Deciphering the actin structure-dependent preferential cooperative binding of cofilin
Figures
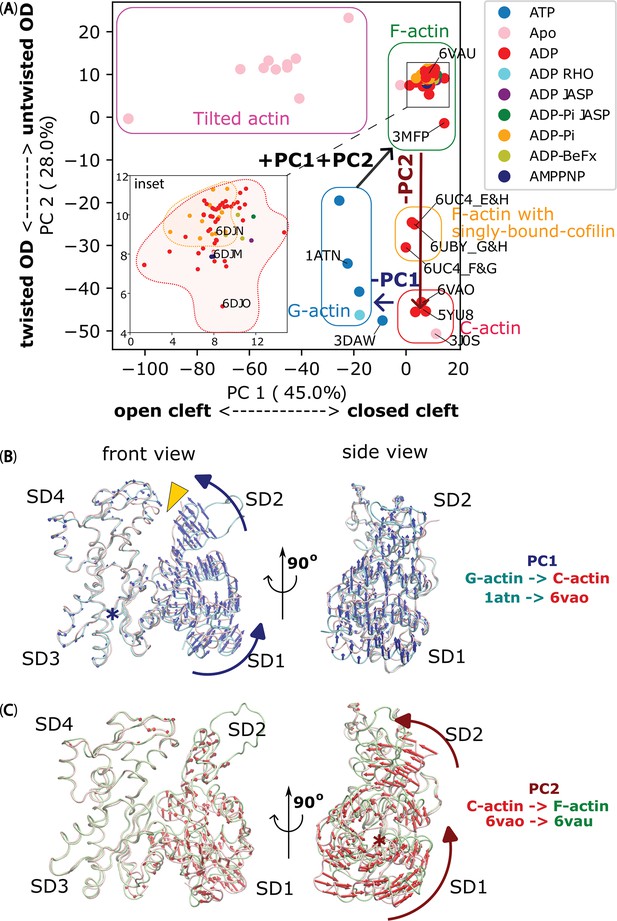
The significant movements of the OD during F-actin to C-actin to G-actin transitions, as revealed by PCA.
(A) PCA of actin chains reported in 46 PDB structures (referred to Table 1) reveals collective transition pathway between different actin conformations: G-actin (soluble monomer – blue box), F-actin (filamentous protomer reported with no cofilin bound – green box), C-actin (filamentous protomer reported within cofilactin after removal of cofilin – red box), tilted actin (reported in 3J8J and 3J8K – magenta box) and single-cofilin-bound actin, where one cofilin molecule bound to an actin protomer within an actin filament (reported in 6UBY – orange box). Protomers at the most barbed end side of a cofilin cluster in 6UC4 also belong to the latter cluster. Notably, a twinfilin-1 bound structure of soluble actin molecule (3DAW) (Paavilainen et al., 2008), previously classified as C-actin (Tanaka et al., 2018), was found between the G-actin and C-actin clusters. PDB entries are coloured with nucleotide molecules present, and structures lacking reported nucleotides are labelled ‘Apo’. The inset shows a magnified region of the F-actin’s PCA cluster (corresponding to the black box in the main graph). (B) The PC1, represented by blue arrows, induces a rotational motion of the OD around hinge region, causing the nucleotide binding cleft between SD2 and SD4 (depicted as a yellow arrowhead) to close during the transition from G-actin (cyan chain) to C-actin (pink chain). (C) The PC2, indicated by red arrows, results in an untwisting motion of the SD2, leading to the flattening of OD during the transformation from C-actin (pink chain) to F-actin (green chain), but without affecting the closing of nucleotide binding cleft (nearly unchanging PC1). The rotational axes in panels B and C were denoted by blue and red asterisks, respectively.
-
Figure 1—source data 1
PC1 and PC2 values obtained from PCA.
These data are presented as origin plots in Figure 1A and Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/95257/elife-95257-fig1-data1-v1.xlsx
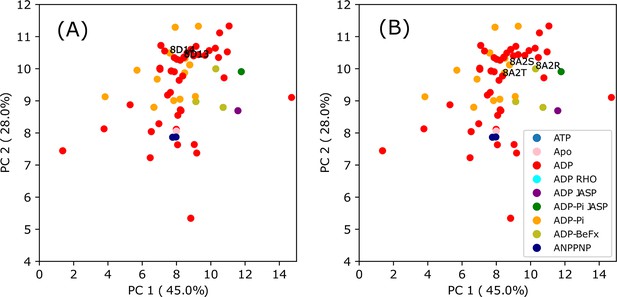
A closer examination of PC1 and PC2 in the PCA clusters of F-actin structures.
Within these PCA clusters of F-actin, similar to the inset of Figure 1A, there is a larger presence of C-actin-like structures in F-ADP-actin compared to F-ADP.Pi-actin. However, the magnified views of the F-actin’s PCA clusters also highlight structural resemblance between F-ADP-actin and F-ADP.Pi-actin, as recently elucidated by Reynolds et al., 2022 (A) and (Oosterheert et al., 2022) (B). Significantly, the F-ADP-actin structure (8D13) and F-ADP.Pi-actin structure (8D14) exhibit remarkably similar PC2 values. Furthermore, the PC2 results demonstrate notable similarities between Mg2+-F-ADP-actin (8A2T) and Mg2+-F-ADP.Pi-actin (8A2S), as well as Mg2+-F-ADP.BeF3-actin (8A2R). These diverse PDB structures may reflect the intrinsic dynamics and polymorphism in actin structure. Regardless of whether a structure is derived from crystallography, Cryo-EM, or NMR data, it is more accurate to refer to it as a structure—one of many possible conformations (Lane, 2023; Miller and Phillips, 2021).
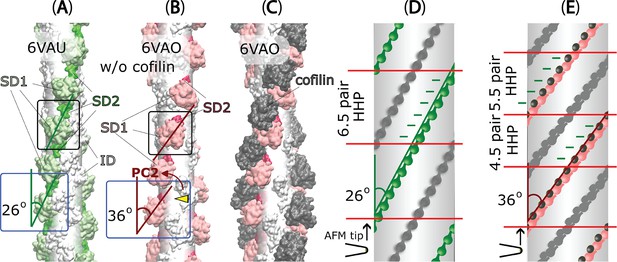
The variations in half helices and the number of protomer pairs per HHP in F-actin, C-actin, and cofilactin structures in relationship with PC2 analysis.
(A–C) Side-views (represented by blue boxes, similar to the right panel of Figure 1C) and outer-views (represented by black boxes) of F-actin, C-actin protomers without and with cofilins along long filaments are depicted. The white translucent cylinders represent the central axes of the filaments. In panel B, a yellow arrowhead highlights a portion of SD2 including D-loop that is buried deeper into the C-actin filament as a result of the reduction of PC2 (represented by the red arrow, corresponding to the red arrow in the right panel of Figure 1C). (D–E) Diagrams of 2D surfaces (unwrapped from imaginary cylinders along the filaments and cut open along imaginary lines scanned by the AFM tip) illustrate the different twisting angles between adjacent protomers in the same strand appears to increase from approximately 26° to around 36° if view from the outer side, resulting in HHPs with varying numbers of detected protomer pairs (marked between red lines). This demonstrates that the global shortened pitch is directly related to the OD twist of the local actin protomers. The green and pink objects represent outer-views of OD domains of the focused chains, along with cofilins shown in black. The opposite chains are depicted in grey.
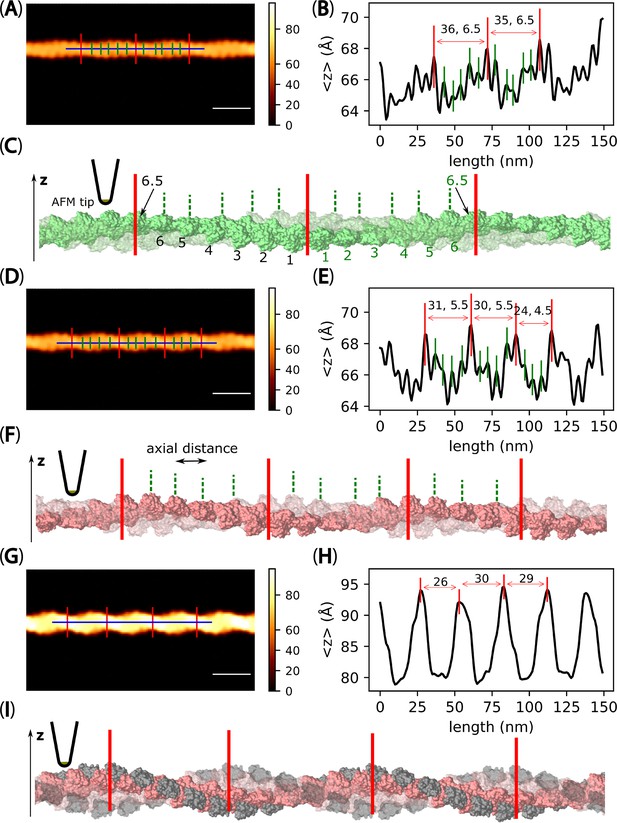
Pseudo-AFM images featuring long filaments representing F-actin and C-actin structures.
It is noted that both filamentous structures share the same length and contain an equal number of actin protomers, specifically 62 protomers. However, the filament representing C-actin exhibits a greater number of half helices than F-actin. This difference can be attributed to the decrease in the number of actin protomers per HHP in C-actin structure. (A) A pseudo-AFM image featuring a long filament representing F-actin structure constructed using the 6VAU. The scale bar represents 25 nm. (B) Longitudinal section profiles of the middle half helices in panel A were used to identify the HHPs (distances between major peaks marked in red, reported in nm) and the numbers of actin protomer pairs per HHP (as counted major and small peaks marked in red and green within each HHP subtracted by 0.5). The offset values of the height profile were set to zero by referring to virtual mica surface, where filament was placed in the pseudo-AFM images. The MADs were computed and shown in brackets. The HHP, the number of protomers per HHP, and MAD were shown accordingly. (C) A side-view of the long actin filament imaged in panel A. To enhance visibility of protomer pairs, one of the chains was depicted as translucent. (D–F) Panels D-F follow a similar format as panels A-C but display the results obtained for a long filament representing C-actin structure (cofilins are removed from initial cofilactin structure reported in 6VAO). The HHP, the number of protomers per HHP, and axial distance (AD) were shown in E accordingly.(G–I) Panels G-I also followed a similar format as panels A-C, but showcased the results obtained for a long cofilactin filament (constructed from 6VAO). Accurately detecting actin pairs in this case was challenging due to cofilin decoration and the tip-sample dilation effect, as explained by Ando, 2022.
