Ligand response of guanidine-IV riboswitch at single-molecule level
Figures
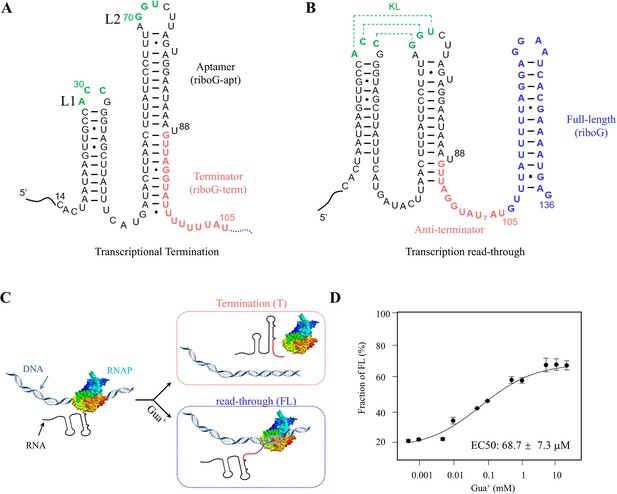
Characteristics of the C. botulinum guanidine-IV riboswitch.
(A, B) The secondary structures of the transcriptional termination and read-through states of the guanidine-IV riboswitch. The full-length of the guanidine-IV riboswitch contains the aptamer domain (black), terminator (red) and the extended sequence (blue). The green nucleotides are involved in forming a KL in the anti-terminator state. (C) A transcriptional model of the guanidine-IV riboswitch. The DNA is shown in blue ribbons. The colors in the RNA are encoded as in (A) and (B). And the addition of Gua+ facilitates transcription read-through. (D) The percentages of the transcription read-through plotted with 0.5 μM–20.0 mM Gua+ at 6.0 mM Mg2+ in the three independent transcription termination reactions. Data represent average ± SD from three replicates (n=3).
-
Figure 1—source data 1
Data for the graphs shown in Figure 1D.
- https://cdn.elifesciences.org/articles/94706/elife-94706-fig1-data1-v1.xlsx
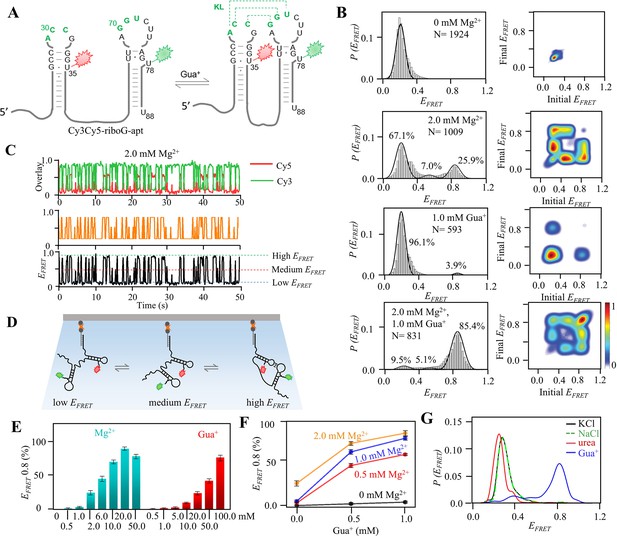
smFRET studies of post-transcriptional Cy3Cy5-riboG-apt at different concentrations of Mg2+ and Gua+.
(A) The secondary structures of Cy3Cy5-riboG-apt at the unfolded (left) and the folded state (right). Cy3 and Cy5 are shown by green and red sparkles, respectively. (B) smFRET histograms and transition density plots for Cy3Cy5-riboG-apt at 0 mM Mg2+, at 2.0 mM Mg2+, at 1.0 mM Gua+, and at 2.0 mM Mg2+ and 1.0 mM Gua+. (C) HMM analysis of smFRET time trace of Cy3Cy5-riboG-apt at 2.0 mM Mg2+, the time resolution is 0.1 s. (D) Schematic diagram for three conformations of Cy3Cy5-labeled RNA in smFRET experiments. The Cy3Cy5-labeled RNA is hybridized with a biotinylated DNA and immobilized on the slides. (E) The percentages of the folded conformation (EFRET ~0.8) of Cy3Cy5-riboG-apt at 0–50.0 mM Mg2+ (cyan columns) and 0.5–100.0 mM Gua+ (red columns). Data represent average ± SD from three replicates (n=3). (F) The percentages of the folded conformation of Cy3Cy5-riboG-apt change with Gua+ at 0, 0.5, 1.0, and 2.0 mM Mg2+. (G) FRET histograms of Cy3Cy5-riboG-apt at 100.0 mM KCl (black curve), NaCl (green curve), urea (red curve), and guanidine (blue curve).
-
Figure 2—source data 1
Data for the graphs shown in Figure 2.
- https://cdn.elifesciences.org/articles/94706/elife-94706-fig2-data1-v1.xlsx
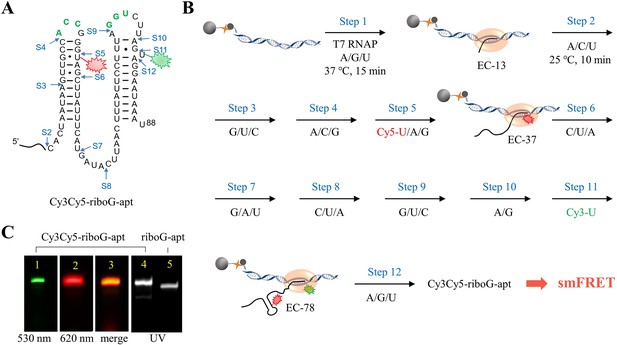
The schematic procedure of preparing Cy3Cy5-riboG-apt by 12 step-PLOR reaction for smFRET study.
(A) The secondary structure of riboG-apt. The positions of donor (Cy3) and acceptor (Cy5) are shown by green and red sparkles at U78 and U35, respectively. The sites started at each step of the PLOR-synthesis were marked by blue arrows. (B) The scheme of 12-step PLOR reaction for preparing Cy3Cy5-riboG-apt for smFRET study. The reagent usages for the 12-step reaction are listed in Supplementary file 1, table S3. (C) The PAGE images of Cy3Cy5-riboG-apt. The Lanes 1, 2, and 4 were irradiated by 530 nm, 620 nm fluorescence and 260 nm UV, respectively. Lane 3 is the merged image of lanes 1 and 2. The unlabeled counterpart, riboG-apt was loaded at lane 5. Cy3Cy5-riboG-apt (lane 4) migrated more slowly than riboG-apt (lane 5), indicating the bulky fluorophores were successfully introduced to Cy3Cy5-riboG-apt.
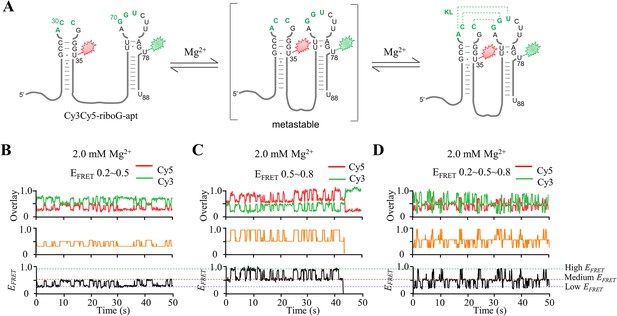
smFRET measurements of Cy3Cy5-riboG-apt at 2.0 mM Mg2+.
(A) The secondary structures of the unfolded (left), pre-folded (medium) and folded (right) states of riboG-apt. (B–D) HMM analysis of the representative single-molecule trajectories with the transitions between EFRET ~0.2 and EFRET ~0.5 (B), EFRET ~0.5 and~0.8 (C) and among EFRET ~0.2, 0.5, and 0.8 (D). The time resolution for smFRET is 100 ms.
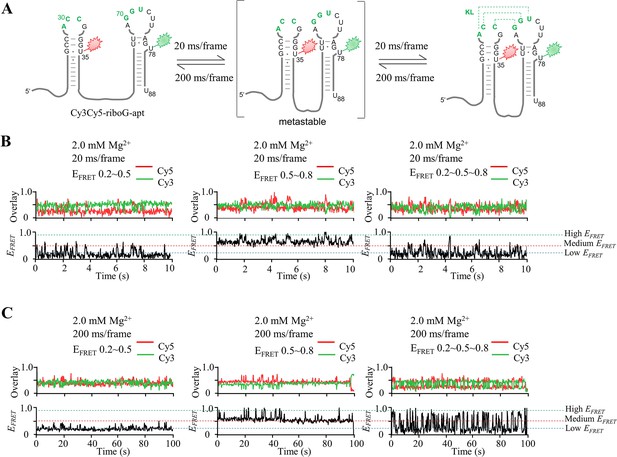
smFRET measurements of Cy3Cy5-riboG-apt at different time resolution.
(A) The secondary structures of the unfolded (left), pre-folded (medium) and folded (right) states of riboG-apt. (B–C) The representative single-molecule trajectories and FRET curves with dynamic transitions. The data collection was performed with time resolution of 20 ms (B) and 200 ms (C).
