An arms race between 5’ppp-RNA virus and its alternative recognition receptor MDA5 in RIG-I-lost teleost fish
Figures
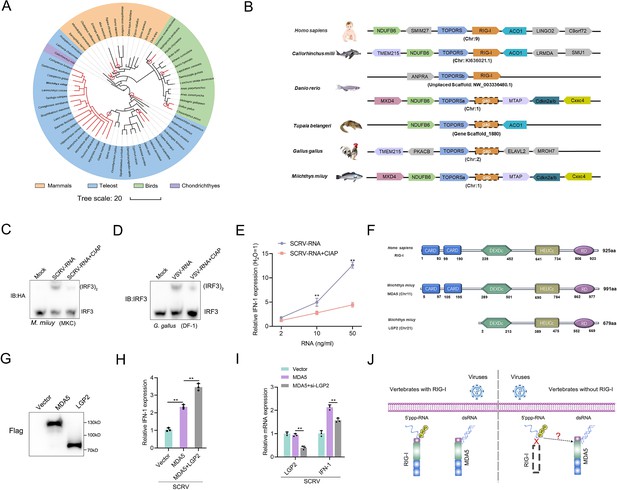
A genetic compensation response model for RIG loss.
(A) Loss of RIG-I in vertebrates. Each branch tip represents one species. Red lines show lineages where loss of RIG-I. The red circle indicates the occurrence of an independent loss event. Branch lengths scale in millions of years (MYA). (B) Comparative analysis of gene synteny of RIG-I in vertebrate genomes. (C and D) SCRV-RNA and VSV-RNA were extracted from SCRV and VSV virus and dephosphorylated them with Calf Intestinal Alkaline Phosphatase (CIAP). Then, MKC cells were transfected with SCRV-RNA and SCRV-RNA +CIAP, and DF-1 cells were transfected with VSV-RNA and VSV-RNA +CIAP. IRF3 dimerization was analyzed by native gel electrophoresis. (E) MKC cells were transfected with different concentrations of SCRV-RNA and SCRV-RNA +CIAP, then the expression of IFN-1 was detected by qRT-PCR. As a control, the expression level of IFN-1 in MKC cells transfected with the same volume of water was set to ‘1’. (F) Predicted protein structures of MDA5 and LGP2 of M. miiuy and RIG-I of H. sapiens. (G) The expression of MDA5 and LGP2 plasmids of M. miiuy were detected by western Blot. (H) MKC cells were co-transfected with MDA5 plasmids and LGP2 plasmids for 24 hr and then treated with SCRV (MOI = 5) for 24 hr. The expression of IFN-1 was determined by qPCR. (I) MKC cells were co-transfected with MDA5 plasmids and si-LGP2 for 24 hr and then treated with SCRV (MOI = 5) for 24 hr. The expression of LGP2 and IFN-1 was determined by qPCR. (J) A genetic compensation response model for RIG loss. All data presented as the means ± SE from three independent triplicated experiments. **, p<0.01, as determined by Student’s t test.
-
Figure 1—source data 1
Excel files of data for Figure 1.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig1-data1-v1.zip
-
Figure 1—source data 2
Raw unedited gels for Figure 1.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig1-data2-v1.zip
-
Figure 1—source data 3
Uncropped and labeled gels for Figure 1.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig1-data3-v1.zip
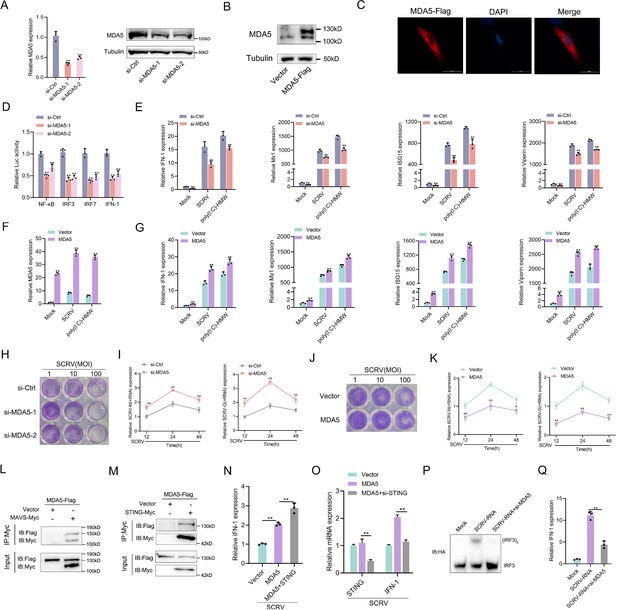
MDA5 promotes host antiviral innate immunity.