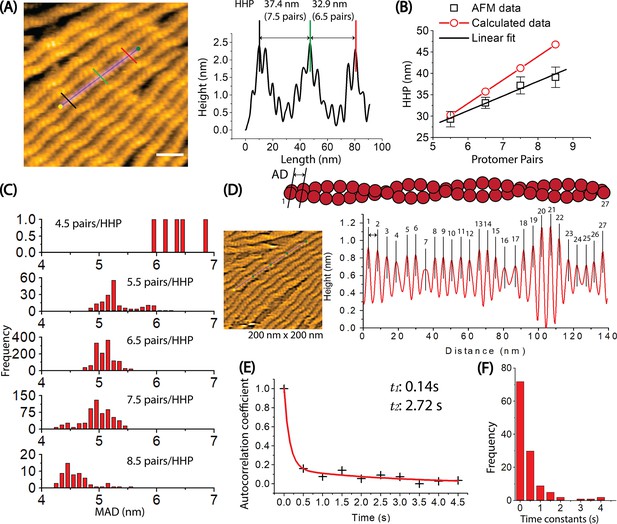
Exploring the dynamics of helical structures of long actin filaments using HS-AFM.
(A) The provided image shows actin filaments with varying numbers of protomer pairs in one half helix. To measure the HHPs and the number of protomer pairs per HHP, a longitudinal section profile of two consecutive half helices was taken as depicted, following the previous method (Hayakawa et al., 2023). Actin filaments were gently immobilized on lipid membrane composed of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-3-trimethylammonium-propane (DPTAP) (90/10 wt%) and imaged in F1 buffer containing 0.5 mM ATP. The offset values were set to zero with reference to the height of the lowest position along the longitudinal section lines. The scale bar represents 25 nm. For the detail, see Methods. (B) The correlation between the counted protomer pairs from image A and the HHP probed by HS-AFM is shown. Black symbols represent the experimental data, with the linear fit (y=10.71 + 3.43 x, R=0.9903). The calculated data (red symbols) also demonstrate a correlation between the number of protomer pairs per HHP and the HHP, which is calculated as 5.5 nm multiplied by the number of counted protomer pairs per HHP. (C) The distribution of MADs is shown for half helices composed of different number of counted protomer pairs. The overall MAD (mean ± SD) is 5.1±0.3 nm (n=15021). (D) The axial distance (AD) between two adjacent actin protomers along the same long-pitch strand in individual bare actin filaments were directly measured as the peak-to-peak distance in four consecutive HHPs. Each protomer pair within the filament was assigned a number ranging from 1 to 27. The ADs were measured in a time series, with the imaging rate of 0.5 s per frame and a total of 258 frames. These AD values for 27 protomer pairs within the actin filament in a time series of 258 frames were utilized to determine the autocorrelation coefficients. The still AFM image was shown after processing with Laplacian of Gaussian filter. (E) An example of the ACF plot displays the autocorrelation coefficients on the y-axis against the time lags on the x-axis. The plotted data was fitted using a second-order exponential decay function with the time constants of (t1, t2), represented by the red curve. (F) A histogram was constructed using the time constants obtained from many similar ACF plots and fittings obtained from different actin filaments (n=61), as showcased in (D, E). The number of data points was 122. These time constants are associated with the overall trend of time decay in autocorrelation of axial distance between two adjacent protomers in the same protofilament.
-
Figure 3—source data 1
Exploring the dynamics of helical structures of long actin filaments using HS-AFM.
These data are presented as origin plots in Figure 3A–F.
- https://cdn.elifesciences.org/articles/95257/elife-95257-fig3-data1-v1.xlsx
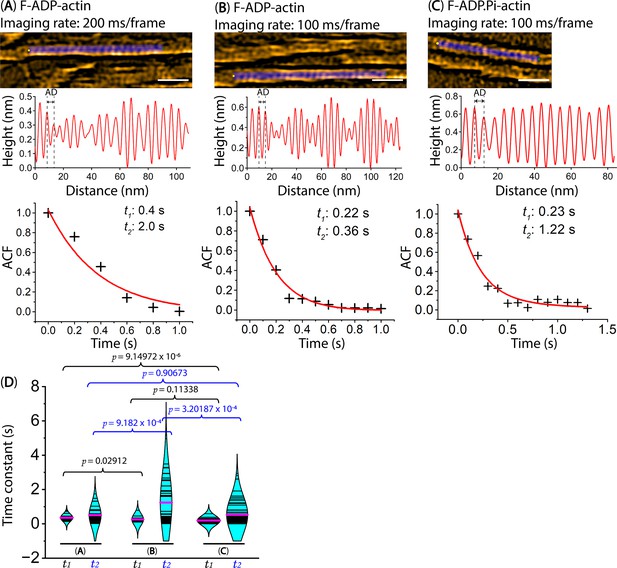
Exploring the dynamics of axial distance (AD) between two adjacent protomers within the same actin filaments.
Actin filaments were gently immobilized on a lipid membrane composed of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-3-trimethylammonium-propane (DPTAP) (90/10 wt%). The still AFM images are shown after processing with Laplacian of Gaussian filter. Scale bars: 25 nm. (A, B) Longitudinal section profiles and distributions of AD between two adjacent protomers within actin segments bound with ADP (F-ADP-actin), captured at high temporal and spatial resolutions using a Quantum AC-10-Super-Sharp high density carbon tip (Nanotools, Germany). F-ADP-actin segments were prepared as described in the Methods section and imaged in F1 buffer containing 1 mM ADP, 5 U/ml hexokinase and 10 mM glucose. (C) Longitudinal section profiles and distributions of AD between two adjacent protomers within actin segments bound with ADP.Pi (F-ADP.Pi-actin), captured at high temporal and spatial resolutions using the same cantilever. F-ADP.Pi-actin segments were prepared as described in the Methods section and imaged in F1 buffer containing 1 mM ATP and 10 mM Pi (K2HPO4/KH2PO4, pH 6.8 buffer). (D) The examples of the ACF plots display the autocorrelation coefficients on the y-axis against the time lags on the x-axis. The plotted data was fitted using a second-order exponential decay function with the time constants of (t1, t2), represented by the red curve. The t1 and t2 values are associated with first and second time decay of AD between two adjacent actin protomers within F-ADP-actin and F-ADP.Pi-actin segments, recorded experimentally at high temporal resolution using HS-AFM. A two-population t-test is employed for statistical analysis to evaluate the significant differences (p-values) between the two populations (mean ± SD). We implemented running frame averages (3 frames) in the ACF analyses. See Table 4.
-
Figure 3—figure supplement 1—source data 1
Exploring the dynamics of axial distance (AD) between two adjacent protomers.
These data are presented as origin plots in Figure 3—figure supplement 1A–D.
- https://cdn.elifesciences.org/articles/95257/elife-95257-fig3-figsupp1-data1-v1.xlsx
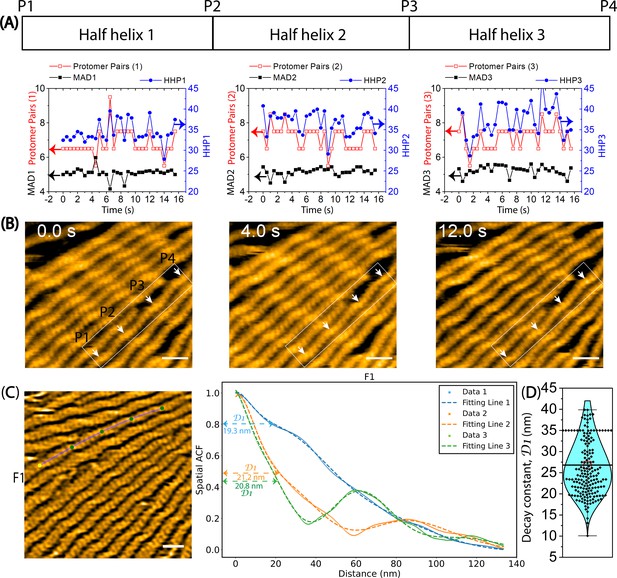
The variation of helical twists measured by HHPs, the number of protomer pairs per HHP, and the MADs in long actin filaments through HS-AFM.
Actin filaments were gently immobilized on lipid membrane composed of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-3-trimethylammonium-propane (DPTAP) (90/10 wt%) and imaged in F1 buffer containing 0.5 mM ATP. (A) The graph demonstrates the changes in HHPs, the number of protomer pairs per HHP, and the MADs over time. The statistical analysis using two-population t-test at a significance level of p≤0.05 revealed that the difference between MAD1 (5.1±0.3 nm, n=32) and MAD2 (5.2±0.2 nm, n=32) is not statistically significant (p=0.2365). However, MAD1 and MAD3 (5.2±0.3 nm, n=32) exhibit a slight difference (p=0.02895). (B) Representative still images obtained from AFM are presented. Arrows indicate the peaks (P1, P2, P3, P4) identified in the raw AFM images. HHP1, HHP2, and HHP3 were measured between P1-P2, P2-P3, and P3-P4, respectively. Scale bars: 25 nm. (C) The spatial ACF curves were produced for a canonical actin filament (F1) at various time points (i.e. Data 1, 2, 3). The curves were fitted using an exponential decaying sinusoidal function (Equation 2), and the resulting decay constant (D1) values from individual curves were presented. The still AFM image is shown after processing with Laplacian of Gaussian filter. Bar: 25 nm. (D) The D1 values were determined through a similar fitting of spatial ACF curves, as illustrated in (C). This examination was conducted at different time points (with an imaging rate of 0.5 s per frame and a total of 50 frames for each canonical filament (F1–F4) using the data, as shown in Figure 4—figure supplement 2A). Subsequently, the distribution of D1 values was analyzed, yielding the mean ± SD value of 26.8±6.8 nm, based on a sample size of 200.