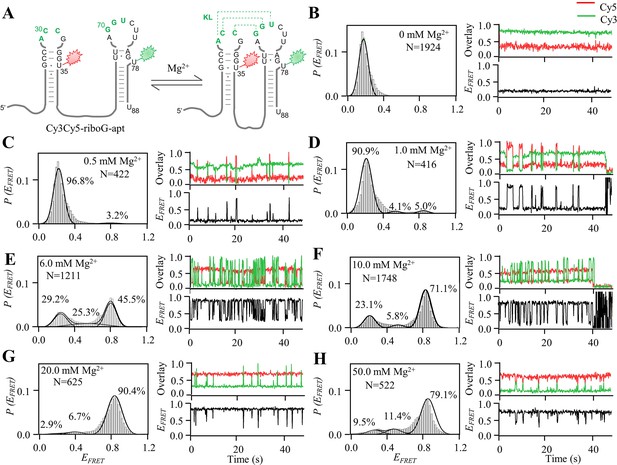
smFRET measurements of Cy3Cy5-riboG-apt at 0–50.0 mM Mg2+.
(A) The secondary structures of the unfolded (left) and folded (right) states of riboG-apt. (B–H) smFRET histograms, representative single-molecule trajectories and FRET curves of Cy3Cy5-riboG-apt at 0 mM Mg2+ (B), 0.5 mM Mg2+ (C), 1.0 mM Mg2+ (D), 6.0 mM Mg2+ (E), 10.0 mM Mg2+ (F), 20.0 mM Mg2+ (G) and 50.0 mM Mg2+ (H).
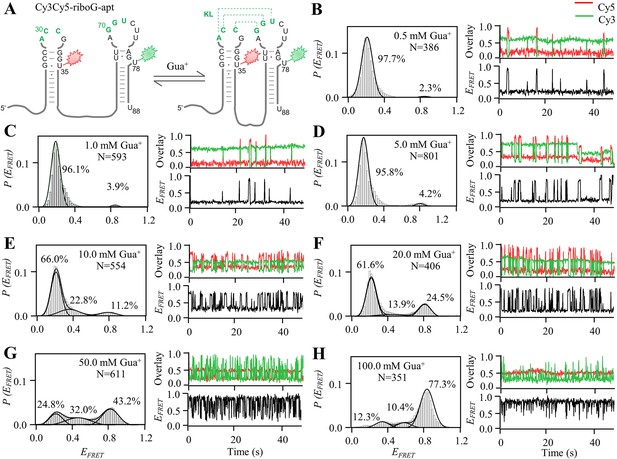
smFRET measurements of Cy3Cy5-riboG-apt at 0.5–100.0 mM Gua+.
(A) The secondary structures of the unfolded (left) and folded (right) states of riboG-apt. (B–H) smFRET histograms, representative single-molecule trajectories and FRET curves of Cy3Cy5-riboG-apt at 0.5 mM Gua+ (B), 1.0 mM Gua+ (C), 5.0 mM Gua+ (D), 10.0 mM Gua+ (E), 20.0 mM Gua+ (F), 50.0 mM Gua+ (G) and 100.0 mM Gua+ (H).
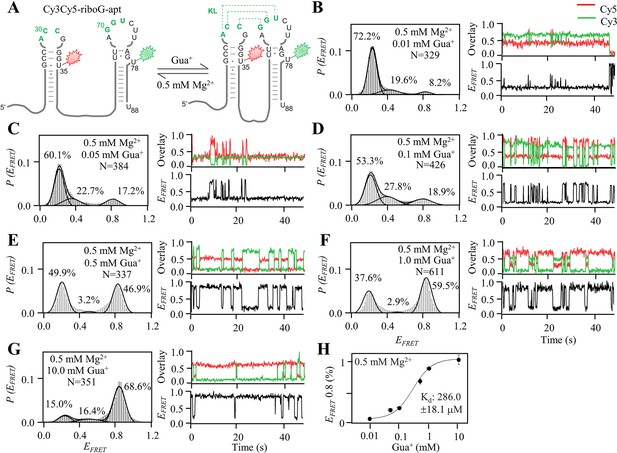
smFRET measurements of Cy3Cy5-riboG-apt at 0.01–10.0 mM Gua+ in the presence of 0.5 mM Mg2+.
(A) The secondary structures of the unfolded (left) and the folded (right) states of riboG-apt. (B–G) smFRET histograms, representative single-molecule trajectories and FRET curves of Cy3Cy5-riboG-apt at 0.01 mM Gua+ (B), 0.05 mM Gua+ (C), 0.1 mM Gua+ (D), 0.5 mM Gua+ (E), 1.0 mM Gua+ (F) and 10.0 mM Gua+ (G) in the presence of 0.5 mM Mg2+. (H) The percentages of the folded state (EFRET ~0.8) of Cy3Cy5-riboG-apt were plotted with the concentrations of Gua+ at 0.5 mM Mg2+, with an apparent Kd of 286.0±18.1 μM in three independent experiments.
-
Figure 2—figure supplement 6—source data 1
Data for the graphs shown in Figure 2—figure supplement 6H.
- https://cdn.elifesciences.org/articles/94706/elife-94706-fig2-figsupp6-data1-v1.xlsx
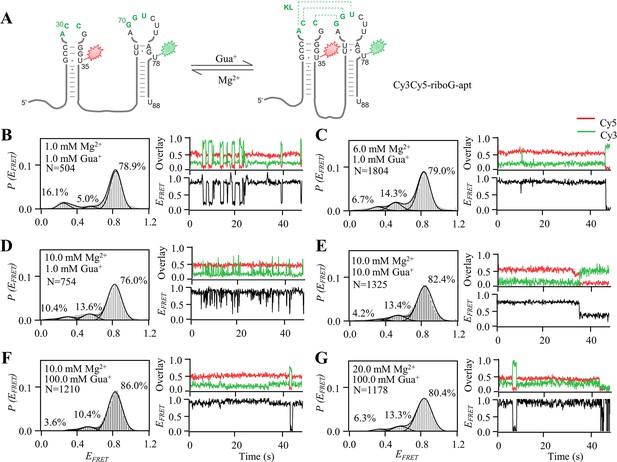
smFRET measurements of Cy3Cy5-riboG-apt at different Gua+ and Mg2+.
(A) The secondary structures of the unfolded (left) and the folded (right) states of riboG-apt. (B–G) smFRET histograms, representative single-molecule trajectories and FRET curves of Cy3Cy5-riboG-apt at 1.0 mM Gua+ and 1.0 mM Mg2+ (B), 1.0 mM Gua+ and 6.0 mM Mg2+ (C), 1.0 mM Gua+ and 10.0 mM Mg2+ (D), 10.0 mM Gua+ and 10.0 mM Mg2+ (E), 100.0 mM Gua+ and 10.0 mM Mg2+ (F) and 100.0 mM Gua+ and 20.0 mM Mg2+ (G).
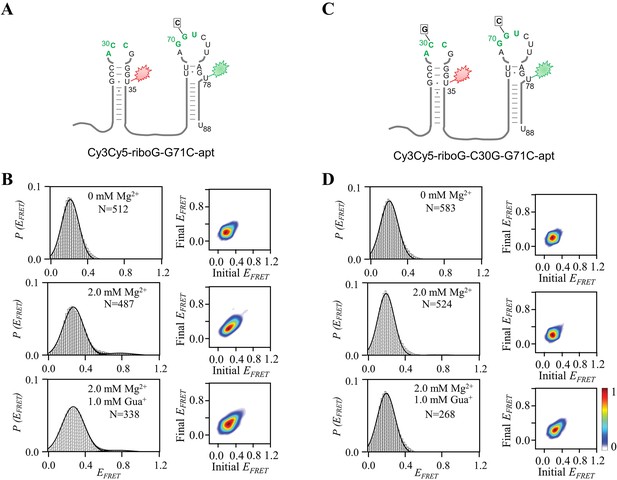
smFRET measurements of Cy3Cy5-riboG-G71C-apt and Cy3Cy5-riboG-C30G-G71C-apt.
(A) The secondary structure of Cy3Cy5-riboG-G71C-apt. (B) smFRET histograms and transition density plots for Cy3Cy5-riboG-G71C-apt at 0 mM Mg2+, at 2.0 mM Mg2+, and at 2.0 mM Mg2+ and 1.0 mM Gua+. (C) The secondary structure of Cy3Cy5-riboG-C30G-G71C-apt. (D) smFRET histograms and transition density plots for Cy3Cy5-riboG-C30G-G71C-apt at 0 mM Mg2+, at 2.0 mM Mg2+, and at 2.0 mM Mg2+ and 1.0 mM Gua+.
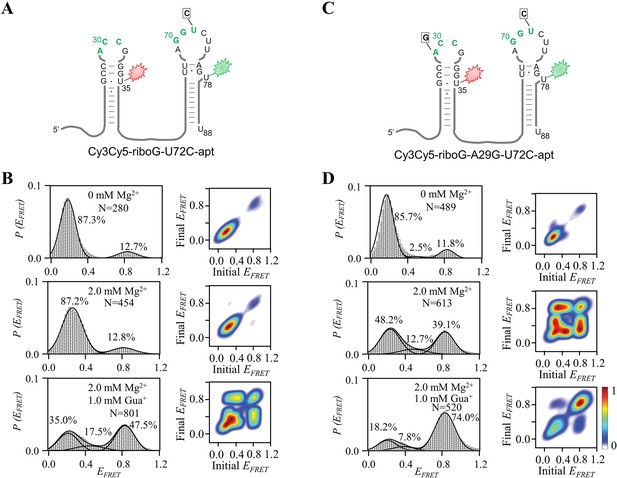
smFRET measurements of Cy3Cy5-riboG-U72C-apt and Cy3Cy5-riboG-A29G-U72C-apt.
