The relationship between gut and nasopharyngeal microbiome composition can predict the severity of COVID-19
Figures
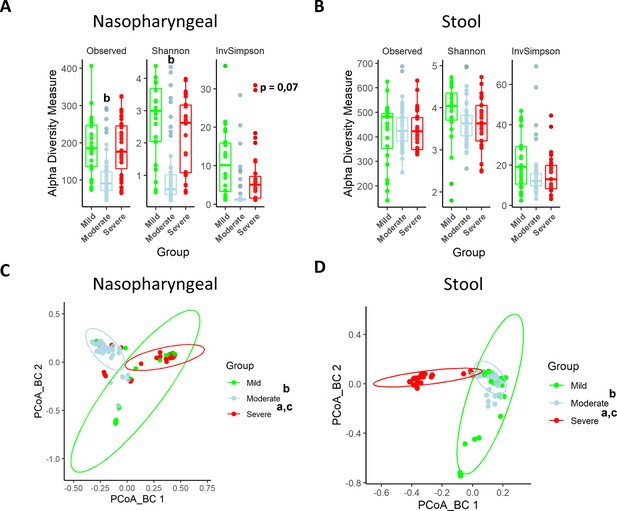
Nasopharyngeal and gut microbiota composition is modified depending on the severity of COVID-19 symptoms.
(A) Alpha diversity index for nasopharyngeal swab samples microbiota. (B) Alpha diversity index for stool samples microbiota. (C) Principal Components Analysis (PCoA) for Bray–Curtis index of nasopharyngeal swab microbiota. (D) PCoA for Bray–Curtis index of stool samples microbiota. Values are represented as mean ± SD. Differences were displayed as: ap<0.05 severe vs. mild; bp<0.05 moderate vs. mild; cp<0.05 severe vs. moderate. PERMANOVA test was employed to determine Bray–Curtis significance differences.
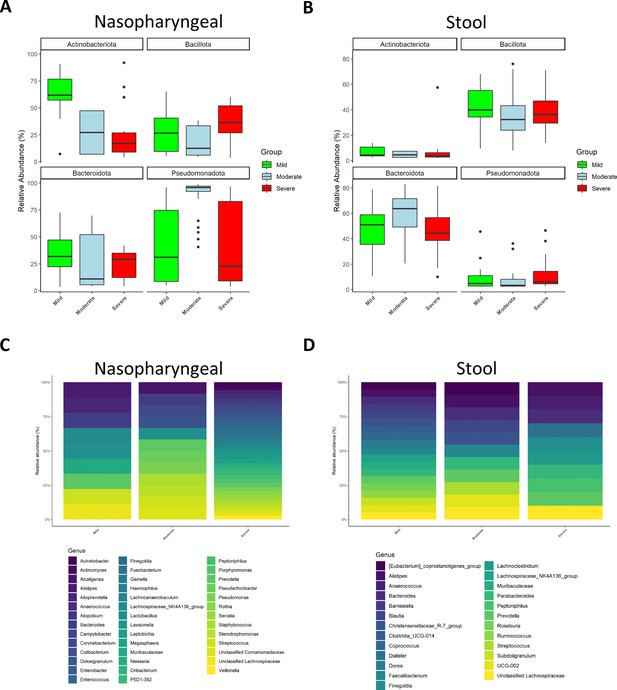
Microbiota composition of nasopharyngeal and stool samples at phylum level is slightly modified by COVID-19 symptoms severity.
In contrast, at genus level, severity increases the total amount of detected bacteria in nasopharyngeal swabs while in stool samples it is promoting a reduction. (A) Representation of the relative abundance of the main phyla in nasopharyngeal swab samples. (B) Representation of the relative abundance of the main phyla in stool samples. (C) Relative abundance of the principal genera detected in nasopharyngeal swab samples. (D) Relative abundance of the principal genera detected in nasopharyngeal swab samples. For (C) and (D) only those amplicon sequence variants (ASVs) with a median higher to 0.5 were chosen.