-
Figure 4—source data 1
The variation of helical twists in long actin filaments.
These data are presented as origin plots in Figure 4A, C and D.
- https://cdn.elifesciences.org/articles/95257/elife-95257-fig4-data1-v1.xlsx
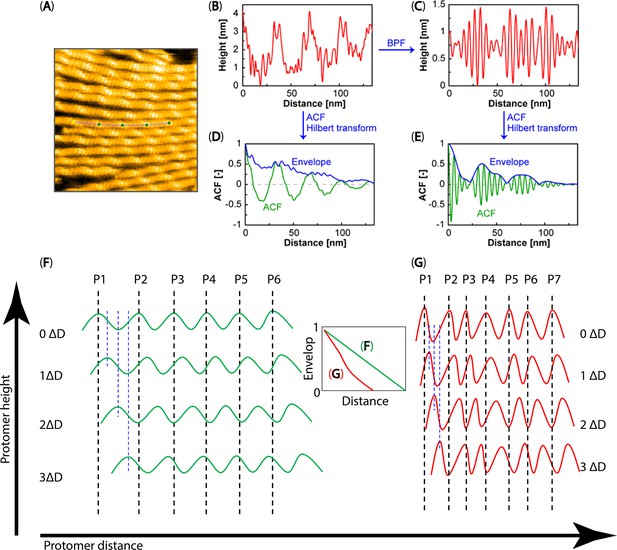
A schematic overview of the spatial ACF analysis.
(A) An HS-AFM image is depicted, with a line profile drawn along an actin filament. (B, C) The height profiles before and after applying the bandpass filter (BPF), respectively. (D, E) The spatial ACF or envelope curves before and after BPF processing and after applying the Hilbert transform are displayed, respectively. (F, G) Illustrating the surface topography of the actin protomers’ heights along filaments, which were used in making the spatial ACF curve (as depicted by the envelope curve). The green waves in (F) represent flatter slopes in the heights between protomers, while the red waves in (G) depict the steeper slopes in the heights between protomers. In theory, the envelope curve (green) versus distance in (F) should linearly decrease. In contrast, the envelope curve (red) versus distance in (G) shows a more pronounced decrease. During the actual analysis, the selected space intervals (ΔD) for measuring the degree to which height values near each other in space are similar to one another in F-actin and hybrid cofilactin/bare actin segments were 0.4 nm and 0.3 nm, respectively. The measurement process involved shifting these space intervals from left to right and up to down, with increasing interval values from the initial position (0 ΔD). P1-P7 denote the peak positions of individual protomers.
-
Figure 4—figure supplement 1—source data 1
A schematic overview of the spatial ACF analysis.
These data are presented as origin plots in Figure 4—figure supplement 1B–E.
- https://cdn.elifesciences.org/articles/95257/elife-95257-fig4-figsupp1-data1-v1.xlsx
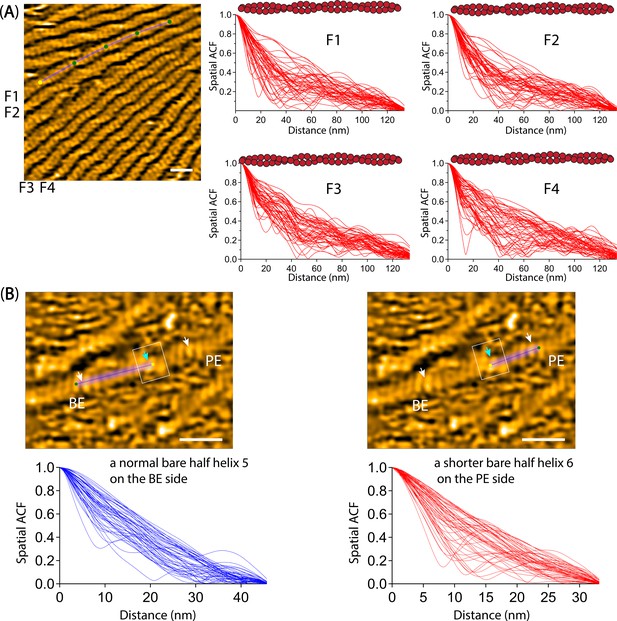
Analysis of spatial ACF curves.
(A) Spatial ACF curves were depicted for canonical actin filaments. This analysis was conducted at different time points (the imaging rate of 0.5 s per frame and a total of 50 frames for each filament (F1–F4)). The still AFM image is shown after processing with Laplacian of Gaussian filter. Scale bar: 25 nm. Related to Figure 4C–D. (B) Spatial ACF curves were depicted for a hybrid cofilactin/bare actin filament, featuring a bare half helix near a cofilin cluster on either the PE or BE side. This analysis was conducted at different time points (the imaging rate of 0.5 s per frame and a total of 50 frames for each half helix on the PE or BE side). Based on our previous study (Ngo et al., 2015), we judged that the bare half helix with the shorter HHP adjacent to the cofilin cluster is on the PE side and that the normal HHP is located on the BE side. The still AFM images are shown after processing with Laplacian of Gaussian filter. Scale bars: 25 nm. Related to Figure 6C.
-
Figure 4—figure supplement 2—source data 1
Analysis of spatial ACF curves.
These data are presented as origin plots in Figure 4—figure supplement 2A–B.
- https://cdn.elifesciences.org/articles/95257/elife-95257-fig4-figsupp2-data1-v1.xlsx
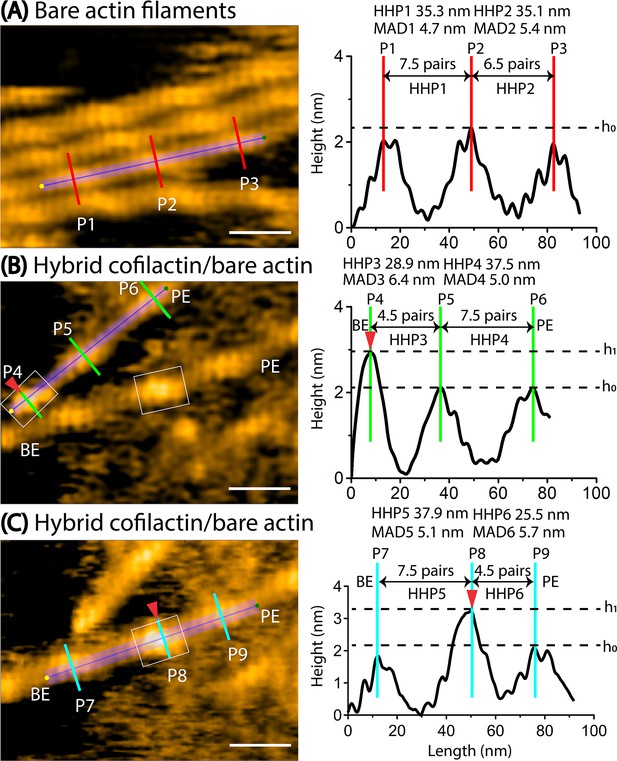
Asymmetric helical twisting on both sides of a small cofilin cluster.
Red arrowheads indicate small area of cofilin clusters (in boxes). Scale bars: 25 nm. Based on our previous study (Ngo et al., 2015), we judged that the bare half helix with the shorter HHP adjacent to the cofilin cluster is on the PE side and that the normal HHP is located on the BE side. (A) A still AFM image shows normal actin filaments, along with a longitudinal section profile representing two consecutive half helices 1 and 2 (denoted HHP1, HHP2). (B) A still AFM image displays a hybrid segment consisting of cofilactin and bare actin areas, with a longitudinal section profile representing two consecutive half helices. A short filament was observed where a small cofilin cluster cut filament at cofilactin/bare actin boundary on the BE side, followed by two consecutive bare half helices 3 and 4 (denoted HHP3, HHP4) on the PE side. The shortened bare half helix 3 and normal bare half helix 4 are presented. The hybrid cofilactin/bare actin filaments, priorly made in solution, were strongly immobilized on lipid membrane composed of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-3-trimethylammonium-propane (DPTAP) (50/50 wt%) and imaged in F1 buffer containing 0.5 mM ATP (see Methods). (C) A still AFM image depicts a hybrid segment comprising cofilactin and bare actin areas, along with a longitudinal section profile representing two consecutive half helices 5 and 6 (denoted HHP5, HHP6). In this case, a short filament contained a small cofilin cluster (estimated as 2–4 molecules) in the middle, configured by a normal bare half helix 5 on the BE side and a shortened bare half helix 6 on the PE side. We estimated the size of a cofilin cluster by measuring its height and width within the white box around P8, distinguishing it with the control bare section lacking cofilin bindings (P7, P9). Despite the invisibility of cofilin molecules below, we made the assumption that cofilin bound uniformly to two strands at a molar ratio of 1:1, enabling us to account for their presence.
-
Figure 5—source data 1
Asymmetric helical twisting on both sides of a small cofilin cluster.
These data are presented as origin plots in Figure 5A–C.