(A) The secondary structure of Cy3Cy5-riboG-U72C-apt. (B) smFRET histograms and transition density plots for Cy3Cy5-riboG-U72C-apt at 0 mM Mg2+, at 2.0 mM Mg2+, and at 2.0 mM Mg2+ and 1.0 mM Gua+. (C) The secondary structure of Cy3Cy5-riboG-A29G-U72C-apt. (D) smFRET histograms and transition density plots for Cy3Cy5-riboG-A29G-U72C-apt at 0 mM Mg2+, at 2.0 mM Mg2+, and at 2.0 mM Mg2+ and 1.0 mM Gua+.
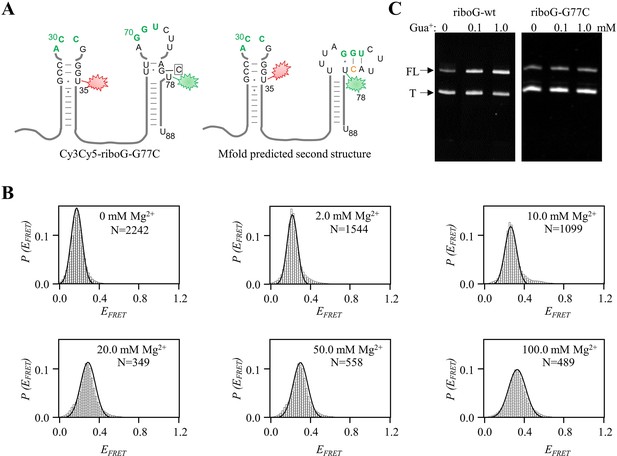
The G77C mutation perturbs the folding and function of riboG-apt.
(A) The secondary structures of Cy3Cy5-riboG-G77C-apt, with the structure predicted by Mfold on the right. (B) smFRET histograms of Cy3Cy5-riboG-G77C-apt at 0–100.0 mM Mg2+. (C) Denaturing PAGE images of transcription termination assays of WT (left) and riboG-G77C, respectively. The FL and T represent the full-length and terminated RNA.
-
Figure 2—figure supplement 10—source data 1
Raw gels for Figure 2—figure supplement 10C.
- https://cdn.elifesciences.org/articles/94706/elife-94706-fig2-figsupp10-data1-v1.zip
-
Figure 2—figure supplement 10—source data 2
Gels with labeled lanes used in Figure 2—figure supplement 10C.
- https://cdn.elifesciences.org/articles/94706/elife-94706-fig2-figsupp10-data2-v1.zip
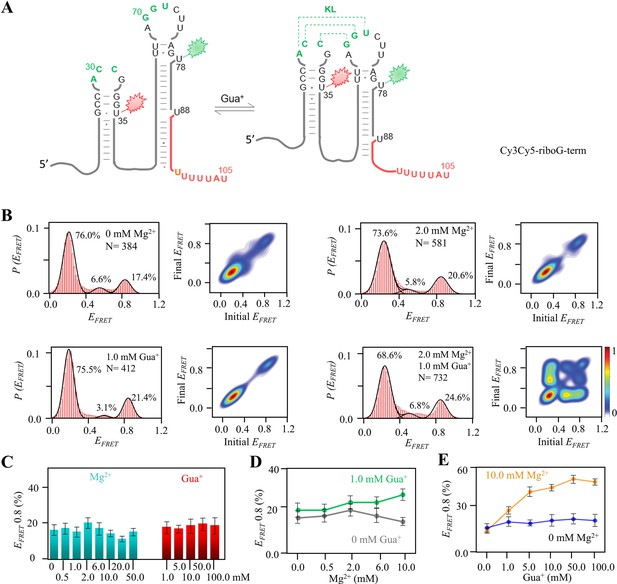
smFRET studies of post-transcriptional riboG-term at different concentrations of Mg2+ and Gua+.
(A) The secondary structures of Cy3Cy5-riboG-term at the unfolded (left) and the folded state (right). (B) smFRET histograms and transition density plots of Cy3Cy5-riboG-term at 0 mM Mg2+, at 2.0 mM Mg2+, at 1.0 mM Gua+, and at 2.0 mM Mg2+ and 1.0 mM Gua+. (C) The percentages of the folded conformation (EFRET ~0.8) of Cy3Cy5-riboG-term at 0–50.0 mM Mg2+ (cyan columns) and 1–100.0 mM Gua+ (red columns). (D) The percentages of the folded conformation of Cy3Cy5-riboG-term change with Mg2+ at 0 (black) and 1.0 mM Gua+ (green). (E) The percentages of the folded conformation of Cy3Cy5-riboG-term change with Gua+ at 0 (blue) and 10.0 mM Mg2+ (orange). Data represent average ± SD from three replicates (n=3).
-
Figure 3—source data 1
Data for the graphs shown in Figure 3.
- https://cdn.elifesciences.org/articles/94706/elife-94706-fig3-data1-v1.xlsx
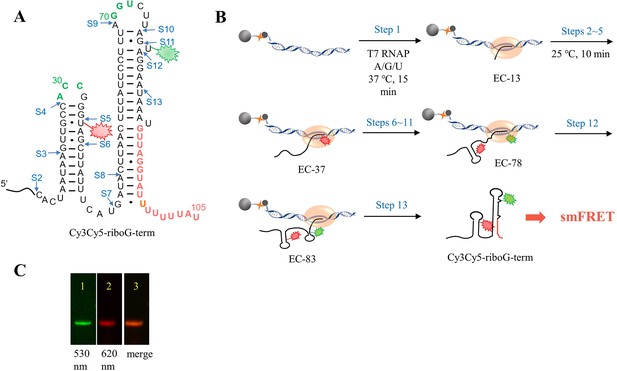
The schematic procedure of preparing Cy3Cy5-riboG-term by 13 step-PLOR reaction for smFRET study.
(A) The secondary structure of riboG-term. The positions of donor (Cy3) and acceptor (Cy5) are shown by green and red sparkles at U78 and U35, respectively. The sites started at each step of the PLOR-synthesis were marked by blue arrows. (B) 13-step PLOR reaction for preparing Cy3Cy5-riboG-term for smFRET study. The reagent usages for the 13-step reaction are listed in Supplementary file 1, table S4. (C) The PAGE images of Cy3Cy5-riboG-term. The Lanes 1 and 2 were irradiated by 530 nm and 620 nm fluorescence, respectively. Lane 3 is the merged image of lanes 1 and 2.
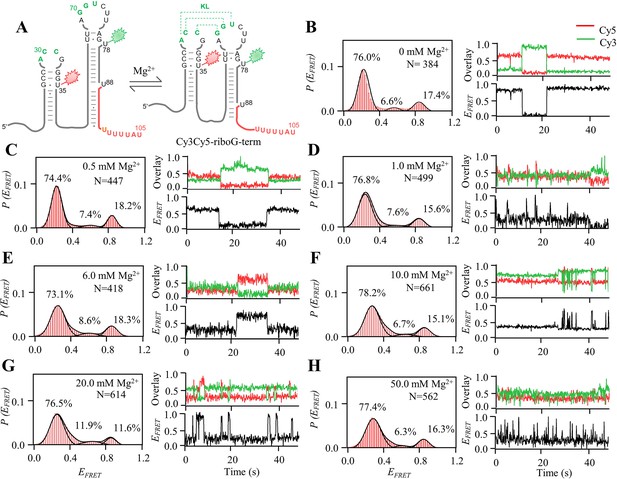
smFRET measurements of Cy3Cy5-riboG-term at 0–50.0 mM Mg2+.
(A) The secondary structures of the unfolded (left) and folded (right) states of riboG-term. (B–H), smFRET histograms, dynamic single-molecule trajectories and FRET curves of Cy3Cy5-riboG-term at 0 mM Mg2+ (B), 0.5 mM Mg2+ (C), 1.0 mM Mg2+ (D), 6.0 mM Mg2+ (E), 10.0 mM Mg2+ (F), 20.0 mM Mg2+ (G) and 50.0 mM Mg2+ (H). Approximately 1.4%, 1.1%, 6.2%, 7.4%, 10.3%, 6.2%, and 2.4% of dynamic single-molecule trajectories were detected in (B), (C), (D), (E), (F), (G), and (H) respectively.
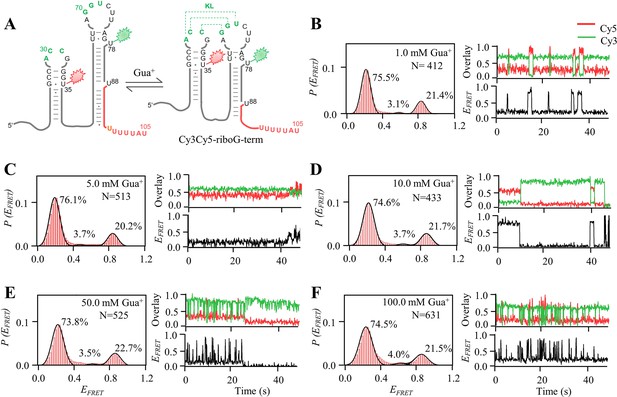
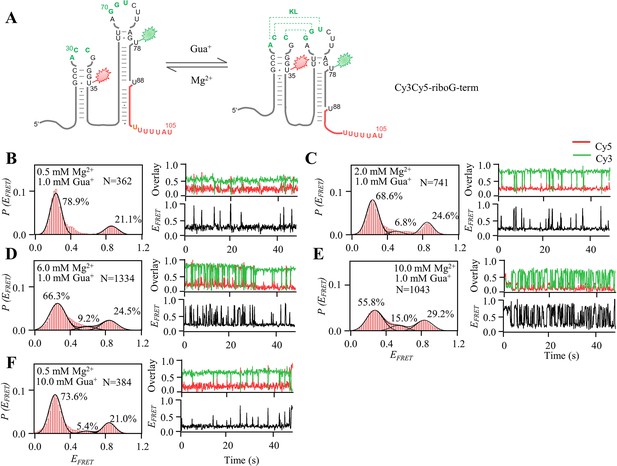
smFRET measurements of Cy3Cy5-riboG-term at 1.0–100.0 mM Gua+.