(A) Silence efficiency of si-MDA5 measured by qRT-PCR and Western blotting. Two siRNAs of MDA5 were transfected into MPC cells for 48 hr, respectively. (B) MKC cells were transfected with MDA5 plasmids for 48 hr, then the overexpression efficiency was detected through MDA5 endogenous antibodies. (C) Subcellular localization of MDA5 in MKCs by immunofluorescence. (D) MDA5 knockdown suppresses NF-κB, IRF3, IRF7, and IFN-1 signaling. MPC cells were transfected with pRL-TK Renilla luciferase plasmid, luciferase reporter genes, together with MDA5 expression plasmid. At 48 hr post-transaction, the luciferase activity was measured and normalized to renilla luciferase activity. (E) Knockdown of MDA5 attenuates the expression of antiviral genes. MPC cells were transfected with si-Ctrl or si-MDA5 for 24 hr and then treated with SCRV (MOI = 5) or poly(I:C)-HMW for 24 hr. The expression of IFN-1, Mx1, ISG15, and Viperin were determined by qPCR. (F and G) Overexpression of MDA5 promotes the expression of antiviral genes. MKC cells were transfected with vector or MDA5 expression plasmids for 24 hr and then treated with SCRV (MOI = 5) or poly (I:C)-HMW for 24 hr. The expression of MDA5 (F), and antiviral genes, including IFN-1, Mx1, ISG15, and Viperin (G) were determined by qPCR. (H and J) MPC cells transfected with si-Ctrl or si-MDA5 and MKC cells transfected with pcDNA3.1 vector or MDA5 plasmid 24 hr and then treated with SCRV at the dose indicated for 48 hr. Then, cell monolayers were fixed with 4% paraformaldehyde and stained with 1% crystal violet. (I and K) MDA5 inhibits SCRV replication. MPC cells were transfected with si-Ctrl or si-MDA5 and MKC cells were transfected with pcDNA3.1 vector or MDA5 expression plasmid for 24 hr, then infected with SCRV (MOI = 5) for 12, 24, or 48 hr. The qPCR analysis was conducted for SCRV-M and SCRV-G RNA levels. (L and M) MDA5 immunoprecipitates with MAVS (L) and STING (M). EPC cells (~1 × 107) were co-transfected with MAVS-Myc or STING-Myc and MDA5-Flag expression plasmids for 24 hr, followed by immunoprecipitation (IP) with anti-Myc. (N and O) MKC cells were transfected with MDA5 plasmids and STING plasmids (N) or si-STING (O) for 24 hr and then treated with SCRV (MOI = 5) for 24 hr. The expression of IFN-1 was determined by qPCR. (P and Q) MKC cells were transfected with si-MDA5 for 24 hr and then transfected with SCRV-RNA for 24 hr, then the IRF3 dimerization and IFN-1 levels were analyzed by native gel electrophoresis (P) and qPCR (Q), respectively. All data presented as the means ± SE from three independent triplicated experiments. **, p<0.01; *, p<0.05, as determined by Student’s t test.
-
Figure 2—source data 1
Excel files of data for Figure 2.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig2-data1-v1.zip
-
Figure 2—source data 2
Raw unedited gels for Figure 2.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig2-data2-v1.zip
-
Figure 2—source data 3
Uncropped and labeled gels for Figure 2.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig2-data3-v1.zip
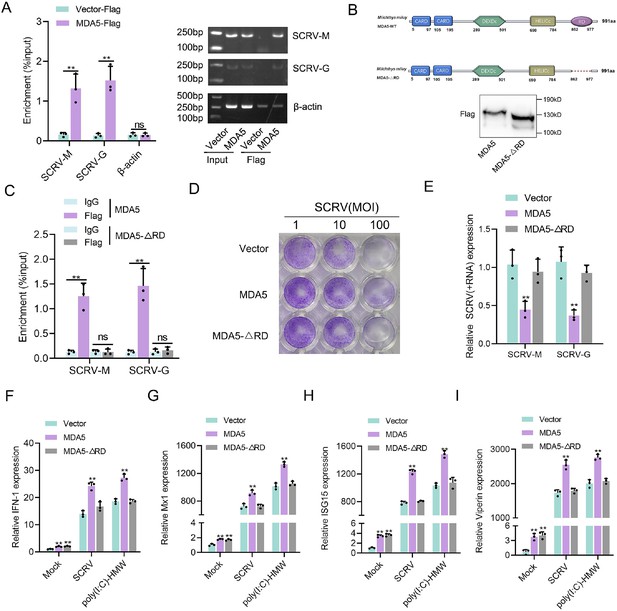
RD domain is required for MDA5 to recognize SCRV RNA.