- https://cdn.elifesciences.org/articles/95257/elife-95257-fig5-data1-v1.xlsx
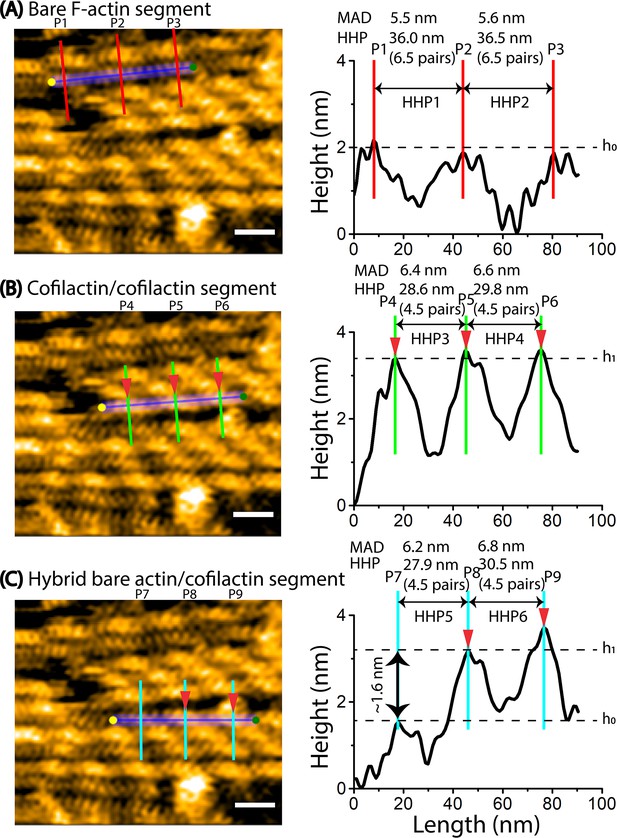
The propagation of the supertwisted half helices induced by cofilin clusters results in the shortening of a bare half helix beyond the cofilactin/bare actin boundary.
The HHPs and peak heights were determined using methods similar to our previous reports (Hayakawa et al., 2023; Ngo et al., 2015). Scale bars: 25 nm. Arrowheads indicated the peaks of cofilin clusters. h0 and h1 represented the peak heights in two consecutive HHPs of bare actin and cofilactin segments. The offset values were set to zero with reference to the height of the lowest position along the longitudinal section lines. The height and width differences were utilized to determine the bare actin and cofilactin segments. The bare actin, cofilactin, and hybrid cofilactin/bare actin filaments, priorly made in solution, were strongly immobilized on lipid membrane composed of DPPC/DPTAP (50/50 wt%) and imaged in F1 buffer containing 0.5 mM ATP. See Methods. (A) A still AFM image presents a normal actin filament, accompanied by a longitudinal section profile across two consecutive half helices to measure the HHP1 and HHP2 and the number of actin protomer pairs per HHP accordingly. The MADs were computed from AFM analysis and shown. (B) A still AFM image depicts a cofilactin filament, accompanied by a longitudinal section profile across two consecutive half helices to measure the HHP3 and HHP4 and the numbers of actin protomer pairs per HHP accordingly. The MADs were calculated from our AFM analysis and shown. (C) A still AFM image showcases a hybrid bare actin/cofilactin filament, accompanied by a longitudinal section profile across two consecutive half helices to measure the shortened bare HHP5 and shortened cofilactin HHP6 and the number of actin protomers per HHP. The MADs were calculated from our AFM analysis and shown. It should be emphasized that only MADs obtained in the bare regions without cofilin decoration were used to make Histograms in Figure 5—figure supplement 2E.
-
Figure 5—figure supplement 1—source data 1
The propagation of the supertwisted half helices induced by cofilin clusters.
These data are presented as origin plots in Figure 5—figure supplement 1A–C.
- https://cdn.elifesciences.org/articles/95257/elife-95257-fig5-figsupp1-data1-v1.xlsx
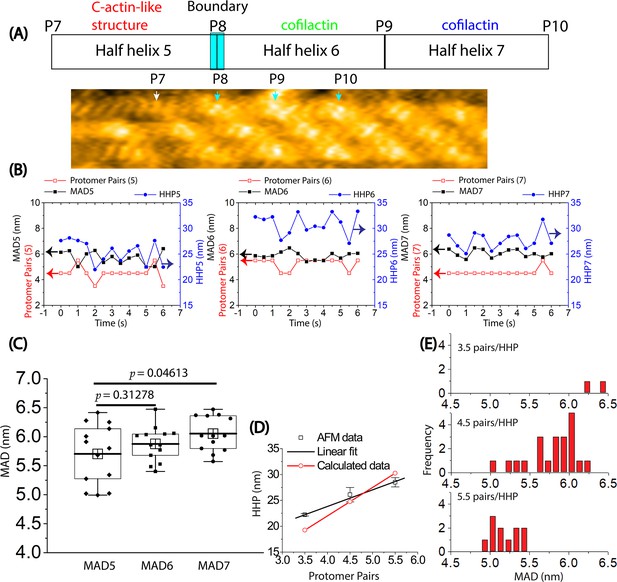
The analysis of the variation in helical twisting by measuring HHPs, the number of protomer pairs per HHP, and MADs in a hybrid bare actin/cofilactin segment using HS-AFM.
(A) Representative still AFM image of a hybrid bare actin/cofilactin segment in Figure 5—figure supplement 1C is provided. The peaks (P7, P8, P9, P10) in the raw AFM image are indicated by white and cyan arrows for the bare actin and cofilactin regions, respectively. HHP5, HHP6, and HHP7 and the number of protomer pairs within each HHP were measured between P7-P8, P8-P9, and P9-P10, respectively and MAD5, MAD6, and MAD7 were computed accordingly. Scale bars: 25 nm. (B) The variation of HHPs, the number of protomer pairs per HHP, and MADs in shortened bare and cofilactin regions were examined over time. (C) A comparison between MAD5 (5.7±0.5 nm) observed in the shortened bare area and MAD6 (5.9±0.3 nm) along with MAD7 (6.1±0.3 nm) within the cofilactin region was presented. A two-population t-test with a sample size of n=13 was used. (D) The relationship between the HHP and the number of protomer pairs per HHP measured for C-actin-like structures within the shortened bare half helix adjacent to cofilin cluster, as explored through HS-AFM, was illustrated using black symbols. The collected data was subjected to fitting with a linear equation (y=10.99 + 3.21 x, R=0.9975) in the region of the shortened bare half helix situated close to the boundary of cofilactin and bare actin. Calculated data, depicted with red symbols, presented the correlation between the number of protomer pairs and the HHP that was calculated as 5.5 nm multiplied by the number of protomer pairs within each HHP. (E) The histograms depict the distribution of MADs for C-actin-like structures within the shortened bare half helices adjacent to cofilin clusters consisting of different number of protomer pairs.
-
Figure 5—figure supplement 2—source data 1
The variation in helical twisting in a hybrid bare actin/cofilactin segment.
These data are presented as origin plots in Figure 5—figure supplement 2B–E.
- https://cdn.elifesciences.org/articles/95257/elife-95257-fig5-figsupp2-data1-v1.xlsx
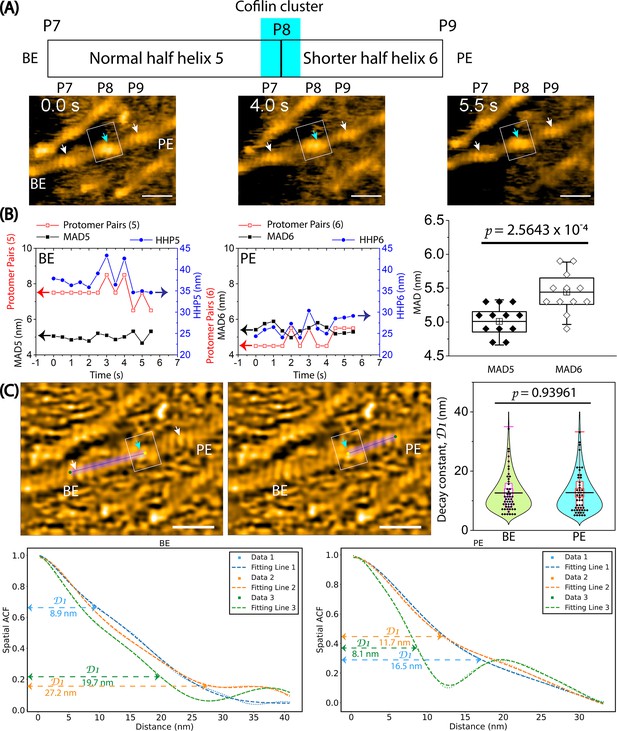
The variation of half helices, the number of protomer pairs per HHP, and the MADs in a hybrid cofilactin/bare actin filament probed by HS-AFM.
Based on our previous study (Ngo et al., 2015), we judged that the bare half helix with the shorter HHP adjacent to the cofilin cluster is on the PE side and that the normal HHP is located on the BE side. (A) Representative still images obtained from AFM in Figure 5C are displayed. Arrows indicate the peaks (P7, P8, P9) identified in the raw AFM images. A normal bare half helix 5 (denoted HHP5) and a shortened bare half helix 6 (denoted HHP6) were measured between P7-P8 and P8-P9, respectively. Cyan and white arrows indicate the peak positions within cofilin clusters and bare actin peaks, respectively. Boxes denote the cofilin clusters. Scale bars are 25 nm. (B) The graph depicts the analysis of half helices 5 and 6, the number of protomer pairs in each half helix (Protomer Pairs (5) and Protomer Pairs (6)), and the MAD5 and MAD6 over time. Two-populations t-test was used to validate the statistical difference between MAD5 (5.0±0.2 nm) and MAD6 (5.4±0.3 nm). The sample size was 12. (C) The spatial ACF curves were generated for a normal half helix 5 and a shorter half helix 6 near a cofilin cluster on either the PE or BE side in a hybrid cofilactin/bare actin filament at various time points (i.e. Data 1, 2, 3). The curves were fitted using an exponential decaying sinusoidal function (Equation 2), and the resulting decay constant (D1) values from individual curves were presented. This examination was conducted at different time points (with an imaging rate of 0.5 s per frame and a total of 50 frames for each half helix on the PE or BE side using the data, as shown in Figure 4—figure supplement 2B). The D1 values were determined through a similar fitting of spatial ACF curves. Subsequently, the distribution of D1 values was analyzed, yielding the mean ± SD values for each half helix on the PE and BE side of 12.7±7.0 nm and 12.6±6.6 nm, respectively. The sample size of each case was 50. The statistical analysis using two-population t-test at a significance level of p≤0.05 revealed that the difference between two mean values was not significant. The still AFM images are shown after processing with Laplacian of Gaussian filter. Scale bars: 25 nm.