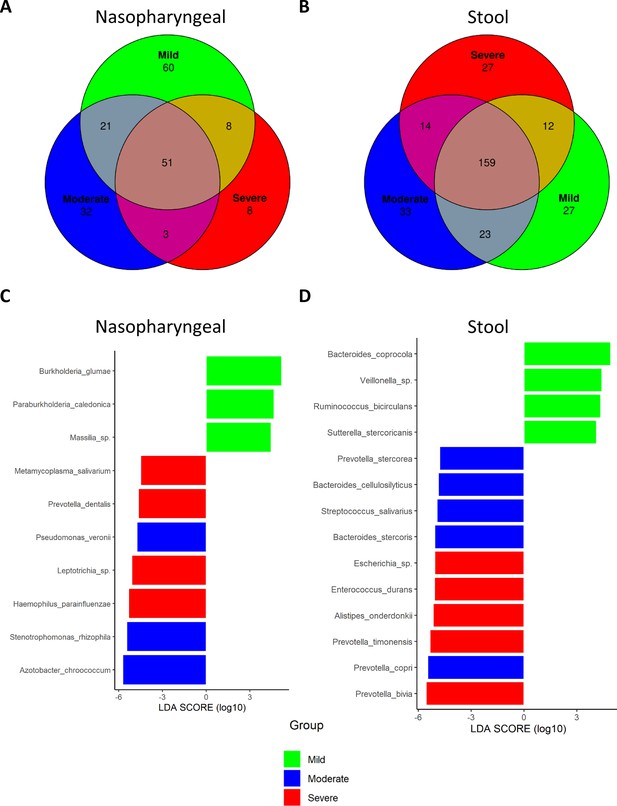
Differential analysis expression of microbiota composition from nasopharyngeal and stool samples revealed the presence of specific bacteria related to COVID-19 severity index.
(A) Venn diagram showing amplicon sequence variants (ASVs) distribution in nasopharyngeal swab samples. (B) Venn diagram showing ASVs distribution in stool samples. (C) LEfSe plot of taxonomic biomarkers present in nasopharyngeal swab samples (p-value=0.01 and linear discriminant analysis [LDA] value = 4). (D) LEfSe plot of taxonomic biomarkers present in stool samples (p-value=0.01 and LDA value = 4). Venn diagrams were acquired with the following parameters: detection level = 0.01 and prevalence level = 0.01.
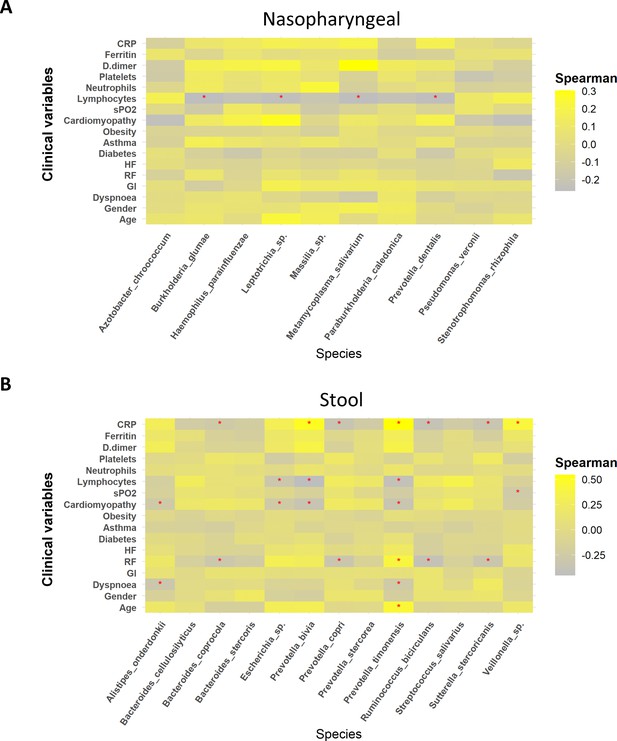
Whereas mild biomarkers showed negative correlations towards clinical variables, severe biomarkers presented positive correlations.