(A) The association of MDA5 proteins with SCRV. (B) Schematics and expression of MDA5 and truncated MDA5 (MDA5-△RD). (C) The association of MDA5 and MDA5-△RD proteins with SCRV. (D) MKC cells transfected with pcDNA3.1 vector, MDA5, or MDA5-△RD plasmids for 24 hr and then treated with SCRV at the dose indicated for 48 hr. Then, cell monolayers were fixed with 4% paraformaldehyde and stained with 1% crystal violet. (E) MKC cells were transfected with pcDNA3.1 vector, MDA5 or MDA5-△RD expression plasmids for 24 hr, then infected with SCRV for 24 hr, then the qPCR analysis was conducted for SCRV-M and SCRV-G RNA levels. (F–I) Expression of IFN-1 (F), Mx1 (G), ISG15 (H), and Viperin (I) in Mock-infected, SCRV-infected, and poly(I:C)-HMW-stimulated MKC cells transfected with pcDNA3.1 vector, MDA5, or MDA5-△RD expression plasmids. All data presented as the means ± SE from three independent triplicated experiments. **, p<0.01, as determined by Student’s t test.
-
Figure 3—source data 1
Excel files of data for Figure 3.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig3-data1-v1.zip
-
Figure 3—source data 2
Raw unedited gels for Figure 3.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig3-data2-v1.zip
-
Figure 3—source data 3
Uncropped and labeled gels for Figure 3.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig3-data3-v1.zip
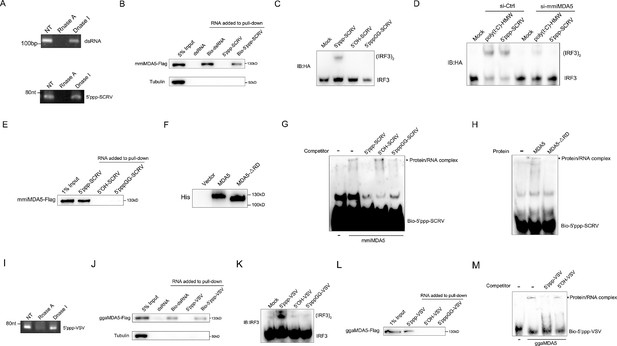
MDA5 recognizes 5’ppp-RNA in M. miiuy and G. gallus.
(A) Schematic representation of 5’ppp-SCRV and dsRNA. The product of in vitro transcription runs as a single product degraded by RNase I. (B) Pull-down of biotinylated or non-biotinylated dsRNA and 5’ppp-SCRV by mmiMDA5. MKC cells were transfected with mmiMDA5-Flag plasmid, and the input and immunoprecipitated MDA5 proteins were analyzed by Western blot. (C) MKC cells were transfected with 5’ppp-SCRV, 5’OH-SCRV (5’ppp-SCRV dephosphorylated by CIAP), or 5’pppGG-SCRV (5’ppp-SCRV capped by m7G cap analog), then IRF3 dimerization was analyzed by native gel electrophoresis. (D) WT and si-MDA5 MKC cells were transfected with 5’ppp-SCRV and poly(I:C)-HMW, then IRF3 dimerization was analyzed by native gel electrophoresis. (E) The cytoplasmic fraction of MKC cells transfected with Flag-tagged mmiMDA5 was incubated with biotinylated 5’ppp-SCRV, 5’OH-SCRV, or 5’pppGG-SCRV. RNA-protein complexes were pulled down using streptavidin affinity beads. Input and pull-down samples were analyzed by SDS-PAGE and immunoblotting using anti-Flag antibody. (F) Purity of recombinant mmiMDA5 and mmiMDA5-△RD were determined by SDS-PAGE and immunoblotting using anti-His antibody. (G) EMSA of 5’ppp-SCRV with mmiMDA5. For binding competition, indicated non-biotinylated RNAs (50-fold molar excess over the probe) were included. The asterisk marks the binding bands between mmiMDA5 protein and different RNA probes. (H) EMSA of 5’ppp-SCRV with mmiMDA5 or mmiMDA5-△RD. The asterisk marks the binding bands between mmiMDA5 or mmiMDA5-△RD proteins and 5’ppp-SCRV RNA probes. (I) Schematic representation of 5’ppp-VSV. The product of in vitro transcription runs as a single product degraded by RNase I. (J) Pull-down of dsRNA and 5’ppp-VSV by ggaMDA5. DF-1 cells were transfected with ggaMDA5-Flag plasmids, and the input and immunoprecipitated MDA5 proteins were analyzed by Western blot. (K) DF-1 cells were transfected with 5’ppp-VSV, 5’OH-VSV (5’ppp-VSV dephosphorylated by CIAP), or 5’pppGG-VSV (5’ppp-VSV capped by m7G cap analog), then IRF3 dimerization was analyzed by native gel electrophoresis. (L) The cytoplasmic fraction of DF-1 cells transfected with Flag-tagged ggaMDA5 was incubated with biotinylated 5’ppp-VSA, 5’OH-VSV, or 5’pppGG-RNA. RNA-protein complexes were pulled down using streptavidin affinity beads. Input and pull-down samples were analyzed by SDS-PAGE and immunoblotting using anti-Flag antibody. (M) EMSA of 5’ppp-RNA with ggaMDA5. For binding competition, indicated non-biotinylated RNAs (50-fold molar excess over the probe) were included. The asterisk marks the binding bands between ggaMDA5 protein and different RNA probes.