-
Figure 6—source data 1
The analysis of a hybrid cofilactin/bare actin filament.
These data are presented as origin plots in Figure 6B–C.
- https://cdn.elifesciences.org/articles/95257/elife-95257-fig6-data1-v1.xlsx
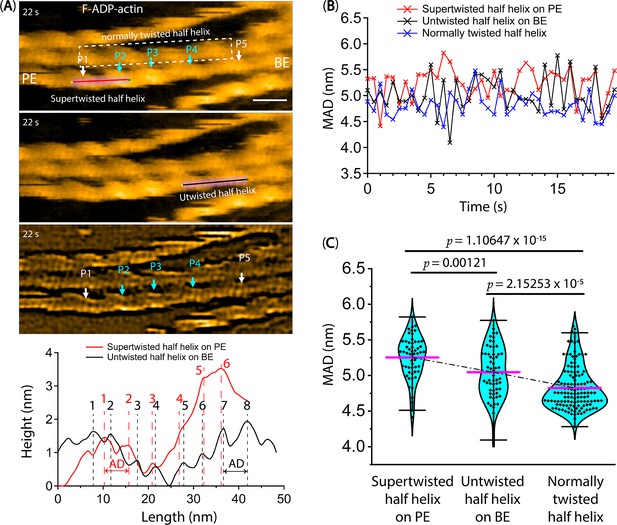
The fluctuation of the mean axial distance (MAD) between two adjacent actin protomers along the same long-pitch strand of F-ADP-actin segments as a function of time.
(A) The longitudinal section profiles and distributions of axial distance (AD) within barely supertwisted and untwisted half helices neighboring cofilin clusters on the PE and BE sides, respectively. White and cyan arrows indicate the peak positions (P1–P5) within bare actin and cofilin clusters, respectively. Notably, P4 was completely decorated with cofilin clusters, resulting in a supertwisted HHP measured between P3 and P4 (Video 5). This supertwisted structure persists even after cofilin clusters are removed during imaging. This structure, containing fewer protomer pairs, closely resembles the bare HHP measured between P1 and P2 on the PE side, as opposed to the normal HHP between P4 and P5 on the BE side. The number of protomer pairs per half helix determined by longitudinal section profiles is presented and calculated by subtracting 0.5 from the total number of peaks (see Methods). F-ADP-actin segments were prepared as described in the Methods section and imaged in F1 buffer containing 1 mM ADP, 5 U/ml hexokinase, 10 mM glucose, and 300 nM cofilin. Actin filaments were gently immobilized on a lipid membrane composed of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-3-trimethylammonium-propane (DPTAP) (90/10 wt%) and captured at high temporal and spatial resolutions using a Quantum AC-10-Super-Sharp high-density carbon tip. There was minimal nonspecific binding of cofilin to this lipid membrane surface. Based on our previous study (Ngo et al., 2015), we judged that the bare half helix with the shorter HHP adjacent to the cofilin cluster is on the PE side and that the normal HHP is located on the BE side. Scale bar: 25 nm. (B, C) The MADs were calculated as described in the Methods to follow the time course of the variation in MADs in barely supertwisted (5.3±0.3, N=55) on the PE side, untwisted (5.0±0.3, N=57) on the BE side, and normally twisted (4.8±0.3, N=129) half helices, for comparison between them. A two-population t-test was employed for statistical analysis to evaluate significant differences (p-values) between the two populations (mean ± SD).
-
Figure 6—figure supplement 1—source data 1
The fluctuation of the mean axial distance (MAD) of F-ADP-actin segments.
These data are presented as origin plots in Figure 6—figure supplement 1A–C.
- https://cdn.elifesciences.org/articles/95257/elife-95257-fig6-figsupp1-data1-v1.xlsx
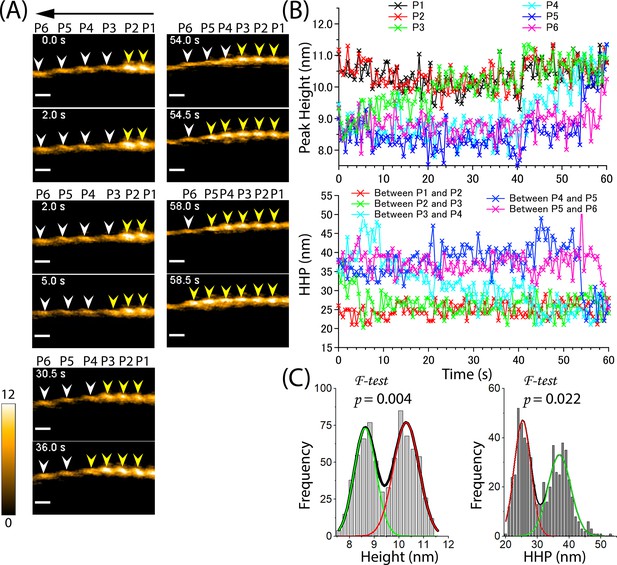
Propagation of supertwisted half helices within the cofilactin region beyond the boundary of cofilactin and bare actin, leading to the pre-shortening of a bare half helix and subsequent recruitment of cofilin cluster growth.
(A) Still AFM images show the time-dependent growth of cofilin clusters beyond the boundary. The arrow indicates the direction of cofilin cluster growth. The peaks of cofilactin and bare actin segments are marked by yellow and white arrowheads, respectively. Peak heights (P1–P6) were determined in five consecutive half helices. Actin filaments were loosely immobilized on a lipid membrane composed of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-3-trimethylammonium-propane (DPTAP) (90/10 wt%) and imaged in an F1 buffer containing 300 nM cofilin, 1 mM ATP and 10 mM Pi. Scale bars: 25 nm. (B) Examination of changes in peak heights and HHPs in the cofilactin and bare actin regions over time. Peak heights and HHPs were measured as described previously (Ngo et al., 2015). (C) Histograms of peak heights (8.6±0.4 nm, 10.3±0.5 nm, n=726) and HHPs (25.3±2.6 nm, 36.9±3.6 nm, n=605) were obtained from the data in (B). The significant difference between the peaks in double Gaussian fitting was statistically examined using an F-test. The green and red curves denote the fractions of F-actin and cofilactin structures, respectively.
-
Figure 7—source data 1
Propagation of supertwisted half helices within the cofilactin region beyond the boundary of cofilactin and bare actin.
These data are presented as origin plots in Figure 7B–C.
- https://cdn.elifesciences.org/articles/95257/elife-95257-fig7-data1-v1.xlsx
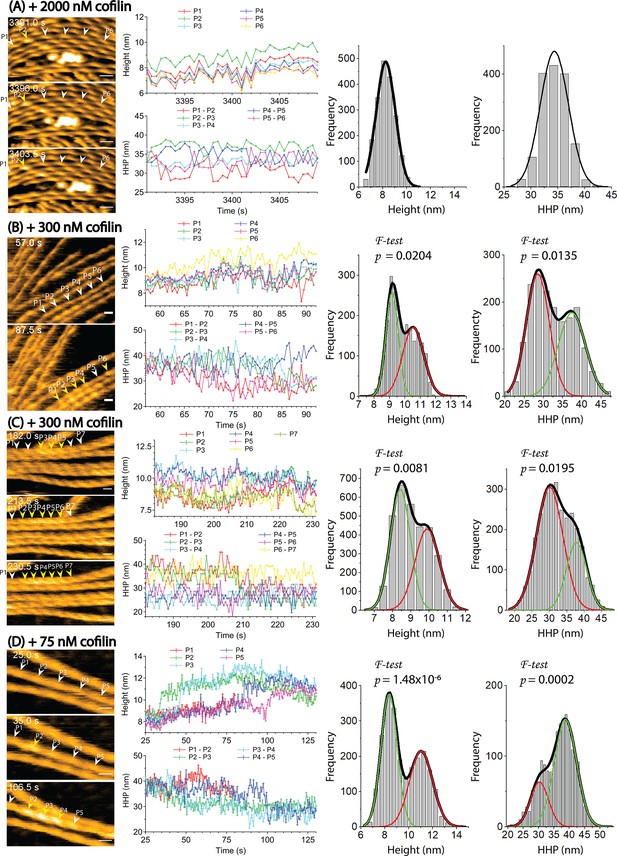
Cooperative binding of cofilin is heavily dependent on the dynamic and cooperative conformational changes involving helical twisting within actin filaments.
(A) The cooperative binding of cofilin to actin filaments was significantly inhibited when the torsional motion during helical twisting within the filaments was suppressed. The filaments were strongly attached onto a highly positively charged lipid membrane containing 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-3-trimethylammonium-propane (DPTAP) (75/25 wt%) and imaged in F1 buffer containing 2000 nM cofilin and 0.5 mM ATP. (B, C, D) The normal cooperative bindings of cofilin to actin filaments were observed when the filaments were loosely immobilized on weakly positively charged lipid membrane comprising DPPC/DPTAP (90/10 wt%) and imaged in F1 buffer under different conditions: 300 nM cofilin +0.5 mM ATP (B), 300 nM cofilin +1 mM ADP (C), and 75 nM cofilin +1 mM ATP +10 mM Pi (D). The changes in peak heights and HHPs of a representative actin filament under each condition were analyzed over time. Histograms were constructed by analyzing the changes in peak heights and HHPs of all actin filaments in the observed field during the growth of the cofilin cluster at various time intervals after adding cofilin. The statistical significance of the peaks in the histograms was examined using an F-test. The green and red curves denote the fractions of F-actin and cofilactin structures, respectively. Time labels in the still AFM images indicate the elapsed time after adding cofilin to the imaging buffer. Peaks of bare actin and cofilactin are denoted by white and yellow arrowheads, respectively. Scale bars: 25 nm.