(A) Correlation plot of nasopharyngeal swab biomarkers and clinical variables. (B) Correlation plot of stool samples biomarkers and clinical variables. RR: respiratory rate; HR: heart rate; GI: gastrointestinal alterations. Correlation was calculated by taking into account abundance levels of each bacteria against the clinical variable of severe ill patients. Red asterisks stand for p-values<0.05 and correlations ≥0.3 or ≤–0.25.
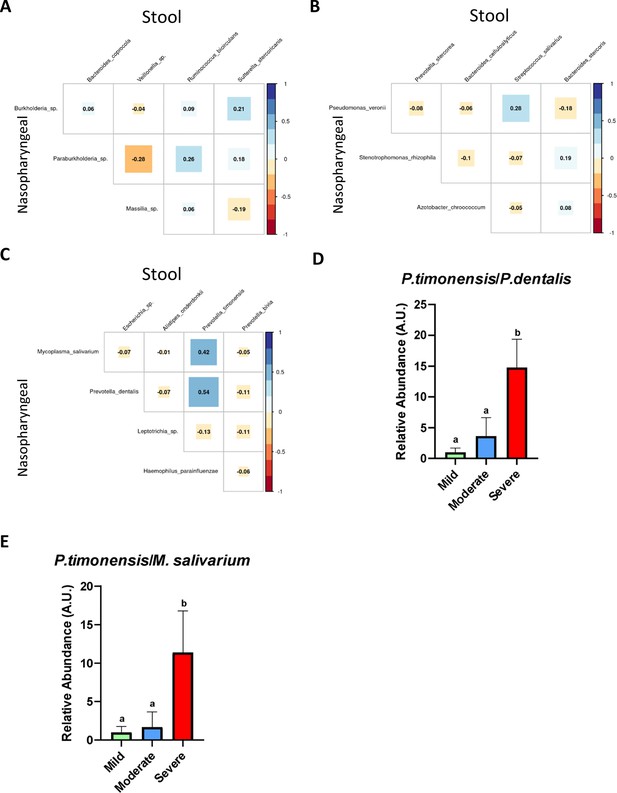
The existence of a relationship between the abundance of nasopharyngeal severe biomarkers and stool severe biomarkers allows the employment of an abundance ratio between them as a new tool for predicting COVID-19 severity.
(A) Correlation plot among biomarkers found in nasopharyngeal swab and stool samples in mild condition. (B) Correlation plot among biomarkers found in nasopharyngeal swab and stool samples in moderate condition. (C) Correlation plot among biomarkers found in nasopharyngeal swab and stool samples in severe condition. (D) Ratio of the abundance between P. timonensis (stool) and M. salivarium biomarkers. (E) Ratio of the abundance between P. timonensis (stool) and P.dentalis (nasopharyngeal swab) biomarkers. ap<0.05 severe vs. mild; bp<0.05 moderate vs. mild.
Tables
Clinical data description of enrolled patients.
Differences were displayed as: ap<0.05 severe vs. mild; bp<0.05 moderate vs. mild; cp<0.05 severe vs. moderate. ANOVA or Kruskal were employed for numerical variables and Fisher’s test for categorical variables.