-
Figure 4—source data 1
Raw unedited gels for Figure 3.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig4-data1-v1.zip
-
Figure 4—source data 2
Uncropped and labeled gels for Figure 3.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig4-data2-v1.zip
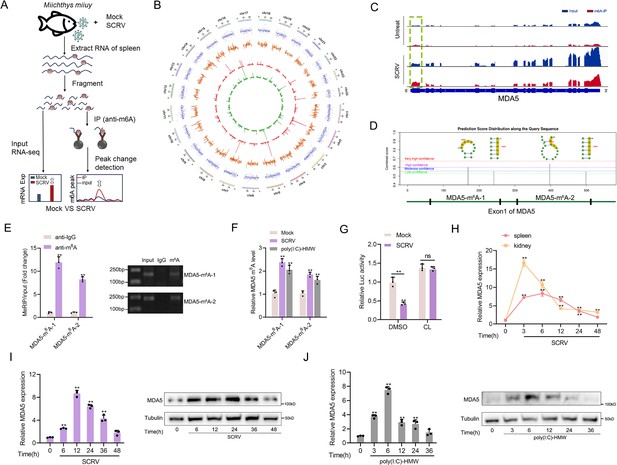
Increased m6A modification and expression of MDA5 upon SCRV infection.
(A) Schematic of the MeRIP-seq protocol used to identify differential m6A methylation following infection of M. miiuy spleen tissues with SCRV. (B) Circos plots showing differentially m6A-methylated peaks identified from normal and SCRV-infected spleen tissues of M. miiuy. The green and red lines on the innermost ring represented the m6A peaks identified from normal and SCRV-infected spleen tissues of M. miiuy, respectively. The orange lines represent the log2 fold change values for each differentially m6A peak. The blue spots represent the fold change of gene expression. Chromosomes were shown on the outermost ring, the ruler on which represented the physical distance is in millions of bases (Mb). (C) The m6A abundance in MDA5 mRNA transcripts detected by MeRIP-seq, the m6A peak of MDA5 is circled in the green box. (D) The exon1 sequence of MDA5 was submitted to the SRAMP website, and then the predicted m6A site was displayed. Four predicted methylation sites located on the exon1 of MDA5 were marked by red box, then we designed two m6A specific primers (MDA5-m6A-1 and MDA5-m6A-2). (E) m6A abundance on MDA5 exon1 detected by MeRIP-qPCR and emiquantitative PCR in MKC cells. (F) MeRIP-qPCR analysis of relative m6A level of MDA5 in Mock, SCRV-infected, and poly(I:C)-HMW-stimulated MKC cells. (G) Relative luciferase activity of pmirGLO-MDA5-exon1 firefly luciferase reporters in Mock and SCRV-infected (MOI = 5) MKC cells that treated with DMSO or Cycloleucine (CL, 20 mM). (H) mRNA levels of MDA5 in spleen and kidney tissues measured by qRT-PCR at indicated time after SCRV infection (MOI = 5). (I and J) mRNA and protein levels of MDA5 in MKC cells measured by qRT-PCR and Western blotting at indicated time after SCRV infection (MOI = 5) (I) and poly(I:C)-HMW stimulation (J). All data presented as the means ± SE from three independent triplicated experiments. **, p<0.01, as determined by Student’s t test (E-G) and one-way ANOVA with Dunnett’s multiple comparisons test (H-J).
-
Figure 5—source data 1
Excel files of data for Figure 5.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig5-data1-v1.zip
-
Figure 5—source data 2
Raw unedited gels for Figure 5.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig5-data2-v1.zip
-
Figure 5—source data 3
Uncropped and labeled gels for Figure 5.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig5-data3-v1.zip
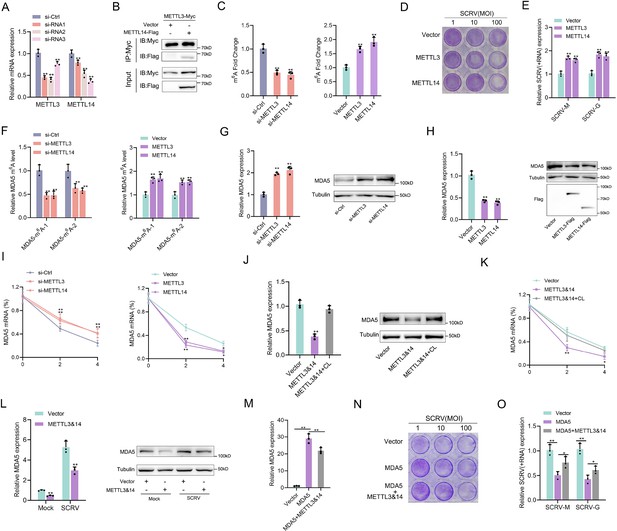
m6A-modification weakens MDA5 mRNA stability and antiviral ability.