-
Figure 8—source data 1
Cooperative binding of cofilin involving helical twisting within actin filaments.
These data are presented as origin plots in Figure 8A–D.
- https://cdn.elifesciences.org/articles/95257/elife-95257-fig8-data1-v1.xlsx
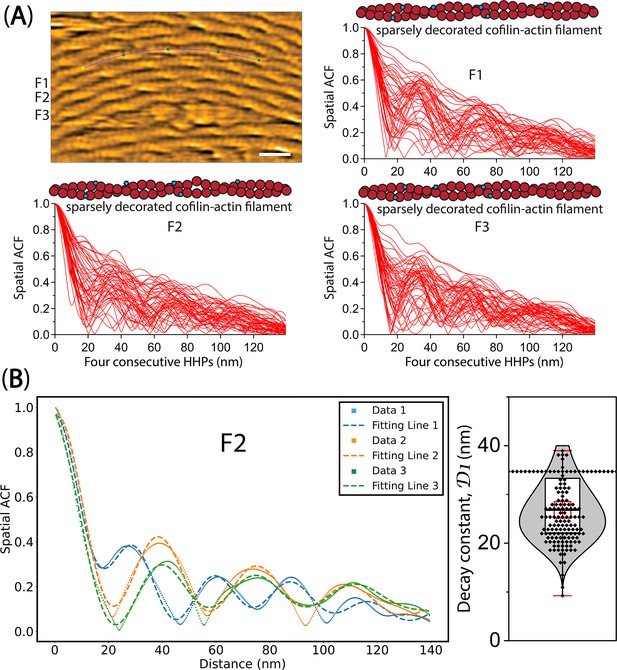
The spatial ACF curve analysis.
(A) The spatial ACF curves were produced for sparsely decorated cofilin-actin filaments (F1, F2, F3), strongly attached on lipid surface, at various time points (i.e., Data 1, 2, 3). The curves were fitted using an exponential decaying sinusoidal function (Equation 2). The still AFM image in Figure 8A is shown after processing with Laplacian of Gaussian filter. Bar: 25 nm. (B) The decay constant (D1) values were determined through a similar fitting of spatial ACF curves, as illustrated in Figure 4C. This examination was conducted at different time points (with an imaging rate of 0.5 seconds per frame and a total of 50 frames). Subsequently, the distribution of D1 values was analyzed, yielding the mean ± SD value of 26.8±6.3 nm, based on a sample size of 147.
-
Figure 8—figure supplement 1—source data 1
The spatial ACF curve analysis.
These data are presented as origin plots in Figure 8—figure supplement 1A–B.
- https://cdn.elifesciences.org/articles/95257/elife-95257-fig8-figsupp1-data1-v1.xlsx
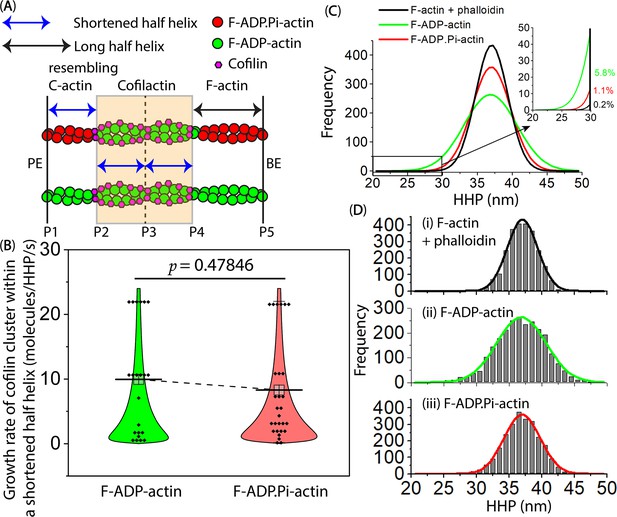
Comparison of the growth rate of cofilin clusters within the shortened bare half helices that resemble the C-actin structure adjacent to cofilin clusters on the PE side, for both F-ADP-actin and F-ADP.
Pi-actin.(A) In this cartoon representation, hybrid cofilactin/bare actin filaments are depicted, consisting of four half helices. Two of these half helices are fully decorated with cofilin clusters in the middle, followed by a shortened bare half helix and a normal long bare half helix on the PE and BE sides, respectively. P1-P5 were labels indicating peak heights at individual crossover points. To make a meaningful comparison, the lag time measurements were initiated exclusively when a bare half helix between P1 and P2 experienced shortening, and they continued until a newly matured cluster emerged in P1 within the shortened bare half helix (P1–P2) adjacent to the preformed cofilin cluster (P2). Each fully decorated supertwisted half helix contains 11 cofilin molecules (referred to Figure 2—figure supplement 1I). To determine the mean growth rate of the cofilin cluster within a shortened bare half helix adjacent to the cofilin cluster, 11, the number of cofilin molecules in one half helix, was divided by the lag time in seconds. (B) This part presents a comparison of the growth rate of cofilin clusters within shortened bare half helices that resemble the C-actin structure adjacent to cofilin clusters on the PE side. This comparison was made when actin filaments were incubated with the same cofilin concentration (i.e. 300 nM) in the F1 buffer containing 1 mM ADP +5 U/ml hexokinase +10 mM glucose or 1 mM ATP +10 mM Pi, resulting in filamentous protomers representing F-ADP-actin or F-ADP.Pi-actin structures. The growth rates (mean ± SD) were measured for 22 shortened bare half helices in F-ADP-actin (10.0±8.3 molecules/HHP/s) and 28 shortened bare half helices in F-ADP.Pi-actin (8.3±7.7 molecules/HHP/s). Using the two-population t-test, the results showed that the difference was not significant (p=0.47846) at the p≤0.05 level. (C, D) Histograms illustrate the distribution of the HHP (mean ± SD) in control actin filaments for three conditions: (i) phalloidin-stabilized F-actin (37.0±2.3 nm), (ii) F-ADP-actin (36.9±3.8 nm), and (iii) F-ADP.Pi-actin (37.0±2.8 nm), with each condition having 2487 data points. For comparison, single Gaussian fits in D were integrated and displayed in C. The inset within C exhibited fractions of the naturally supertwisted half helices with HHPs shorter than 30 nm. Utilizing one-way ANOVA with a significance level of p≤0.05, there were significant differences observed between (i) and (ii) (p=0.00837, F=6.95754) as well as between (ii) and (iii) (p=0.04080, F=4.18633), primarily attributable to the statistical difference in the variance of HHP between condition (ii) vs. (i) and (iii). The difference observed between (i) and (iii) was not significant (p=0.58272, F=0.30189).
-
Figure 9—source data 1
The growth rate of cofilin clusters within the shortened bare half helices for both F-ADP-actin and F-ADP. Pi-actin.
These data are presented as origin plots in Figure 9B–D.
- https://cdn.elifesciences.org/articles/95257/elife-95257-fig9-data1-v1.xlsx
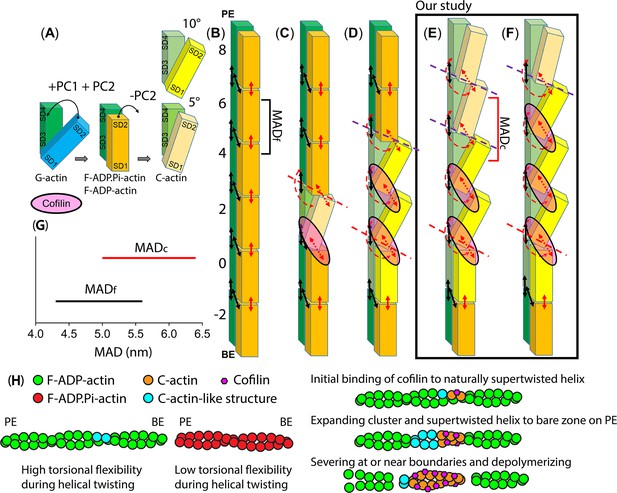
The hypothesis regarding the actin structure-dependent preferential cooperative binding of cofilin.
For simplicity, a single actin protofilament comprising six protomers labelled from –2 to 8 is shown, both without and with cofilin. The black and red arrows represent stable bonds and unstable bonds between domains of two adjacent actin protomers in the same strand, respectively. The broken red arrows indicate completely disrupted bonds between domains of two adjacent actin molecules. Additionally, broken red and purple lines show positions sensitive to being severed, with the purple lines indicating a stronger severing activity.(A) The conformational transition between G-actin, F-actin (bound with ADP.Pi, ADP), and C-actin structures. In the lower model of C-actin, the binding of a single cofilin molecule to an actin filament induces a small movement of SD2, causing a rotational shift of approximately 5° in the OD relative to the ID. In the upper one, when the cofilin clusters bind, they induce a larger rotation of approximately 10° in the OD (Galkin et al., 2011; Tanaka et al., 2018). (B) A single protofilament of an actin filament. Upon polymerization, OD rotates relative to the ID to close nucleotide binding cleft and untwist the OD to flatten actin molecule. The orange color corresponds to OD (SD1 + SD2), while green corresponds to ID (SD3 + SD4; Oda et al., 2009). (C) A single protofilament of an actin filament bound to a single cofilin (Huehn et al., 2020; Tanaka et al., 2018). (D) A single protofilament of an actin filament bound to cofilin clusters. Actin conformational changes induced by cofilin clusters are local and limited to protomers directly contacting bound cofilin. The cofilin bindings accelerate a rapid transition from F-ATP-actin to F-ADP.Pi-actin and F-ADP-actin states of protomers on the PE side. Cofilin tunes adjacent protomers, resembling F-ADP-actin structure on the PE side, to expand the clusters. The cooperative binding of cofilin requires a critical size of a cofilin cluster set at 2. Notably, the severing of the actin filament is predominantly observed at the junction between cofilactin and bare actin (Huehn et al., 2020; Suarez et al., 2011; Tanaka et al., 2018). (E–F) A single protofilament of an actin filament bound to cofilin clusters in the authors' previous study (Ngo et al., 2015) and in the current study. Our hypothesis supports that actin conformational changes induced by cofilin clusters are not local and not limited to protomers directly contacting bound cofilin (C-actin) on the PE side. Cofilin clusters have the ability to shorten a half helix by reducing the number of protomers. Additionally, cofilin cluster not only has the capacity to convert F-actin to C-actin within cofilactin region but also allosterically expands the C-actin structure from the bound to unbound region on the PE side (E). The MADc in the undecorated C-actin-like structure region are elongated compared to MADf in F-actin region. The shortening of half helices and the elongated MADc play a pivotal role in facilitating the preferential cooperative binding of cofilin to actin protomers via F-actin to C-actin transition pathway. If cofilin does not promptly attach to protomers in shortened bare half helix region (as depicted in E-F), the protomers within C-actin-like structure regions can readily open the nucleotide binding clefts and transition back to G-actin state via C-actin to G-actin transition, ultimately severing the filament at or near boundaries (broken lines). (G) HS-AFM measurements of MAD values measured in half helices of filaments representing F-actin (MADf) and undecorated C-actin-like structure (MADc) adjacent to cofilin cluster on the PE side. (H) A hypothesis describing how the function of cofilin is regulated by the torsional flexibility during helical twisting of actin filaments. The F-ADP-actin has increased the fractions of naturally supertwisted helical structures and the torsional flexibility during helical twisting, thereby creating some proposed C-actin-like structures to facilitate the initial binding and cluster expansion of cofilin, compared to F-ADP.Pi-actin. To emphasize the inhibitory effect of Pi on the initial binding of cofilin, we depict a simplified model of F-ADP.Pi-actin, featuring very infrequent structures resembling C-actin. As a result, the initial binding of cofilin and growing of critical cluster set at 2–4 cofilin molecules are more challenging compared to F-ADP-actin.