(A) The secondary structures of the unfolded (left) and folded (right) states of riboG-term. (B–F), smFRET histograms, dynamic single-molecule trajectories and FRET curves of Cy3Cy5-riboG-term at 1.0 mM Gua+ (B), 5.0 mM Gua+ (C), 10.0 mM Gua+ (D), 50.0 mM Gua+ (E) and 100.0 mM Gua+ (F). Approximately 1.3%, 3.4%, 3.1%, 3.0%, and 11.5% of dynamic single-molecule trajectories were detected in (B), (C), (D), (E), and (F), respectively.
smFRET measurements of Cy3Cy5-riboG-term at different Gua+ and Mg2+.
(A) The secondary structures of the unfolded (left) and folded (right) states of riboG-term. (B–F), smFRET histograms, representative single-molecule trajectories and FRET curves of Cy3Cy5-riboG-term at 1.0 mM Gua+ and 0.5 mM Mg2+ (B), 1.0 mM Gua+ and 2.0 mM Mg2+ (C), 1.0 mM Gua+ and 6.0 mM Mg2+ (D), 1.0 mM Gua+ and 10.0 mM Mg2+ (E), and 10.0 mM Gua+ and 0.5 mM Mg2+ (F).
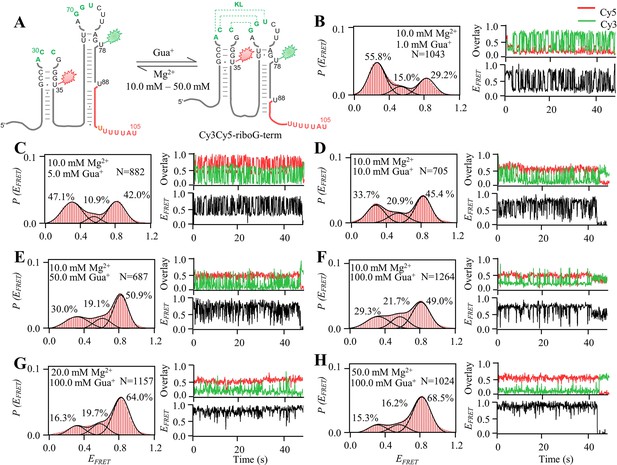
smFRET measurements of Cy3Cy5-riboG-term at different Gua+ and Mg2+.
(A) The secondary structures of the unfolded (left) and folded (right) states of riboG-term. (B–H), smFRET histograms, representative single-molecule trajectories and FRET curves of Cy3Cy5-riboG-term at 1.0 mM Gua+ and 10.0 mM Mg2+ (B), 5.0 mM Gua+ and 10.0 mM Mg2+ (C), 10.0 mM Gua+ and 10.0 mM Mg2+ (D), 50.0 mM Gua+ and 10.0 mM Mg2+ (E), 100.0 mM Gua+ and 10.0 mM Mg2+ (F), 100.0 mM Gua+ and 20.0 mM Mg2+ (G), and 100.0 mM Gua+ and 50.0 mM Mg2+ (H).
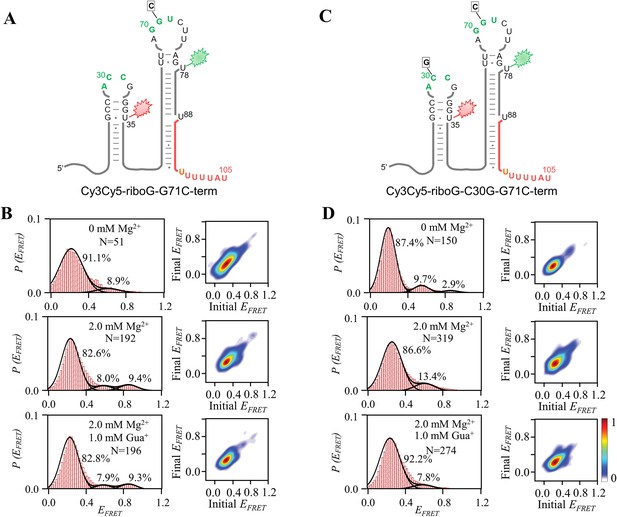
smFRET measurements for Cy3Cy5-riboG-G71C-term and Cy3Cy5-riboG-C30G-G71C-term.
(A) The secondary structure of Cy3Cy5-riboG-G71C-term. (B) smFRET histograms and transition density plots for Cy3Cy5-riboG-G71C-term at 0 mM Mg2+, at 2.0 mM Mg2+, and at 2.0 mM Mg2+ and 1.0 mM Gua+. (C) The secondary structure of Cy3Cy5-riboG-C30G-G71C-term. (D) smFRET histograms and transition density plots for Cy3Cy5-riboG-C30G-G71C-term at 0 mM Mg2+, at 2.0 mM Mg2+, and at 2.0 mM Mg2+ and 1.0 mM Gua+.
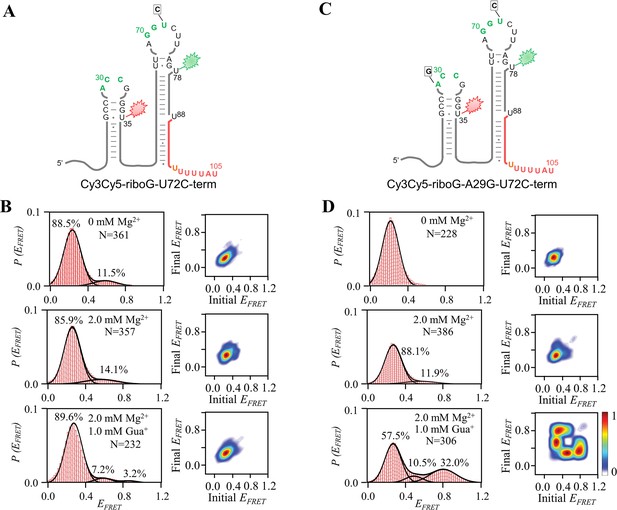
smFRET measurements for Cy3Cy5-riboG-U72C-term and Cy3Cy5-riboG-A29G-U72C-term.
(A) The secondary structure of Cy3Cy5-riboG-U72C-term. (B) smFRET histograms and transition density plots for Cy3Cy5-riboG-U72C-term at 0 mM Mg2+, at 2.0 mM Mg2+, and at 2.0 mM Mg2+ and 1.0 mM Gua+. (C) The secondary structure of Cy3Cy5-riboG-A29G-U72C-term. (D) smFRET histograms and transition density plots for Cy3Cy5-riboG-A29G-U72C-term at 0 mM Mg2+, at 2.0 mM Mg2+, and at 2.0 mM Mg2+ and 1.0 mM Gua+.
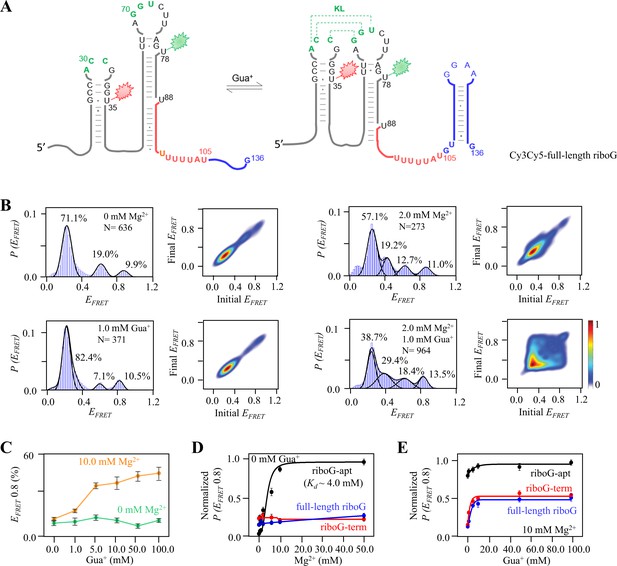
smFRET studies of the isolated full-length riboG at different concentrations of Mg2+ and Gua+.
(A) The secondary structures of Cy3Cy5-ful-length riboG at the unfolded (left) and the folded state (right). The full-length riboG contains the aptamer domain (black), terminator (red) and the extended sequence (blue). Cy3 and Cy5 are shown by green and red sparkles, respectively. (B) smFRET histograms and transition density plots for Cy3Cy5-full-length riboG at 0 mM Mg2+, at 2.0 mM Mg2+, at 1.0 mM Gua+, and at 2.0 mM Mg2+ and 1.0 mM Gua+. (C) The percentages of the folded conformation of Cy3Cy5-full-length riboG change with Gua+ at 0 (green) and 10.0 mM Mg2+ (orange). (D–E) The normalized percentages of the folded conformation of Cy3Cy5-riboG-apt (black), Cy3Cy5-riboG-term (red) and Cy3Cy5-full-length riboG (blue) at 0–50 mM Mg2+ (D) and 0–100.0 mM Gua+ in the presence of 10.0 mM Mg2+ (E).Data represent average ± SD from three replicates (n=3).
-
Figure 4—source data 1
Data for the graphs shown in Figure 4.
