Molecular basis of neurodegeneration in a mouse model of Polr3-related disease
Figures
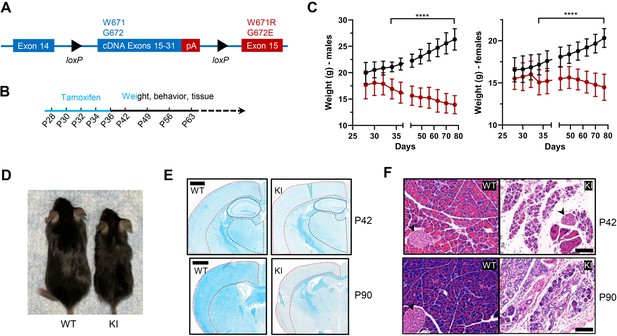
Body weight, length, and histological characteristics of Polr3a-tamKI mice.
(A) Schematic of the modified Polr3a locus showing floxed WT sequences encoding exons 15–31 with adjacent sequences for termination and polyadenylation (pA) and Polr3a exon15 containing the leukodystrophy mutations. (B) Tamoxifen injections and experimental timeline. (C) Lower body weights of Polr3a-tamKI (red) versus WT (black) mice (males n=18/group, nested t test p<0.0001****, females n=20/group, nested t test p<0.0001****). Data show mean ± SD. (D) Body length differences at P56. Images are representative. (E) LFB-staining of WT and KI brain sections at P42 and P90. The cerebral cortex, hippocampus, and thalamus are marked by red, blue, and black lines, respectively. Scale bar, 1000 µm. (F) H&E-stained WT and KI pancreata at P42 and P90 shows a dramatic loss of acinar cells. Pancreatic islets are marked with a black arrowhead. Scale bar,100 µm.
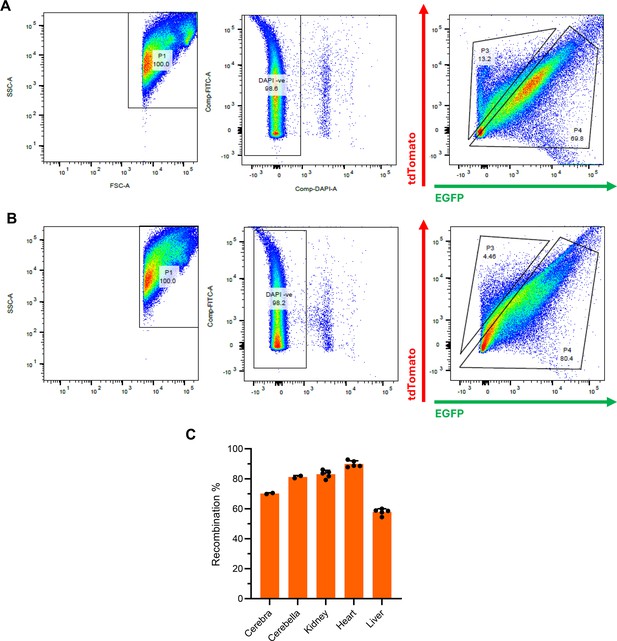
Analysis of recombination frequency in Polr3a-tamKI mice.
(A) Flow cytometry analysis of a cerebral homogenate from tamoxifen-treated Polr3a-tamKI mice carrying a dual tdTomato-EGFP recombination reporter (Muzumdar et al., 2007). Five tamoxifen injections (6 mg/40 g, i.p) were administered every other day for 5 days starting at P28. Mice were sacrificed at P42. The plots show side scatter (SSC) and forward scatter (FSC) for area (A) of individual cells, DAPI staining for viability and gating of tdTomato and EGFP based on single color controls. (B) Flow cytometry analysis of a cerebellar homogenate from the same mouse as in panel A. (C) Recombination frequencies in different tissues. Data for cerebra and cerebella were determined by flow cytometry of two mice treated as described in panel A. Data for kidney, heart, and liver were determined by fluorescence microscopy with manual counting of tdTomato and EGFP-positive cells. Cell counts were performed on multiple sections with >150 cells scored per section. Results show the mean ± SD.
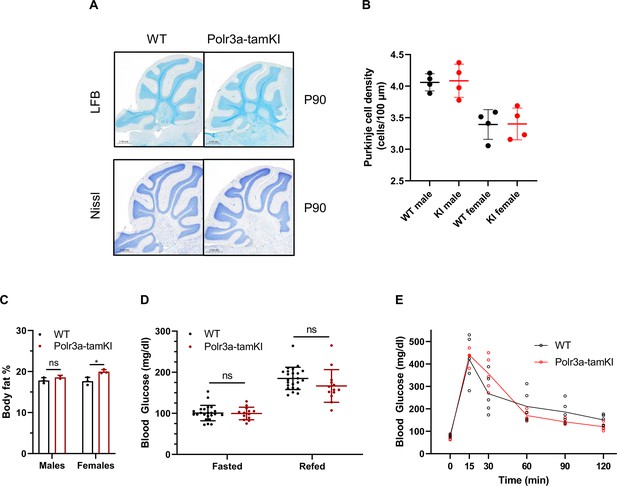
Cerebellar histology and glucose homeostasis of Polr3a-tamKI mice.
