doi.org/10.1111/tpj.15687 | KR93 | |
| Strain, strain background (Synechocystis sp. PCC6803) | SigA-3FLAG-KmR | https://doi.org/10.1111/tpj.15687 | KR94 | |
| Strain, strain background (Synechocystis sp. PCC6803) | ∆cyabrb2::KmR | In this study | KR340 | The genome of GT strain was manipulated by the transformation of the plasmid VK203 |
| Strain, strain background (Synechocystis sp. PCC6803) | cyAbrB(sll0359)–3xFLAG-KmR | In this study | KR338 | The genome of GT strain was manipulated by the transformation of the plasmid VK200 |
| Strain, strain background (Synechocystis sp. PCC6803) | cyAbrB2(sll0822)–3xFLAG-KmR | In this study | KR339 | The genome of GT strain was manipulated by the transformation of the plasmid VK201 |
| Strain, strain background (Synechocystis sp. PCC6803) | ∆cyabrB2::KmR ∆sigE::CmR | In this study | KR359 | The genome of G50 strain was manipulated by the transformation of the plasmid VK82 |
| Recombinant DNA reagent | sigE∆CmR | In this study | VK82 | Plasmid backbone:pTA2 (Toyobo), available upon request |
| Recombinant DNA reagent | AbrB1-3F-KmR | In this study | VK200 | Plasmid backbone:pTA2 (Toyobo), available upon request |
| Recombinant DNA reagent | AbrB2-3F-KmR | In this study | VK201 | Plasmid backbone:pTA2 (Toyobo), available upon request |
| Recombinant DNA reagent | cyabrB2∆KmR | In this study | VK203 | Plasmid backbone:pTA2 (Toyobo), available upon request |
| Antibody | Anti-FLAG | Sigma-aldrich | F1804 | RRID:AB_262044 For immunoprecipitation |
| Antibody | Anti-FLAG (alkaline phosphatase conjugated) | Sigma-aldrich | A9469 | RRID:AB_439699 For western blot (1:20,000) |
Additional files
-
Supplementary file 1
Oligonucleotides used in this study and the summary of NGS analysis.
(a) Oligonucleotides used in this study. (b) Numbers and percentages of NGS reads passed the processes. (c–e) Log2FC, LogCPM, LR, p-value, and false discovery rate (FDR) calculated by edgeR lrt method. (c) Processed data from time-course transcriptome for GT strain. (d) Processed data from the comparison between GT and sigE∆ strain in each timepoints. (e) Processed data from the comparison between GT and cyabrb2∆ strain in each timepoints. (f) List of SigE binding summit in the aerobic condition from ChIP-seq data. (g) List of SigE binding summit in the microoxic condition from ChIP-seq data. (h) List of SigA binding summit in the aerobic condition from ChIP-seq data. (i) List of SigA binding summit in the microoxic condition from ChIP-seq data. (j) List of cyAbrB2 binding region in the aerobic condition from ChIP-seq data. (k) List of cyAbrB2 binding region in the microoxic condition from ChIP-seq data. (l) List of cyAbrB1 binding region in the aerobic condition from ChIP-seq data. (m) Raw result of gene set enrichment analysis of time-course transcriptome (vs the aerobic condition).
- https://cdn.elifesciences.org/articles/94245/elife-94245-supp1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/94245/elife-94245-mdarchecklist1-v1.docx
-
Source data 1
Uncropped images for Figure 3—figure supplement 2 and 3, Figure 5—figure supplement 1 and 2, and Figure 7—figure supplement 2.
- https://cdn.elifesciences.org/articles/94245/elife-94245-data1-v1.zip