(A) Silence efficiency of si-METTL3 and si-METTL14 measured. Three siRNAs of METTL3 and METTL14 were transfected into MPC cells for 48 hr, respectively. (B) METTL3 immunoprecipitates with METTL14. EPC cells (1×107) were co-transfected with METTL3-Myc and METTL14-Flag expression plasmids for 24 hr, followed by immunoprecipitation (IP) with anti-Myc. (C) METTL3 and METTL14 significantly increased the m6A content. MPCs were transfected with si-Ctrl or si-METTL3 or si-METTL14 and MKCs were transfected with vector or METTL3 or METTL14 plasmids for 48 hr, then the m6A level was measured by colorimetry. (D) METTL3 and METTL14 overexpressed MKC cells seeded in 48-well plates overnight were treated with SCRV at the dose indicated for 48 hr. Then, cell monolayers were fixed with 4% paraformaldehyde and stained with 1% crystal violet. (E) MKC cells were transfected with pcDNA3.1 vector and METTL3 or METTL14 expression plasmid for 24 hr, then infected with SCRV (MOI = 5) for 24 hr. The qPCR analysis was conducted for SCRV-M and SCRV-G RNA levels. (F) The m6A level alteration of MDA5 upon METTL3 or METTL14 knockdown or overexpression was examined by MeRIP-qPCR. MPC cells were transfected with si-Ctrl or si-METT3 and MKC cells were transfected with vector or METTL3 or METTL14 plasmids for 48 hr. (G and H) MPC cells were transfected with si-Ctrl, si-METTL3 or METTL14 and MKC cells were transfected with vector, METTL3, or METTL14 plasmids for 48 hr, the expression of MDA5 was detected by qRT-PCR and western blotting. (I) MPC cells were transfected with si-Ctrl, si-METTL3, or si-METTL14, and MKC cells were transfected with vector, METTL3, or METTL14 plasmids, then 5 µg/ml actinomycin D was added to the cells for 0 hr, 2 hr, and 4 hr. The half-life of MDA5 was analyzed by qRT-PCR. (J) MKC cells were transfected with vector or METTL3&14 plasmids for 24 hr and then treated with Cycloleucine (CL) for 24 hr in a final concentration of 20 mM. The expression of MDA5 was detected by qRT-PCR and western blotting. (K) MKC cells were transfected with vector or METTL3&14 plasmids for 24 hr and then treated with CL for 24 hr at 20 mM, then 5 µg/ml actinomycin D was added to the cells for 0 hr, 2 hr, and 4 hr. The half-life of MDA5 was analyzed by qRT-PCR. (L) MKC cells were transfected with vector or METTL3&14 plasmids for 24 hr and stimulated with SCRV (MOI = 5) for 24 hr, then the expression of MDA5 was detected by qRT-PCR and western blotting. (M) MKC cells seeded in 48-well plates overnight were transfected with MDA5 or MDA5 + METTL3&14 plasmids for 48 hr, then the expression of MDA5 was detected by qRT-PCR. (N) MKC cells seeded in 48-well plates overnight were transfected with MDA5 or MDA5 + METTL3&14 plasmids were treated with SCRV at the dose indicated for 48 hr. Then, cell monolayers were fixed with 4% paraformaldehyde and stained with 1% crystal violet. (O) MKC cells were transfected with MDA5 or MDA5 + METTL3&14 plasmids for 24 hr, then infected with SCRV (MOI = 5) for 24 hr. The qPCR analysis was conducted for SCRV-M and SCRV-G RNA levels. All data presented as the means ± SE from three independent triplicated experiments. **, p<0.01; *, p<0.05, as determined by Student’s t test.
-
Figure 6—source data 1
Excel files of data for Figure 6.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig6-data1-v1.zip
-
Figure 6—source data 2
Raw unedited gels for Figure 6.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig6-data2-v1.zip
-
Figure 6—source data 3
Uncropped and labeled gels for Figure 6.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig6-data3-v1.zip
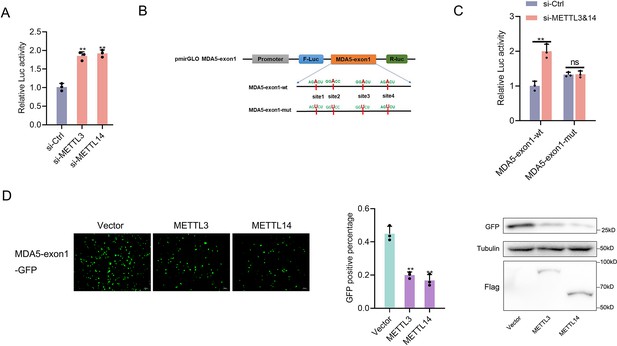
The regulation of m6A writers (METTL3&14) to the MDA5-exon1.