- https://cdn.elifesciences.org/articles/94706/elife-94706-fig4-data1-v1.xlsx
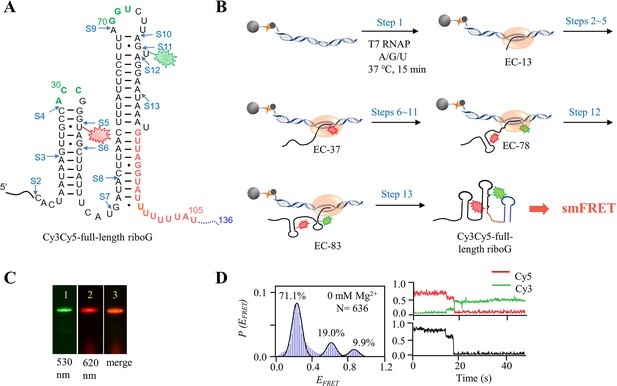
The schematic procedure of preparing Cy3Cy5-full-length riboG by 13 step-PLOR reaction for smFRET study.
(A) The secondary structure of full-length riboG. The positions of donor (Cy3) and acceptor (Cy5) are shown by green and red sparkles at U78 and U35, respectively. The sites started at each step of the PLOR-synthesis were marked by blue arrows. (B) 13-step PLOR reaction for preparing Cy3Cy5-full-length riboG for smFRET study. The reagent usages for the 13-step reaction are listed in Supplementary file 1, table S5. (C) The PAGE images of Cy3Cy5-full-length riboG. The Lanes 1 and 2 were irradiated by 530 nm and 620 nm fluorescence, respectively. Lane 3 is the merged image of lanes 1 and 2. (D) smFRET study of Cy3Cy5-full-length riboG in the absence of Mg2+. The smFRET histogram, dynamic single-molecule trajectory and FRET curve for Cy3Cy5-full-length riboG at 0 mM Mg2+. Approximately 1.4% of dynamic single-molecule trajectories was detected.
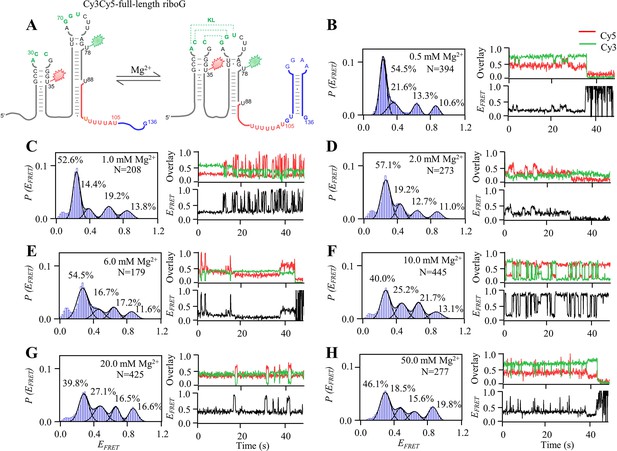
smFRET measurements of Cy3Cy5-full-length riboG at 0.5–50.0 mM Mg2+.
(A) The secondary structures of the unfolded (left) and folded (right) states of full-length riboG. B–H, smFRET histograms, dynamic single-molecule trajectories and FRET curves of Cy3Cy5-full-length riboG at 0.5 mM Mg2+ (B), 1.0 mM Mg2+ (C), 2.0 mM Mg2+ (D), 6.0 mM Mg2+ (E), 10.0 mM Mg2+ (F), 20.0 mM Mg2+ (G) and 50.0 mM Mg2+ (H). Approximately 2.6%, 3.8%, 5.5%, 11.7%, 11.5%, 16.0%, and 28.9% of dynamic single-molecule trajectories were detected (B), (C), (D), (E), (F), (G), and (H), respectively.
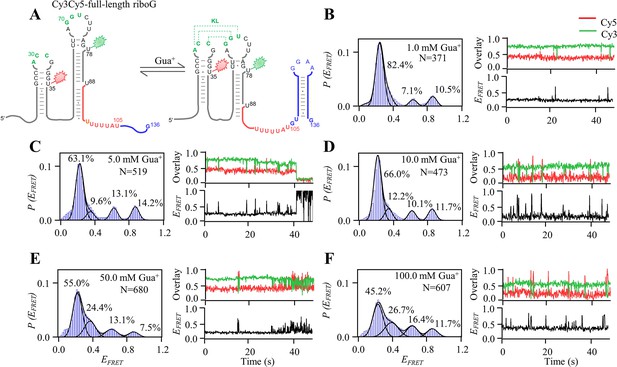
smFRET measurements of Cy3Cy5-full-length riboG at 1.0–100.0 mM Gua+.
(A) The secondary structures of the unfolded (left) and folded (right) states of full-length riboG. (B–F), smFRET histograms, dynamic single-molecule trajectories and FRET curves of Cy3Cy5-full-length riboG at 1.0 mM Gua+ (B), 5.0 mM Gua+ (C), 10.0 mM Gua+ (D), 50.0 mM Gua+ (E) and 100.0 mM Gua+ (F). Approximately 3.0%, 5.6%, 15.9%, 27.9%, and 46.6% of dynamic single-molecule trajectories were detected (B), (C), (D), (E), and (F), respectively.
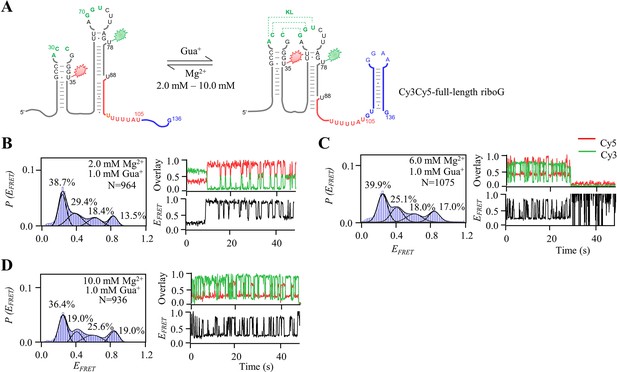
smFRET measurements of Cy3Cy5-full-length riboG at 1.0 mM Gua+ and 2.0–10.0 mM Mg2+.
(A) The secondary structures of the unfolded (left) and folded (right) states of full-length riboG. (B–D), smFRET histograms, dynamic single-molecule trajectories and FRET curves of Cy3Cy5-full-length riboG at 1.0 mM Gua+ and 2 mM Mg2+ (B), 1.0 mM Gua+ and 6.0 mM Mg2+ (C), and 1.0 mM Gua+ and 10.0 mM Mg2+ (D) Approximately 44.3%, 59.5%, and 66.5% of dynamic single-molecule trajectories were detected (B), (C), and (D), respectively.
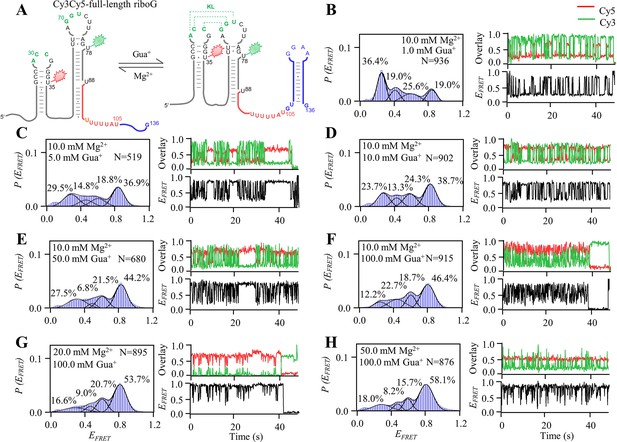
smFRET measurements of Cy3Cy5-full-length riboG at different Gua+ and Mg2+.
(A) The secondary structures of the unfolded (left) and folded (right) states of full-length riboG. (B–H), smFRET histograms, dynamic single-molecule trajectories and FRET curves of Cy3Cy5-full-length riboG at 1.0 mM Gua+ and 10.0 mM Mg2+ (B), 5.0 mM Gua+ and 10.0 mM Mg2+ (C), 10.0 mM Gua+ and 10.0 mM Mg2+ (D), 50.0 mM Gua+ and 10.0 mM Mg2+ (E), 100.0 mM Gua+ and 10.0 mM Mg2+ (F), 100.0 mM Gua+ and 20.0 mM Mg2+ (G), and 100.0 mM Gua+ and 50.0 mM Mg2+ (H). Approximately 66.5%, 83.4%, 65.7%, 79.6%, 66.9%, 63.9%, and 59.9% of dynamic single-molecule trajectories were detected (B), (C), (D), (E), (F), (G), and (H), respectively.
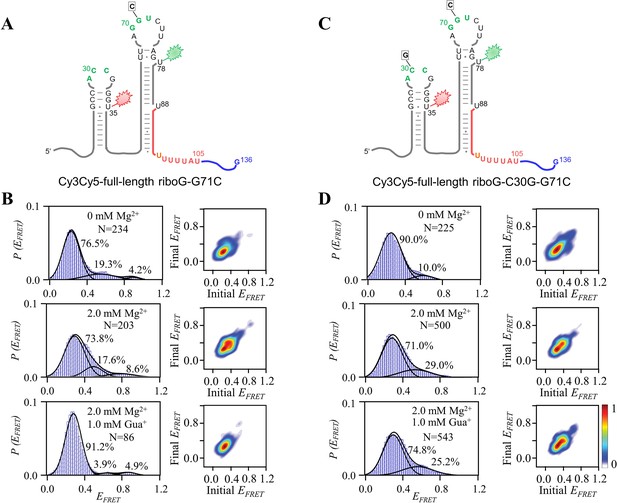
smFRET measurements for Cy3Cy5-full-length riboG-G71C and Cy3Cy5-full-length riboG-C30G-G71C.