Videos
Possible transition pathways from G-actin to C-actin (constructed by +PC1) and C-actin to F-actin (constructed by +PC2), and vice versa, constructed by -PC1 and -PC2, respectively.
The transition from G-actin to F-actin ultimately requires both +PC1+PC2. Related to Figure 1.
HS-AFM imaging to investigate the dynamic structures during helical twisting by measuring the variations of HHPs, the number of actin protomers per HHP, and MADs in F-actin structures within long actin filaments.
Our results provided strong evidence supporting the variable helical twisting and dynamics in MADs within actin filaments. However, the results contrast the existing rigid and inflexible depiction of helical twists in Cryo-EM and X-ray diffraction structures. Actin filaments were gently immobilized on lipid membrane composed of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-3-trimethylammonium-propane (DPTAP) (90/10 wt%). Bars: 25 nm. Imaging rate: 2 frames per second (fps). Video plays at 5 fps. Related to Figure 3.
HS-AFM imaging to investigate the dynamic structures during helical twisting by measuring the variability of MADs in F-ADP-actin and F-ADP.Pi-actin structures within long actin filaments.
Our findings provided strong evidence supporting the dynamics in MADs within these actin filaments, though the current temporal resolution could not distinguish a significant difference between F-ADP-actin and F-ADP.Pi-actin, even at high imaging rates. Actin filaments were gently immobilized on lipid membrane composed of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-3-trimethylammonium-propane (DPTAP) (90/10 wt%) and imaged in F1 buffer containing either 1 mM ADP +5 U/ml hexokinase +10 mM glucose or 1 mM ATP +10 mM Pi. Bars: 25 nm. Imaging was conducted at rates of 5 and 10 fps for F-ADP-actin and at 10 fps for F-ADP.Pi-actin. The video is played at 5 fps. Related to Figure 3—figure supplement 1.
HS-AFM imaging to investigate the dynamic structures during helical twisting by measuring the variations of HHPs, the number of actin protomers per HHP, and MADs in the normal and shorter half helices neighboring a cofilin cluster on the BE and PE sides. Bars: 25 nm. Imaging rate: 2 fps. Video plays at 5 fps. Related to Figures 5–6.
HS-AFM imaging to investigate the dynamic structures during helical twisting by measuring the variations of MADs in the barely supertwisted and untwisted half helices neighboring a cofilin cluster on the PE and BE sides, respectively, as well as in the normally twisted half helices in the same images.
Actin filaments were imaged in an F1 buffer containing 300 nM cofilin and 1 mM ADP, 5 U/ml hexokinase and 10 mM glucose. There was minimal nonspecific binding of cofilin to this lipid membrane surface. Bars: 25 nm. Imaging rate: 2 fps. Video plays at 5 fps. Related to Figure 6—figure supplement 1.
HS-AFM imaging to demonstrate the preferentially cooperative binding of cofilin toward a pre-shortened bare half helix resembling C-actin structures along actin filaments.
Even with 10 mM Pi, the cofilin cluster could supertwist the half helices in the actin filaments once a critical cluster was established. Bars: 25 nm. Imaging rate: 2 fps. Video plays at 5 fps. Related to Figure 7.
Cooperative binding of cofilins along actin filaments was greatly inhibited when actin filaments were strongly immobilized on lipid membrane composed of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-3-trimethylammonium-propane (DPTAP) (75/25 wt%).
The strong immobilization hindered the torsional motion of actin filaments during helical twisting, resulting in suppressing cofilin binding and cluster expansion. Actin filaments were imaged in an F1 buffer containing 2000 nM cofilin and 0.5 mM ATP. Time label denotes the elapsed time after addition of cofilin. Bars: 25 nm. Imaging rate: 2 fps. Video plays at 10 fps. Related to Figure 8A.
Cooperative binding of cofilins along actin filaments was normal when actin filaments were loosely immobilized on lipid membrane composed of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-3-trimethylammonium-propane (DPTAP) (90/10 wt%).
Actin filaments were imaged in an F1 buffer containing 300 nM cofilin and 0.5 mM ATP. Time label denotes the elapsed time after addition of cofilin. Bars: 25 nm. Imaging rate: 2 fps. Video plays at 10 fps. Related to Figure 8B.
Cooperative binding of cofilins along actin filaments was normal when actin filaments were loosely immobilized on lipid membrane composed of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-3-trimethylammonium-propane (DPTAP) (90/10 wt%).
Actin filaments were imaged in an F1 buffer containing 300 nM cofilin, 1 mM ADP, 5 U/ml hexokinase, and 10 mM glucose. Time label denotes the elapsed time after addition of cofilin. Bars: 25 nm. Imaging rate: 2 fps. Video plays at 10 fps. Related to Figure 8C.
Growth of cofilin cluster along actin filaments was normal when actin filaments were loosely immobilized on lipid membrane composed of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-3-trimethylammonium-propane (DPTAP) (90/10 wt%).
Even with 10 mM Pi, the cofilin cluster could supertwist the half helices in the actin filaments once a critical cluster was established. Actin filaments were imaged in an F1 buffer containing 75 nM cofilin and 1 mM ATP +10 mM Pi (see Methods). Time label denotes the elapsed time after addition of cofilin. Bars: 25 nm. Imaging rate: 2 fps. Video plays at 10 fps. Related to Figure 8D.
Tables
List of 46 protein data bank (PDB) structures that were utilized in the PCA, as depicted in Figure 1.
Due to the presence of varying structures among different chains in some PDB structures, each chain was considered as an individual structure. Notably, in the case of 6UC4, chains E and H were combined and renamed as chain E (E&H=E), while chains F and G were combined and renamed as chain F (F&G=F). Similarly, in the case of 6UBY, chains G and H were combined and renamed as chain G (G&H=G).