(A) Relative luciferase activity of pmirGLO-MDA5-exon1 firefly luciferase reporters in MPC cells transfected with si-Ctrl, si-METTL3 or si-METTL14. (B) Schematic diagram of m6A site mutation on exon1 of MDA5. ‘A’ in the predicted methylation motif was replaced by ‘U’. (C) Relative luciferase activity of wild-type or mutant pmirGLO-MDA5-exon1 firefly luciferase reporter in MPC cells transfected with si-Ctrl or si-METTL3&14. (D) METTL3 and METTL14 could downregulate GFP expression of mVenus-MDA5-exon1. EPC cells were co-transfected with the mVenus-MDA5-exon1 and vector, METTL3 or METTL14 plasmids. At 48 h post-transfection, the fluorescence intensity and the GFP expression were evaluated by enzyme-labeled instrument and western blotting, respectively. All data presented as the means ± SE from three independent triplicated experiments. **, p<0.01, as determined by Student’s t test.
-
Figure 6—figure supplement 1—source data 1
Excel files of data for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig6-figsupp1-data1-v1.zip
-
Figure 6—figure supplement 1—source data 2
Raw unedited gels for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig6-figsupp1-data2-v1.zip
-
Figure 6—figure supplement 1—source data 3
Uncropped and labeled gels for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig6-figsupp1-data3-v1.zip
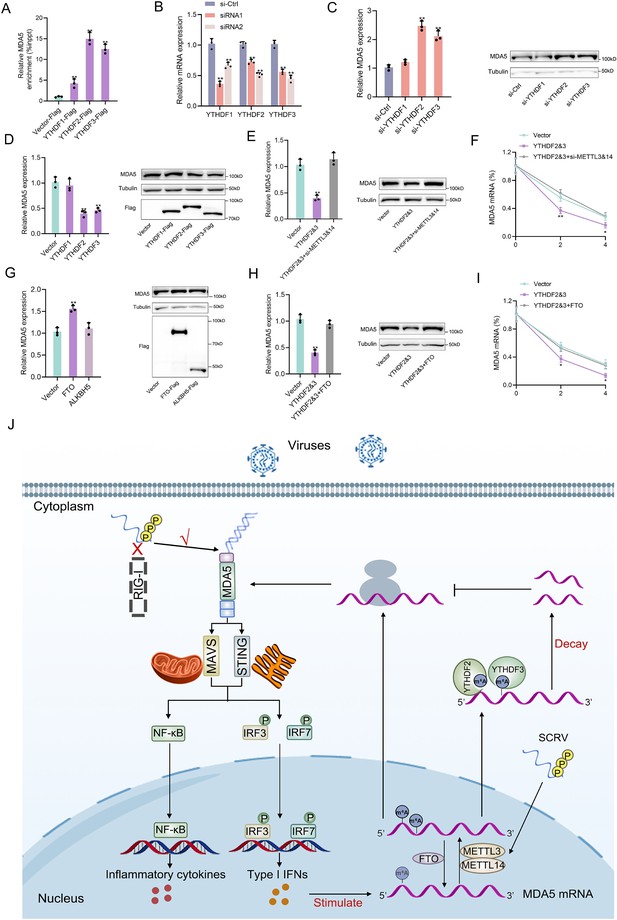
Detailed m6A regulatory mechanism of MDA5.
(A) The binding relationship between YTHDF1, YTHDF2, or YTHDF3 and MDA5 mRNA was validated using a RIP assay. MKC cells were transfected with YTHDF1-Flag, YTHDF2-Flag, YTHDF3-Flag, or pcDNA3.1-Flag for 48 hr. (B) SiRNA silencing effect test of YTHDF1, YTHDF2, and YTHDF3. MPC cells were transfected with si-YTHDF1, si-YTHDF2, or si-YTHDF3 for 48 h. (C and D) MPC cells were transfected with si-YTHDF1, si-YTHDF2, si-YTHDF3 or si-Ctrl (C), and MKC cells were transfected with YTHDF1, YTHDF2, YTHDF3 or vector for 48 hr (D), then the expression of MDA5 was detected by qRT-PCR and western blotting. (E) Relative mRNA and protein levels of MDA5 in MKC cells after co-transfected with YTHDF2&3 plasmids and si-METTL3&14 by qPCR and western blot assays. (F) MKC cells were transfected with YTHDF2&3 plasmids and si-METTL3&14, then 5 µg/ml actinomycin D was added to the cells for 0 hr, 2 hr, and 4 hr. The half-life of MDA5 was analyzed by qRT-PCR. (G) MKC cells were transfected with vector, FTO, ALKBH5 plasmids for 48 hr, then the expression of MDA5 was detected by qRT-PCR and Western blotting. (H) Relative mRNA and protein levels of MDA5 in MKC cells after co-transfected with YTHDF2&3 plasmids and FTO plasmids by qPCR and western blotting. (I) MKC cells were transfected with YTHDF2&3 plasmids and FTO plasmids, then 5 µg/ml actinomycin D was added to the cells for 0 hr, 2 hr, and 4 hr. The half-life of MDA5 was analyzed by qRT-PCR. (J) Schematic diagram of arms race between MDA5 and 5’ppp-RNA virus in M. miiuy. All data presented as the means ± SE from three independent triplicated experiments. **, p<0.01; *, p<0.05, as determined by Student’s t test.