(A) The secondary structure of Cy3Cy5-full-length riboG-G71C. (B) smFRET histograms and transition density plots for Cy3Cy5-full-length riboG-G71C at 0 mM Mg2+, at 2.0 mM Mg2+, and at 2.0 mM Mg2+ and 1.0 mM Gua+. (C) The secondary structure of Cy3Cy5-full-length riboG-C30G-G71C. (D) smFRET histograms and transition density plots for Cy3Cy5-full-length riboG-C30G-G71C at 0 mM Mg2+, at 2.0 mM Mg2+, and at 2.0 mM Mg2+ and 1.0 mM Gua+.
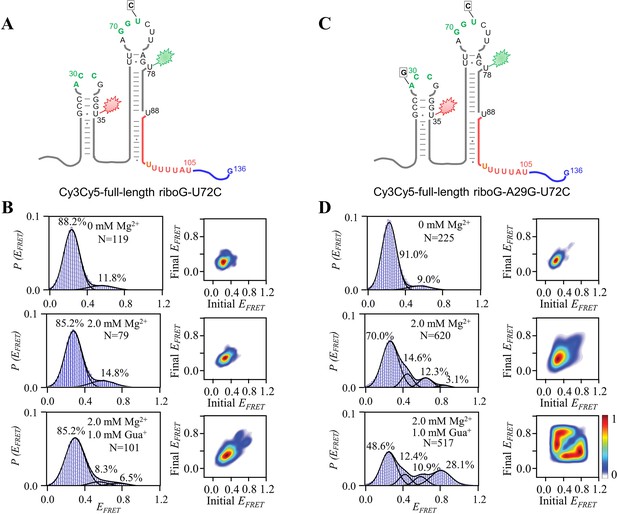
smFRET measurements for Cy3Cy5-full-length riboG-U72C and Cy3Cy5-full-length riboG-A29G-U72C.
(A) The secondary structure of Cy3Cy5-full-length riboG-U72C. (B) smFRET histograms and transition density plots for Cy3Cy5-full-length riboG-U72C at 0 mM Mg2+, at 2.0 mM Mg2+, and at 2.0 mM Mg2+ and 1.0 mM Gua+. (C) The secondary structure of Cy3Cy5-full-length riboG-A29G-U72C. (D) smFRET histograms and transition density plots for Cy3Cy5-full-length riboG-A29G-U72C at 0 mM Mg2+, at 2.0 mM Mg2+, and at 2.0 mM Mg2+ and 1.0 mM Gua+.
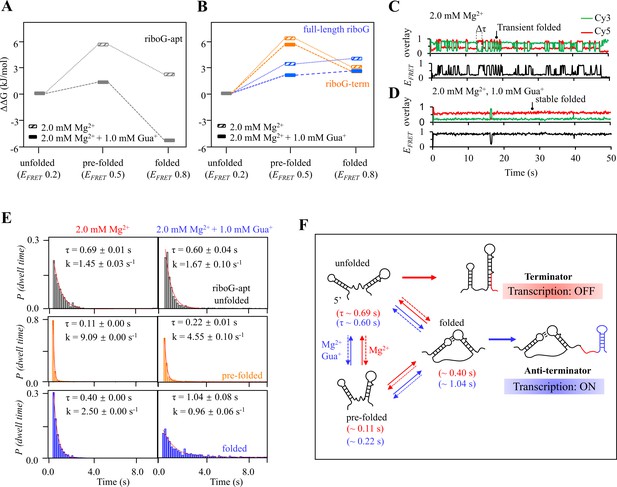
Relative free energy (ΔΔG) and kinetics analysis of isolated riboG-apt, riboG-term and full-length riboG.
(A) The relative free energy of the pre-folded and folded states of riboG-apt in the presence of 0 and 1.0 mM Gua+ at 2.0 mM Mg2+. The free energy of the unfolded state was referred as control. (B) The relative free energy of the pre-folded and folded states of riboG-term (orange) and full-length riboG (blue) in the presence of 0 and 1.0 mM Gua+ at 2.0 mM Mg2+. (C–D) Representative time traces of riboG-apt at 0 (C) and 1.0 mM Gua+ (D) in the presence of 2.0 mM Mg2+. Δτ is the dwell time. (E) The lifetime (τ) and rate constant (k) of unfolded (black), pre-folded (orange) and folded (blue) states of riboG-apt was determined by exponential decays of the dwell time distributions. (F) Schematic diagram of riboG-apt folding and transcriptional processes at 2.0 mM Mg2+ in the presence of 0 mM Gua+ (red) and 1.0 mM Gua+ (blue).
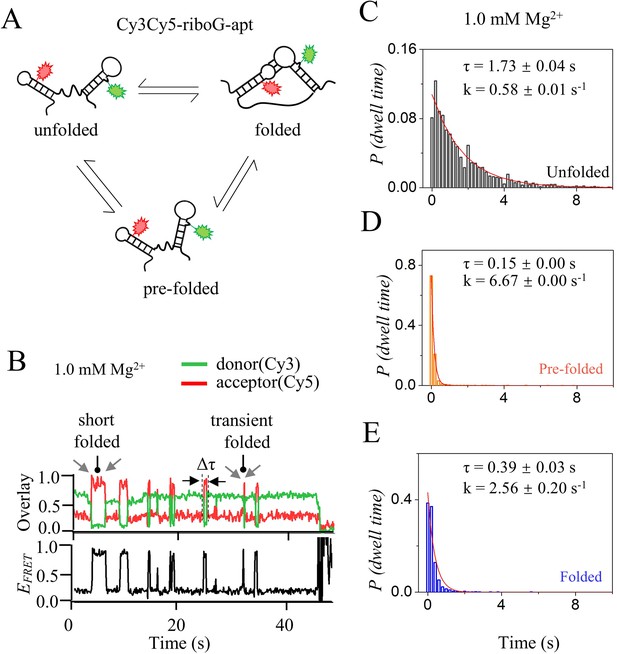
Kinetics analysis of the unfolded, pre-folded and folded states of riboG-apt at 1.0 mM Mg2+.
(A) The structural switching of the unfolded, pre-folded and folded states of riboG-apt. The green and red sparkles represent Cy3 and Cy5, respectively. (B) The representative single-molecule trajectories of riboG-apt at 1.0 mM Mg2+. Δτ is the dwell time. (C–E), Dwell time histograms for the unfolded (C), pre-folded (D) and folded (E) used for determination of transition rate constants at 1.0 mM Mg2+. Plots were fit with exponential decay curves to extract conformation switch kinetics.
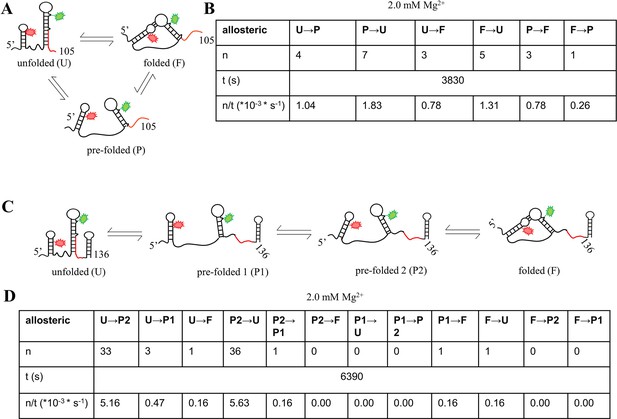
Kinetics analysis of isolated riboG-term and full-length riboG at 2.0 mM Mg2+.
(A) The structural switching of the unfolded (U), pre-folded (P) and folded states (F) of riboG-term. (B) The transitions between two states of riboG-term at 2.0 mM Mg2+. n: number of observed transition events; t: total observation time; n/t: frequency of transition between two states, and n/t is used to the rate constant of transitions (k). (C) The structural switching of unfolded (U), pre-folded 1 (P1), pre-folded 2 (P2) and folded (F) states of full-length riboG. (D) The transitions between two states of full-length riboG at the presence of 2.0 mM Mg2+.
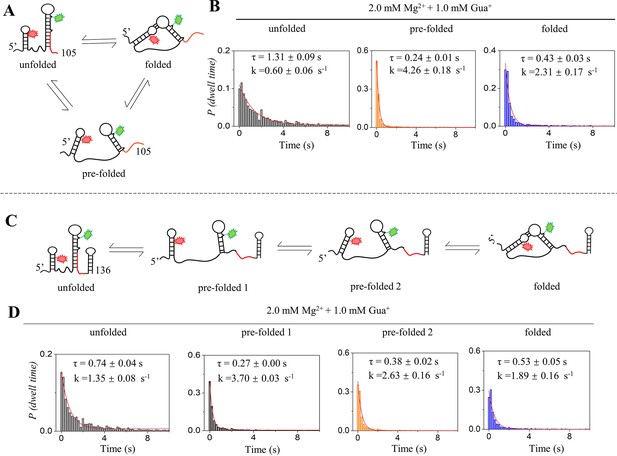
Kinetics analysis of isolated riboG-term and full-length riboG at 2.0 mM Mg2+ and 1.0 mM Gua+.
(A) The structural switching of the unfolded (U), pre-folded (P) and folded states (F) of riboG-term. (B) The histograms of dwell time for the unfolded (gray), pre-folded (orange) and folded (blue) states of riboG-term. (C) The structural switching of unfolded (U), pre-folded 1 (P1), pre-folded 2 (P2) and folded (F) states of full-length riboG. (D) The histograms of dwell time for the unfolded (gray), pre-folded 1 (black), pre-folded 2 (orange) and folded (blue) states of full-length riboG.