| PDBID | List of actin chains | Method | Resolution (Å) | Nucleotide | Protein bound | First author | Year |
|---|---|---|---|---|---|---|---|
| 1ATN | A | X-RAY | 2.8 | ATP | DNASE I | KABSCH | 1990 |
| 1J6Z | A | X-RAY | 1.54 | ADP-RHO | OTTERBEIN | 2001 | |
| 2BTF | A | X-RAY | 2.55 | ATP | PROFILIN | SCHUTT | 1993 |
| 2ZWH | A | X-RAY | 3.3 | ADP | ODA | 2009 | |
| 3DAW | A | X-RAY | 2.55 | ATP | TWINFILIN-1 | PAAVILAINEN | 2008 |
| 3HBT | A | X-RAY | 2.7 | ATP | WANG | 2010 | |
| 3J0S | A, B, C, D, E, F, G, H, I, J, K, L | EM | 9 | COFILIN-2 | GALKIN | 2011 | |
| 3J8I | D, E, F, G, H | EM | 4.7 | ADP | GALKIN | 2015 | |
| 3J8J | A, B, C, D, E, F, G, H, I, J, K | EM | 12 | GALKIN | 2015 | ||
| 3J8K | A, B, C, D, E, F, G, H, I, J | EM | 12 | GALKIN | 2015 | ||
| 3MFP | A | EM | 6.6 | ADP | FUJII | 2010 | |
| 4A7N | A, B, C, D, E | EM | 8.9 | ADP | BEHRMANN | 2012 | |
| 5ONV | A, B, C, D, E | EM | 4.1 | ADP | MERINO | 2018 | |
| 5OOC | A, B, C, D, E | EM | 3.6 | ADP JASP | MERINO | 2018 | |
| 5OOD | A, B, C, D, E | EM | 3.7 | ADP-Pi JASP | MERINO | 2018 | |
| 5OOE | A, B, C, D, E | EM | 3.6 | ANP | MERINO | 2018 | |
| 5OOF | A, B, C, D, E | EM | 3.4 | ADP-BeFx | MERINO | 2018 | |
| 5YU8 | A, B, C, D, E | EM | 3.8 | ADP | COFILIN-2 | TANAKA | 2018 |
| 6BNO | A, B, C, D, E, F, G, H | EM | 5.5 | ADP | GUREL | 2017 | |
| 6BNU | A, B, C, D, E, F, G, H | EM | 7.5 | GUREL | 2017 | ||
| 6DJM | A, B, C, D | EM | 3.1 | AMPPNP | CHOU | 2019 | |
| 6DJN | A, B, C, D | EM | 3.1 | ADP-Pi | CHOU | 2019 | |
| 6DJO | A, B, C, D | EM | 3.6 | ADP | CHOU | 2019 | |
| 6FHL | A, B, C, D, E | EM | 3.3 | ADP-Pi | MERINO | 2018 | |
| 6KLL | A, B, C, D | EM | 3 | ADP | ODA | 2020 | |
| 6KLN | A, B, C, D | EM | 3.4 | ADP | ODA | 2020 | |
| 6UBY | A, B, C, D, E, F, G | EM | 7.5 | ADP | COFILIN-1 | HUEHN | 2020 |
| 6UC0 | A, B, C, D, E, F, G | EM | 7.5 | ADP | COFILIN-1 | HUEHN | 2020 |
| 6UC4 | A, B, C, D, E, F, J, K, L | EM | 9.2 | ADP | COFILIN-1 | HUEHN | 2020 |
| 6VAO | D, A, B, C, E | EM | 3.4 | ADP | COFILIN-1 | HUEHN | 2020 |
| 6VAU | B, A, C, D, E | EM | 3.5 | ADP | HUEHN | 2020 | |
| 7Q8B | A, B, C, D, E | EM | 3.3 | ADP-Pi | KOTILA | 2022 | |
| 7Q8C | A, B, C, D, E | EM | 2.72 | ADP | KOTILA | 2022 | |
| 7Q8S | A, C, D, E, F | EM | 3.4 | ADP | ADF/COFILIN | KOTILA | 2022 |
| 8A2R | C, A, B, D, E | EM | 2.17 | ADP-BeF3 | OOSTERHEERT | 2022 | |
| 8A2S | C, A, B, D, E | EM | 2.22 | ADP-Pi | OOSTERHEERT | 2022 | |
| 8A2T | C, A, B, D, E | EM | 2.24 | ADP | OOSTERHEERT | 2022 | |
| 8A2U | C, A, B, D, E | EM | 2.21 | ADP-BeF3 | OOSTERHEERT | 2022 | |
| 8A2Y | C, A, B, D, E | EM | 2.15 | ADP-Pi | OOSTERHEERT | 2022 | |
| 8A2Z | C, A, B, D, E | EM | 2.15 | ADP | OOSTERHEERT | 2022 | |
| 8D13 | A, B, C | EM | 2.43 | ADP | REYNOLDS | 2022 | |
| 8D14 | A, B, C | EM | 2.51 | ADP-Pi | REYNOLDS | 2022 | |
| 8D15 | A, B, C, D, E, F, G | EM | 3.61 | ADP | REYNOLDS | 2022 | |
| 8D16 | A, B, C, D, E, F, G | EM | 3.71 | ADP-Pi | REYNOLDS | 2022 | |
| 8D17 | A, B, C, D, E, F, G | EM | 3.69 | ADP | REYNOLDS | 2022 | |
| 8D18 | A, B, C, D, E, F, G | EM | 3.66 | ADP | REYNOLDS | 2022 |
The lists of chains and matrices utilized to build the long filaments.
| PDBID | Chains | RMSD (Å) | Transformation matrix as Euler angles |
|---|---|---|---|
| 6VAU | BA | 0.000497 | [166.588953–2.1209466e-01–2.62674783e-02] |
| 8A2S | ED | 0.000704 | [-166.93535314–10.63461097 –5.38436149] |
| 8A2T | AB | 0.000704 | [-167.39029871–13.65820025 –6.3200023] |
| 3MFP | AB | 0.000398 | [166.656064 2.13617418e-05 7.68141187e-05] |
| 6UC4 | EF | 0.619515 | [164.58920564–0.207997407 –0.80958005] |
| 6VAO | CA | 0.000699 | [162.515987–4.24094412e-02–1.82361154e-01] |
| 5YU8 | ED | 0.000561 | [-162.100031–4.00777476e-05 4.03030046e-05] |
| 3J0S | KL | 0.001134 | [-162.12014856–0.17228856 –0.18642638] |
| *For cofilactin filament | |||
| 6VAO* | CAJF | 0.000699 | [162.515987–4.24094412e-02–1.82361154e-01] |
Variation during helical twisting measured by HHPs, the number of protomer pairs per HHP, and the MADf observed for protomers representing F-actin structures within long bare actin filaments, as probed by HS-AFM.
The total number of HHPs observed was 2226. The total number of protomer pairs used for estimating MADf was calculated by multiplying the number of protomer pairs per HHP by the number of HHPs.
| The number of protomer pairs per HHP | HHPs (nm) | MADf (nm) | The number of HHPs (proportion) | The total number of protomer pairs |
|---|---|---|---|---|
| 4.5 | 28.6±1.4 | 6.3±0.3 | 5 (0.2%) | 22.5 |
| 5.5 | 29.3±1.8 | 5.3±0.3 | 182 (8.2%) | 1001 |
| 6.5 | 33.1±1.3 | 5.1±0.2 | 1355 (60.9%) | 8807.5 |
| 7.5 | 37.2±2.0 | 5.0±0.3 | 624 (28.0%) | 4680 |
| 8.5 | 39.1±2.4 | 4.6±0.3 | 60 (2.7%) | 510 |
Quantifying time constants (t1, t2) obtained from fitting individual ACF curves.
The t1 and t2 values are associated with first and second time constants of axial distance (AD) between two adjacent actin protomers within F-ADP-actin and F-ADP.Pi-actin segments. These time constants reflect the time decay in the autocorrelation of AD, recorded experimentally at high temporal resolution using HS-AFM. Related to Figure 3—figure supplement 1.
| Experimental conditions | t1 (s) | t2 (s) |
|---|---|---|
| F-ADP-actin (200ms/frame) | 0.4±0.2 (N=82) | 0.5±0.7 (N=80) |
| F-ADP-actin (100ms/frame) | 0.3±0.3 (N=78) | 1.2±1.8 (N=78) |
| F-ADP.Pi-actin (100ms/frame) | 0.2±0.3 (N=152) | 0.5±1.1 (N=141) |
Variation during helical twisting measured by HHPs, the number of protomer pairs per HHP, and the MADc observed for protomers resembling C-actin structures within shortened bare half helices adjacent to the cofilactin/bare actin boundary, as probed by HS-AFM.
The total number of HHPs observed was 32. The total number of protomer pairs used for estimating MADc was calculated by multiplying the number of protomer pairs by the number of HHPs.
| The number of protomer pairs per HHP | HHP (nm) | MADc (nm) | The number of HHPs (proportion) | The total number of protomer pairs |
|---|---|---|---|---|
| 3.5 | 22.2±0.2 | 6.3±0.1 | 2 (5.9%) | 7 |
| 4.5 | 26.1±1.4 | 5.8±0.3 | 21 (61.8%) | 94.5 |
| 5.5 | 28.5±0.9 | 5.2±0.2 | 9 (32.4%) | 49.5 |
The correlation between the time it takes to shorten a bare HHP beyond the cofilactin/bare actin boundary and to form a newly matured cofilin cluster in a pre-shortened half helix.
The lag time (t) represents the duration required for a newly saturated cofilin cluster to bind and decorate onto a bare half helix adjacent to the preformed cluster after this bare half helix has been shortened by approximately 25%. This table is related to Figure 7.
| The time required to form a newly matured cofilin cluster in a pre-shortened half helix | The duration needed to shorten a bare half helix beyond the cofilactin/bare actin boundary | |||||
|---|---|---|---|---|---|---|
| P1 – P2 | P2 – P3 | P3 – P4 | P4 – P5 | P5 – P6 | ||
| P1 | 0 s | |||||
| P2 | 0 s | 0 s | ||||
| P3 | 5.0 s | 2.0 s (t=3.0 s) | ||||
| P4 | 36 s | 30.5 s (t=5.5 s) | ||||
| P5 | 54.5 s | 54 s (t=0.5 s) | ||||
| P6 | 58.5 s | 58.5 s (t = ~0 s) | ||||
A summary of peak heights and HHPs obtained from the histograms in Figure 8.
Single and double Gaussian fitting methods were applied to the data. The significant difference between the peaks in the Gaussian fitting was statistically evaluated and indicated using an F-test. ‘ND’ indicates cases where the information was not determined.
| Peak Height (nm) | Half Helical Pitch (nm) | Frequency | |||
|---|---|---|---|---|---|
| Peak 1(proportion)[F-test, p] | Peak 2(proportion)[F-test, p] | Peak 1(proportion)[F-test, p] | Peak 2(proportion)[F-test, p] | ||
| (A) 2000 nM cofilin, 0.5 mM ATP/ on DPPC/DPTAP (75/25 wt%) | 8.2±0.8 (100%) | ND | 34.3±2.6 (100%) | ND | Height: 1930 HHP: 1556 |
| (B) 300 nM cofilin, 0.5 mM ATP/ on DPPC/DPTAP (90/10 wt%) | 9.2±0.4 (47.0%) [0.0204] | 10.5±0.7 (53.0%) [0.0204] | 28.5±3.0 (55.1%) [0.0135] | 37.4±3.6 (44.9%) [0.0135] | Height: 2212 HHP: 1780 |
| (C) 300 nM cofilin, 1 mM ADP/ on DPPC/DPTAP (90/10 wt%) | 8.4±0.5 (54.8%) [0.0081] | 9.9±0.7 (45.2%) [0.0081] | 30.1±3.8 (70.3%) [0.0195] | 37.8±2.9 (29.7%) [0.0195] | Height: 3183 HHP: 2779 |
| (D) 75 nM cofilin, 1 mM ATP, 10 mM Pi/ on DPPC/DPTAP (90/10 wt%) | 8.3±0.7 (55.3%) [1.48x10–6] | 11.0±1.0 (44.7%) [1.48x10–6] | 30.3±2.9 (25.2%) [0.0002] | 38.7±3.6 (74.8%) [0.0002] | Height: 2316 HHP: 1865 |