-
Figure 7—source data 1
Excel files of data for Figure 7.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig7-data1-v1.zip
-
Figure 7—source data 2
Raw unedited gels for Figure 7.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig7-data2-v1.zip
-
Figure 7—source data 3
Uncropped and labeled gels for Figure 7.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig7-data3-v1.zip
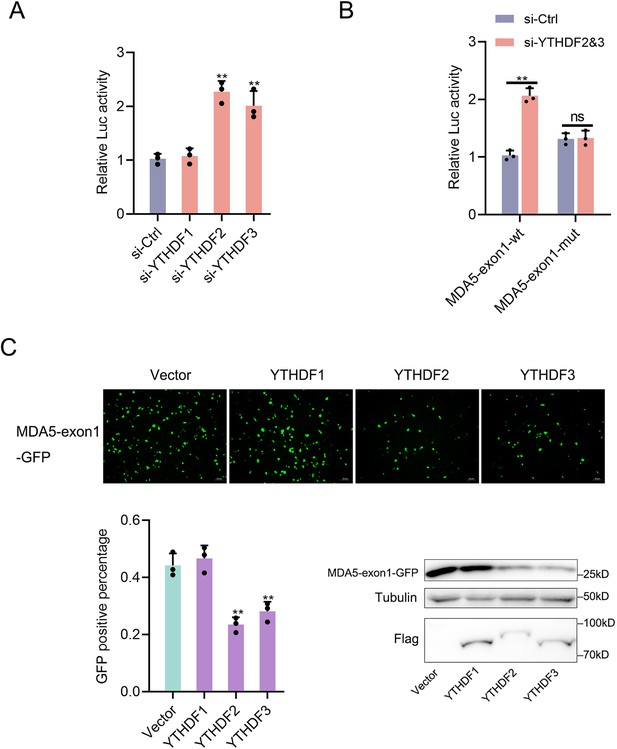
The regulation of YTHDF readers (YTHDF1&2&3) to the MDA5-exon1.
(A) Relative luciferase activity of pmirGLO-MDA5-exon1 firefly luciferase reporters in MPC cells transfected with si-Ctrl, si-YTHDF1, si-YTHDF2 or si-YTHDF3. (B) Relative luciferase activity of wild-type or mutant pmirGLO-MDA5-exon1 plasmids in MPC cells transfected with si-Ctrl or si-YTHDF2&3. (C) YTHDF2 and YTHDF3 could downregulate GFP expression of mVenus-MDA5-exon1. EPC cells were co-transfected with the mVenus-MDA5-exon1 and vector, YTHDF1, YTHDF2, or YTHDF3 plasmids. At 48 hr post-transfection, the fluorescence intensity and the GFP expression were evaluated by enzyme-labeled instrument and western blotting, respectively. All data presented as the means ± SE from three independent triplicated experiments. **, p<0.01, as determined by Student’s t test.
-
Figure 7—figure supplement 1—source data 1
Excel files of data for Figure 7—figure supplement 1.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig7-figsupp1-data1-v1.zip
-
Figure 7—figure supplement 1—source data 2
Raw unedited gels for Figure 7—figure supplement 1.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig7-figsupp1-data2-v1.zip
-
Figure 7—figure supplement 1—source data 3
Uncropped and labeled gels for Figure 7—figure supplement 1.