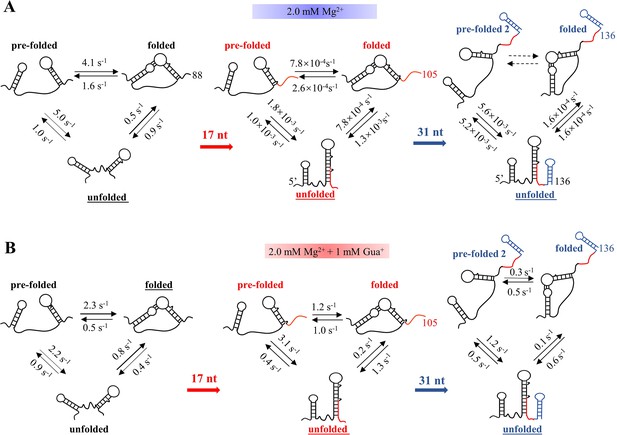
Kinetics analysis of the unfolded, pre-folded or pre-folded 2 and folded states of riboG-apt, riboG-term and full-length riboG without and with guanidine at 2 mM Mg2+.
The stable structures at each stage are underlined.
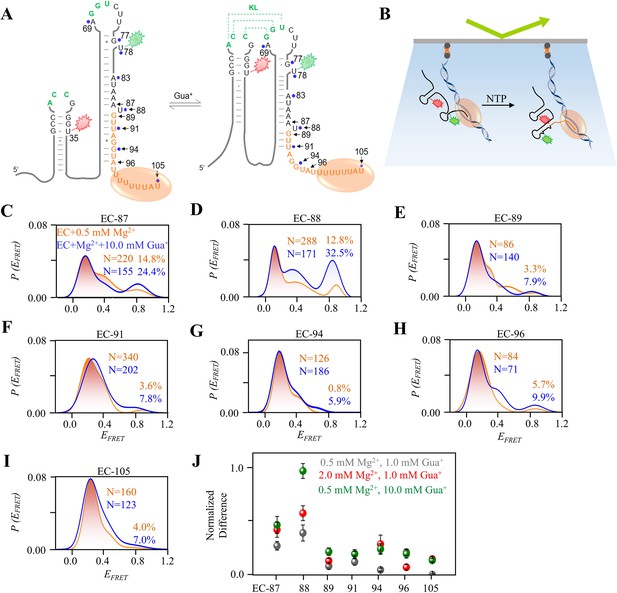
smFRET studies of nascent riboG in ECs without and with Gua+ at 0.5 mM Mg2+.
(A) The ECs containing an RNA at the unfolded state (left) and the folded state (right). The pause sites of EC-87 to EC-105 are marked by black arrows, and the orange ellipse represents the T7 RNA polymerase. (B) Schematic diagram for smFRET experiment for ECs. The ECs were immobilized on the slides by their biotinylated DNA templates. (C–I) smFRET histograms for the EC-87 to EC-105 at 0 mM (orange) and 10.0 mM Gua+ (blue) at 0.5 mM Mg2+. (J) The normalized increase of the folded conformation (EFRET ~0.8) of ECs upon the addition of 1.0 mM Gua+ at 0.5 mM Mg2+ (grey), 10.0 mM Gua+ at 0.5 mM Mg2+ (green) and 1.0 mM Gua+ at 2.0 mM Mg2+ (red). Data represent average ± SD from three replicates (n=3).
-
Figure 6—source data 1
Data for the graphs shown in Figure 6J.
- https://cdn.elifesciences.org/articles/94706/elife-94706-fig6-data1-v1.xlsx
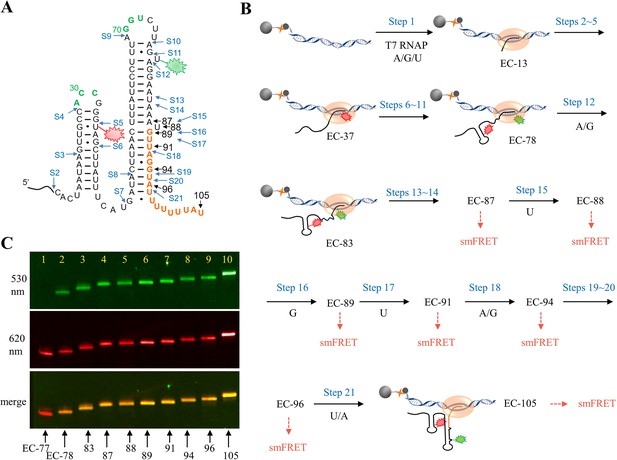
The schematic procedure of preparing EC-87 to EC-105 for smFRET study.
(A) The secondary structure of riboG-term. The start sites at each step of PLOR were marked by blue arrows. The donor, Cy3 (green sparkle) and acceptor, Cy5 (red sparkle) were localized at U78 and U35, respectively. The paused sites at nascent RNA in ECs were marked with black arrows. (B) The schematic procedure of 21-step PLOR reaction for preparing ECs for smFRET study. The reagent usages for the reaction are listed in Supplementary file 1, table S7. (C) PAGE images of EC-77 (lane 1), EC-78 (lane 2), EC-83 (lane 3), EC-87 (lane 4), EC-88 (lane 5), EC-89 (lane 6), EC-91 (lane 7), EC-94 (lane 8), EC-96 (lane 9), and EC-105 (lane 10). The top and middle images were irradiated by 530 and 620 nm fluorescence, respectively. The bottom image was merged of the top and middle images.
-
Figure 6—figure supplement 1—source data 1
Raw gels for Figure 6—figure supplement 1C.
- https://cdn.elifesciences.org/articles/94706/elife-94706-fig6-figsupp1-data1-v1.zip
-
Figure 6—figure supplement 1—source data 2
Gels for Figure 6—figure supplement 1C.
- https://cdn.elifesciences.org/articles/94706/elife-94706-fig6-figsupp1-data2-v1.zip
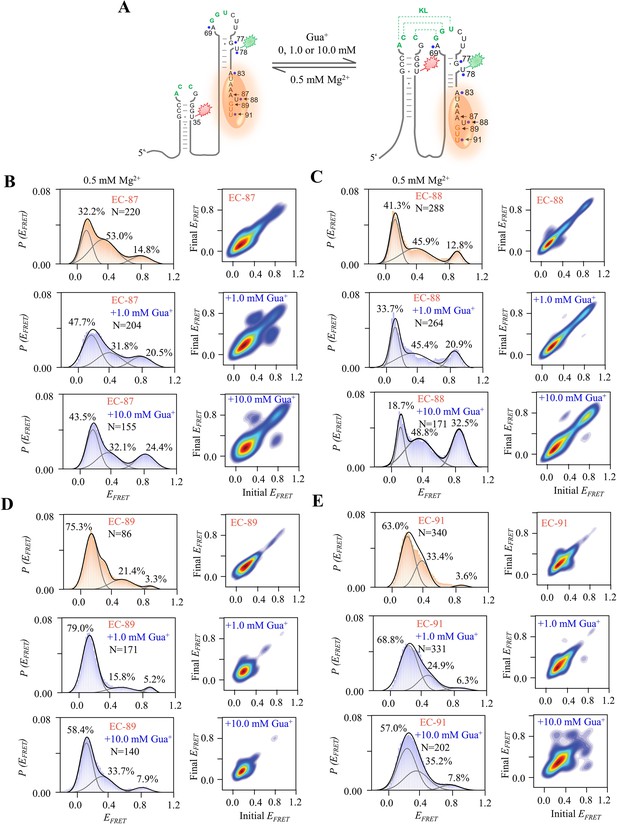
smFRET studies of EC-87, EC-88, EC-89, and EC-91 at 0, 1.0 and 10.0 mM Gua+ in the presence of 0.5 mM Mg2+.
(A) The schematic diagram of ECs with nascent RNA and T7 RNA polymerase. The orange ellipse is RNA polymerase. (B–E) smFRET histograms and transition density plots of EC-87 (B), EC-88 (C), EC-89 (D) and EC-91(E) at 0 mM, 1.0 mM and 10.0 mM Gua+ in the presence of 0.5 mM Mg2+.
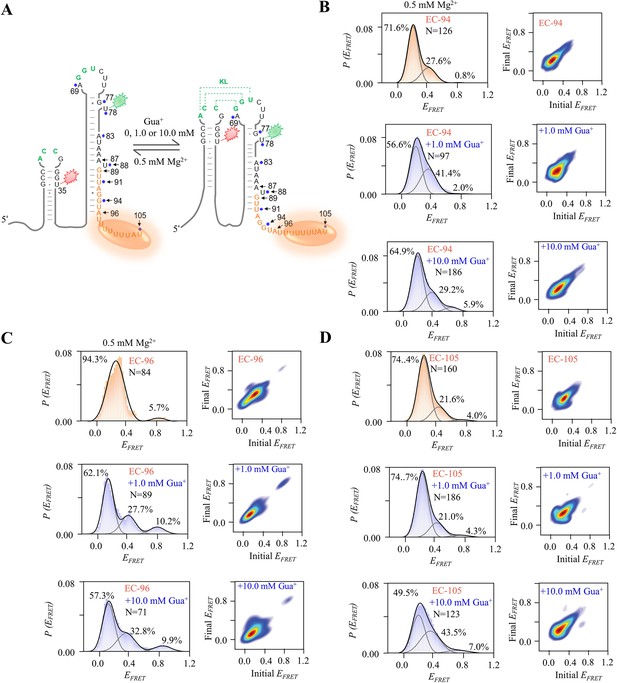
smFRET studies of EC-94, EC-96 and EC-105 at 0, 1.0 and 10.0 mM Gua+ in the presence of 0.5 mM Mg2+.