| Mild(n = 24) | Moderate(n = 51) | Severe(n = 31) | ||||
|---|---|---|---|---|---|---|
| Clinical variables | p-Valuea | p-Valueb | p-Valuec | |||
| Mean age (SD), years | 43±12 | 54±14b | 62±11a,c, | 0.000001 | 0.0041 | 0.017 |
| Male, n (%) | 8 (33) | 24 (47)b | 22 (71)a,c, | 0.0071 | 0.032 | 0.041 |
| Symptoms, n (%) | ||||||
| Presence of dyspnoea | 6 (33) | 38 (75)b | 26 (84)a | 0.00018 | 0.001 | - |
| Presence of gastrointestinal alteration | 5 (26) | 17 (33) | 10 (33) | - | ||
| High respiratory rate | 1 (4) | 11 (22) | 19 (63),a,c | 0.00018 | - | 0.004 |
| Low sPO2 | 6 (33) | 22 (43) | 22 (71)a,c, | 0.001 | - | 0.021 |
| High heart rate | 1 (4) | 14 (27)b | 17 (55)a,c, | 0.0008 | 0.02 | 0.01 |
| Comorbidities, n (%) | ||||||
| Obesity | 5 (26) | 14 (27) | 10 (32) | - | ||
| Diabetes | 4 (20) | 9 (18) | 8 (26) | - | ||
| Asthma | 3 (3) | 3 (6) | 2 (7) | - | ||
| Cardiomyopathy | 3 (3) | 3 (6) | 13 (42)a,c, | 0.02 | 0.001 | - |
| Plasma determinations | ||||||
| Mean lymphocytes (SD), 103/µL | 1.1±0.6 | 1.4±0.6 | 1.2±2.7 | - | ||
| Median neutrophils (IQR), 103/µL | 6 [5.5;6.6] | 6.4 [4.2;8.6] | 7.9 [5.4;10.9] | - | ||
| Mean platelets (SD), 103/µL | 329.6±8.5 | 257.4±115b | 276.4 ± 93a | 0.031 | 0.018 | - |
| Median D-dimer (IQR), mg/L | 0.39 [0.2;0.8] | 0.6 [0.3;1] | 1.6 [0.9;4.3]a,c, | 0.0001 | - | 0.0001 |
| Median ferritin (IQR), ng/L | 157 [126;179] | 487 [274;1027]b | 829 [488;1376]a | 0.0001 | 0.0001 | - |
| Median C reactive protein (IQR), mg/L | 3.4 [2.6;4] | 18.2 [7.8;41.9]b | 162 [65;210]a,c, | 0.0001 | 0.0002 | 0.0001 |
Additional files
-
Supplementary file 1
Relative abundance of phyla found in nasopharyngeal and stool samples.
- https://cdn.elifesciences.org/articles/95292/elife-95292-supp1-v1.docx
-
Supplementary file 2
Relative abundance and exact p-values of genera found in nasopharyngeal samples.
a p<0.05 severe vs. mild; bp<0.05 moderate vs. mild; cp<0.05 severe vs. moderate. ANOVA or Kruskal were employed for numerical variables and Fisher’s test for categorical variables. ‘-’ means that genera were no present in microbiota from that group or that not significant differences were found.
- https://cdn.elifesciences.org/articles/95292/elife-95292-supp2-v1.docx
-
Supplementary file 3
Relative abundance and exact p-values of genera found in stool samples.
a p<0.05 severe vs. mild; b p<0.05 moderate vs. mild; c p<0.05 severe vs. moderate. ANOVA or Kruskal were employed for numerical variables and Fisher’s test for categorical variables. ‘-’ means that genera were no present in microbiota from that group or that not significant differences were found.
- https://cdn.elifesciences.org/articles/95292/elife-95292-supp3-v1.docx
-
Supplementary file 4
Unique ASVs identified for each group in each sample.
- https://cdn.elifesciences.org/articles/95292/elife-95292-supp4-v1.docx
-
Supplementary file 5
Relative abundance of possible biomarkers of stool samples.
- https://cdn.elifesciences.org/articles/95292/elife-95292-supp5-v1.docx
-
Supplementary file 6
Relative abundance of possible biomarkers of nasal swab samples.
- https://cdn.elifesciences.org/articles/95292/elife-95292-supp6-v1.docx
-
Supplementary file 7
Correlation values and exact p-values of the relationship between microbiota and clinical variables.
- https://cdn.elifesciences.org/articles/95292/elife-95292-supp7-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/95292/elife-95292-mdarchecklist1-v1.docx