- https://cdn.elifesciences.org/articles/94898/elife-94898-fig7-figsupp1-data3-v1.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (M. miiuy) | MDA5 | GenBank | PP179381.1 | |
| Gene (M. miiuy) | LGP2 | GenBank | KX351161.1 | |
| Gene (G. gallus) | MDA5 | GenBank | NM_001193638.2 | |
| Strain, strain background (siniperca cheats rhabdovirus) | SCRV virus | This paper | Materials and methods: Fish and challenge | Isolated from mandarin fish |
| Strain, strain background (Vesicular Stomatitis Virus) | VSV virus | BrainCase | VSV31 | |
| Cell line (M. miiuy) | MKC | This paper | Materials and methods: Cell culture | M. miiuy kindey cell line |
| Cell line (M. miiuy) | MPC | This paper | Materials and methods: Cell culture | M. miiuy spleen cell line |
| Cell line (G. gallus) | DF-1 | ATCC | Cat# MZ-2647 RRID:CVCL_0570 | |
| Cell line (Cyprinus carpio) | EPC | ATCC | Cat# CRL-2872 RRID:CVCL_4361 | |
| Cell line (Homo sapiens) | HEK293T | Beyotime | Cat# C6008 RRID:CVCL_0063 | |
| Biological sample (M. miiuy) | Kidney tissue | This paper | Materials and methods: Fish and challenge | Isolated from M. miiuy |
| Biological sample (M. miiuy) | Spleen tissue | This paper | Materials and methods: Fish and challenge | Isolated from M. miiuy |
| Antibody | Anti-MDA5 (Rabbit polyclonal) | Beyotime | Cat# AF7164 | WB (1:500) |
| Antibody | Anti-IRF3 (Rabbit polyclonal) | Boster | Cat# BA4351-2 | WB (1:500) |
| Antibody | Anti-Flag (Mouse polyclonal) | Beyotime | Cat# AF519 | IF (1:300), WB (1:1000) |
| Antibody | Anti-Myc (Mouse polyclonal) | Beyotime | Cat# AF2864 | WB (1:1000) |
| Antibody | Anti-HA (Mouse polyclonal) | Beyotime | Cat# AF2858 | WB (1:1000) |
| Antibody | Anti-Tubulin (Mouse polyclonal) | Beyotime | Cat# AT819 | WB (1:1000) |
| Antibody | Anti-His (Mouse polyclonal) | Beyotime | Cat# AT819 | WB (1:1000) |
| Antibody | HRP-conjugated anti-rabbit IgG | Abbkine | Cat# A25022 | WB (1:5000) |
| Antibody | HRP-conjugated anti-mouse IgG | Abbkine | Cat# A25012 | WB (1:5000) |
| Antibody | Anti-GFP (Mouse monoclonal) | Beyotime | Cat# AG281 | WB (1:1000) |
| Recombinant DNA reagent | PcDNA3.1 | Invitrogen | Cat# V79020 | |
| Recombinant DNA reagent | pmirGLO vector | Promega | Cat# E1330 | |
| Sequence-based reagent | PCR Primers | Synthesized in Genewiz | Listed in Supplementary file 1 | |
| Sequence-based reagent | siRNAs | Synthesized in GenePharma | Listed in section 4.7 | |
| Peptide, recombinant protein | Rnase A | Beyotime | Cat# ST579 | |
| Peptide, recombinant protein | Dnase I | Beyotime | Cat# D7076 | |
| Peptide, recombinant protein | Calf Intestinal Alkaline Phosphatase (CIAP) | Invitrogen | Cat# 18009–019 | |
| Peptide, recombinant protein | Ribo RNAmax-T7 kit | RiboBio | Cat# C11001-2 | |
| Commercial assay or kit | PureBindingRNA-Protein pull-down Kit | Geneseed | Cat# P0202 | |
| Commercial assay or kit | RNA EMSA kit | Beyotime | Cat# GS606 | |
| Commercial assay or kit | Magna RIP RNA-Binding Protein Immunoprecipitation Kit | Millipore | Cat# 17–700 | |
| Commercial assay or kit | m6A RNA Enrichment Kit | Epigentek | Cat# P-9018–24 | |
| Commercial assay or kit | BCA Protein Assay kit | Beyotime | Cat# P0012S | |
| Commercial assay or kit | Endotoxin-Free Plasmid DNA Miniprep Kit | Tiangen | Cat# DP118 | |
| Commercial assay or kit | Dual-Luciferase Reporter Assay System | Promega | Cat# E1980 | |
| Commercial assay or kit | FastQuant RT Kit | Tiangen | Cat# KR106-03 | |
| Commercial assay or kit | SYBR Premix Ex Taq | Takara | Cat# DRR041S | |
| Commercial assay or kit | Lipofectamine RNAiMAX | Invitrogen | Cat# 13778150 | |
| Commercial assay or kit | Lipofectamine 3000 | Invitrogen | Cat# L3000015 | |
| Commercial assay or kit | Ribo RNAmax-T7 Biotin Labeling Transcription Kit | RiboBio | Cat# C11002-2 | |
| Chemical compound, drug | Poly(I:C)-HMW | invivogen | Cat# 31852-29-6 | 10 µg/ml |
| Chemical compound, drug | DAPI | Beyotime | Cat# C1002 | |
| Chemical compound, drug | m7G cap analogue | Yeason | Cat# 10678ES10 | |
| Chemical compound, drug | Cycloleucine | Macklin | Cat# C830431 | 20 mM |
| Chemical compound, drug | Actinomycin D | Solarbio | Cat# A8030-5 | 5 µg/ml |
| Software, algorithm | GraphPad Prism 8 | GraphPad Software | RRID:SCR_002798 | |
| Software, algorithm | TBtools | TBtools | RRID:SCR_023018 | |
| Other | Anti-His beads | Solarbio | Cat# M2300 | Used for protein purification |
Additional files
-
Supplementary file 1
PCR primer information in this study.
- https://cdn.elifesciences.org/articles/94898/elife-94898-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/94898/elife-94898-mdarchecklist1-v1.pdf