(A) The schematic diagram of ECs. The orange ellipse is T7 RNA polymerase. (B–D) The smFRET histograms and transition density plots of EC-94 (B), EC-96 (C) and EC-105 (D) at 0 mM, 1.0 mM and 10.0 mM Gua+ in the presence of 0.5 mM Mg2+.
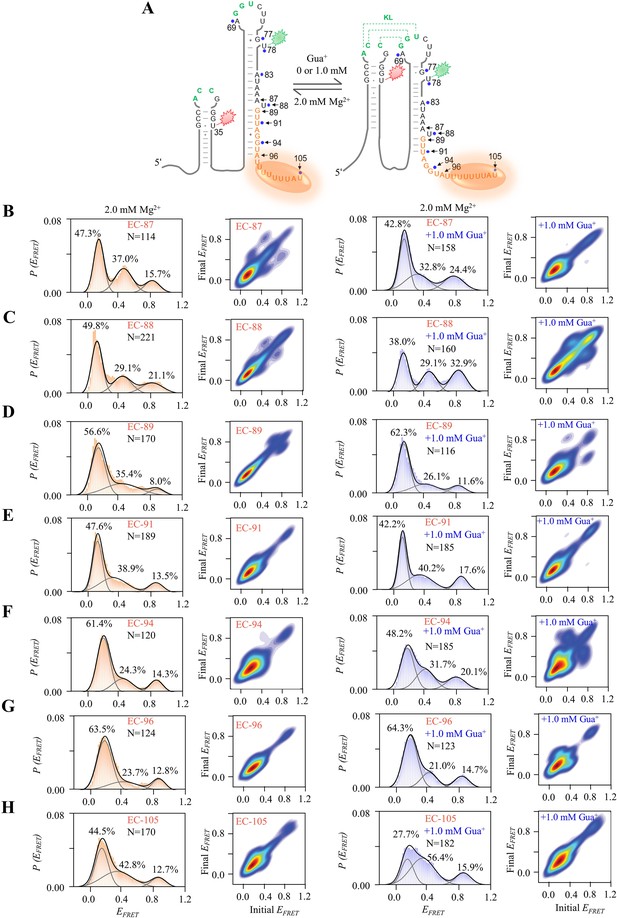
smFRET studies of EC-87, EC-88, EC-89, EC-91, EC-94, EC-96, and EC-105 at 0 and 1.0 mM Gua+ in the presence of 2.0 mM Mg2+.
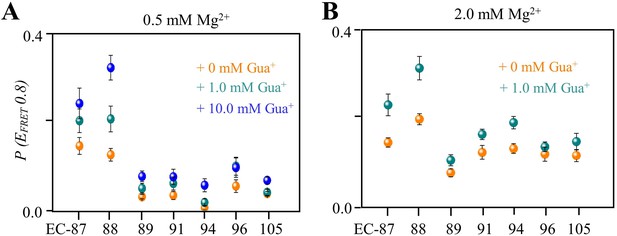
Comparison of the folded-conformation percentages in EC-87, EC-88, EC-89, EC-91, EC-94, EC-96, and EC-105 at different Gua+ in the presence of 0.5 mM Mg2+ (A) and 2.0 mM Mg2+ (B).
The data points of 0 mM, 1.0 mM, and 10.0 mM Gua+ are shown in orange, cyan and blue, respectively.
Ligand-mediated conformational switching of riboG during transcription.
(A) The schematic diagram of synthesizing ECs with nascent RNAs by in vitro transcriptional pause using PLOR reactions. The orange ellipse represents T7 RNA polymerase. (B) The secondary structures of full-length riboG. The halting sites of EC-69, EC-77, EC-78, EC-83, EC-88, EC-91, EC-94, and EC-105 are marked by red dots. (C) PAGE images of the crude products collected at the last step of individual PLOR strategy in the absence and presence of 1.0 mM Gua+. FL and T are represented for the read-through and terminated RNA. The detail strategies were depicted in Figure 7—figure supplement 1B. (D) The transcriptional read-through fractions were plotted with the last halting sites in 9 step-, 11 step-, 12 step-, 13 step-, 14 step-, 15 step-, 16 step-, and 17 step-PLOR. The orange and blue curves are in the absence and presence of 1.0 mM Gua+. Taking into consideration that the 17 step-PLOR reaction exhibited a pause within the terminator region, resulting in a significant amount of terminated product at step 16, crude products from steps 16 and 17 were collected for (C) and (D) of the 17 step-PLOR reaction (Lanes 15 and 16 in C). Data represent average ± SD from three replicates (n=3).
-
Figure 7—source data 1
Raw gels for Figure 7C.
- https://cdn.elifesciences.org/articles/94706/elife-94706-fig7-data1-v1.zip
-
Figure 7—source data 2
Gels for Figure 7C.
- https://cdn.elifesciences.org/articles/94706/elife-94706-fig7-data2-v1.zip
-
Figure 7—source data 3
Data for the graphs shown in Figure 7D.
- https://cdn.elifesciences.org/articles/94706/elife-94706-fig7-data3-v1.xlsx
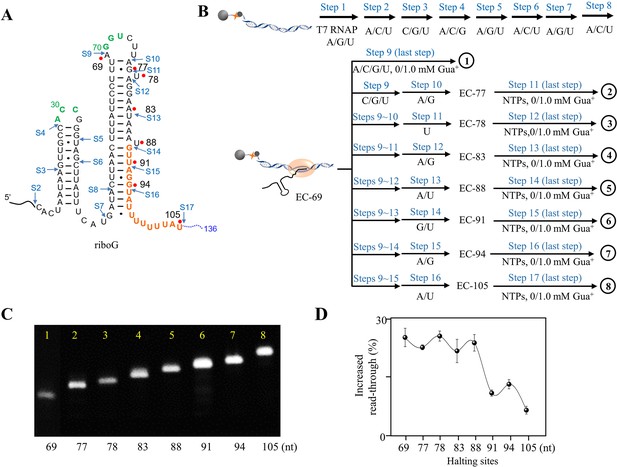
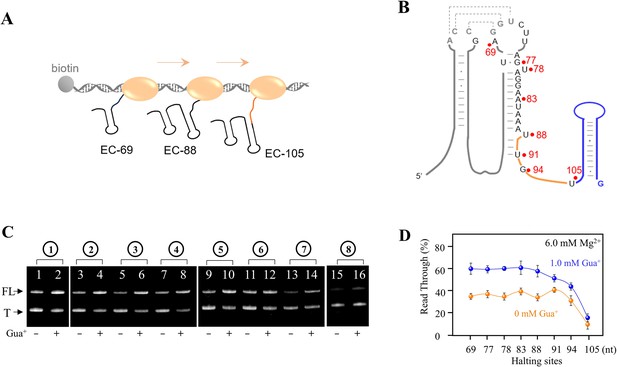
In vitro transcriptional termination assay of the guanidine-IV riboswitch by PLOR.
(A) The full-length guanidine-IV riboswitch can be transcribed by PLOR, halting at specific sites in individual step. The start sites of the 17 step-PLOR reaction were marked by blue arrows. (B) Strategies of 9 step- (1), 11 step- (2), 12 step- (3), 13 step- (4), 14 step- (5), 15 step- (6), 16 step- (7), and 17 step-PLOR (8) reactions for single-round transcriptional termination assay. The first 8 steps were shared among different strategies, and 6.0 mM Mg2+ was added at each step while 1.0 mM Gua+ was added only at the last step. The reagent usages for the 8 reactions are listed in Supplementary file 1, tables S9–16. (C) The denaturing PAGE image of EC-69 to EC-105. The slow migration from EC-69 to EC-105 matches the growing lengths of RNAs. (D) The increase of transcriptional read-through upon the addition of 1 mM Gua+ with the last pause site at 69, 77, 78, 83, 88, 91, 94, and 105, respectively before completing transcription.
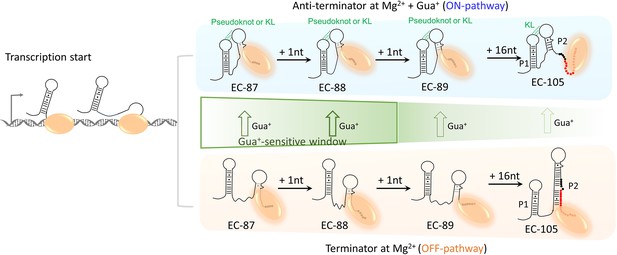
Folding model of riboG in the absence and presence of guanidine.
The folding pathways of anti-terminator conformation in the presence of Gua+ and terminator conformation in the absence of Gua+ are highlighted in blue and orange, respectively. The H-bonds in the KL or pseudoknot are shown as dotted green lines. The nucleotides from EC-87 to EC-88 are shown as black dots, the nucleotides in termination sequence are shown as red dots in EC-89 and EC-105, and DNA templates from EC-87 to EC-105 are not displayed in the model. The T7 RNAP is shown as orange ellipse. The DNA templates are shown as gray ribbons. Green arrows represent the direction of structural switching in native riboG upon the addition of the Gua+. The thicker the green arrow, the higher the switching percentages. And the Gua+-sensitive transcription window of riboG from EC-87 to EC-88 is boxed by green lines.
Additional files
-
Supplementary file 1
Information of DNA/RNA sequences and reagent usages for PLOR reactions.
- https://cdn.elifesciences.org/articles/94706/elife-94706-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/94706/elife-94706-mdarchecklist1-v1.docx