(A) LFB- and Nissl-stained sagittal sections of cerebellum from P90 mice. No differences in staining intensity were noted. (B) Purkinje cell density was quantified in Nissl-stained sections for lobes III, IV/V, VI/VII, and VIII. The results are plotted as the mean ± SD for male and female mice of each genotype. (C) Percent body fat was determined by EchoMRI on mice at P42 after an 8 hr midnight fast and a 2 hr refeed. (D) Blood glucose levels (mean ± SD) at P42 were measured by tail vein bleed after an 8 hr midnight fast, and after a 2 hr refeed (WT n=23, Polr3a-tamKI n=13, multiple t-tests, one-way ANOVA). (E) Glucose tolerance test. Blood glucose was measured after an 8 hr midnight fast and at 15, 30, 60, 90, and 120 min after mice were given a bolus of glucose at 2 g/kg body weight. The lines connect the mean values at each time point. WT and Polr3a-tamKI n=5, multiple t-tests, one-way ANOVA. ns: not significant.
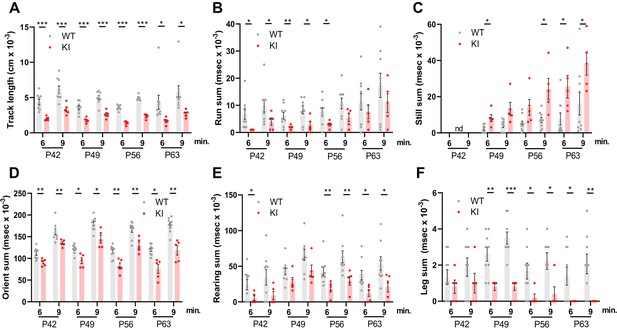
Behavioral studies of WT and Polr3a-tamKI mice.
(A–C) Locomotor activity is reported as the total distance traveled (Track length) and the total time during which the mice are running (Run sum) or standing still (Still sum). (D) Risk assessment (Orient sum) is the sum of three orientation behaviors (see Figure 2—figure supplement 1). (E) Exploratory behavior (Rear sum) is the sum of three rearing behaviors (see Figure 2—figure supplement 1). (F) Leg grooming. Data were collected at weekly intervals beginning at P42 (WT n=9 and KI n=5). Mice were tested for nine minutes with recording in three intervals of 3 min each. Values show the mean ± SEM at 6- and 9-min timepoints. Multiple t tests and one-way ANOVA, * p<0.05, ** p<0.01, *** p<0.001.
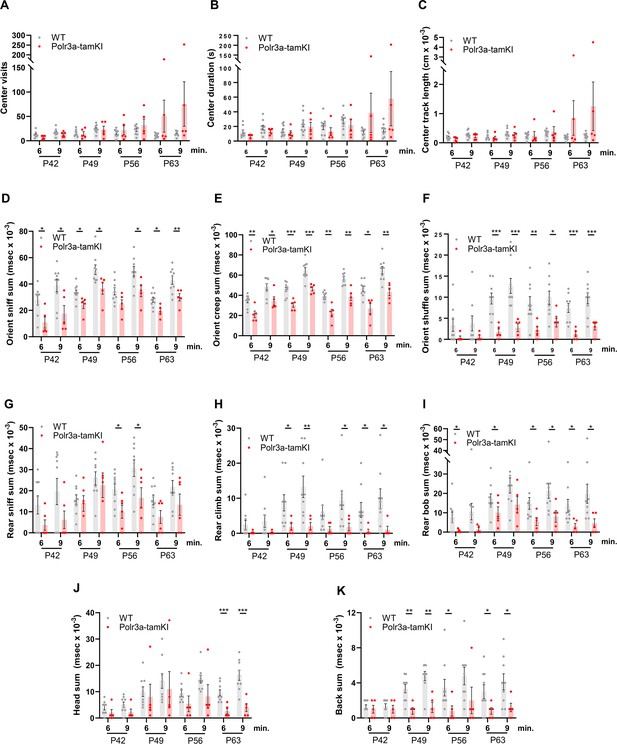
Behavioral spectrometer analysis of WT and Polr3a-tamKI mice.
(A) Center visit counts. (B). Center visit duration. (C). Center track length. Anxiety-like behaviors in panels (A–C) were scored in a 15 cm2 area in the center of the 40 cm2 arena. (D). Orient sniff. (E). Orient creep. (F). Orient shuffle. Risk assessment behavior was monitored in panels (D–F) using three independent metrics. The sum of these behaviors provides an overall measure of risk assessment (see Figure 3D). (G). Rear sniff. (H). Rear climb. (I). Rear bob. Exploratory behavior was monitored in panels (G–I) using three independent metrics. The sum of these behaviors provides an overall measure of exploration (see Figure 3E). (J). Head grooming. (K). Back grooming. Data were collected at weekly intervals beginning at P42 (WT n=9 and Polr3a-tamKI n=5). Mice were tested for 9 min with recording in three intervals of 3 min each. Cumulative data are presented at the 6 and 9 min time points as the mean ± SEM. Multiple t-tests and one-way ANOVA, * p<0.05, ** p<0.01, *** p<0.001.
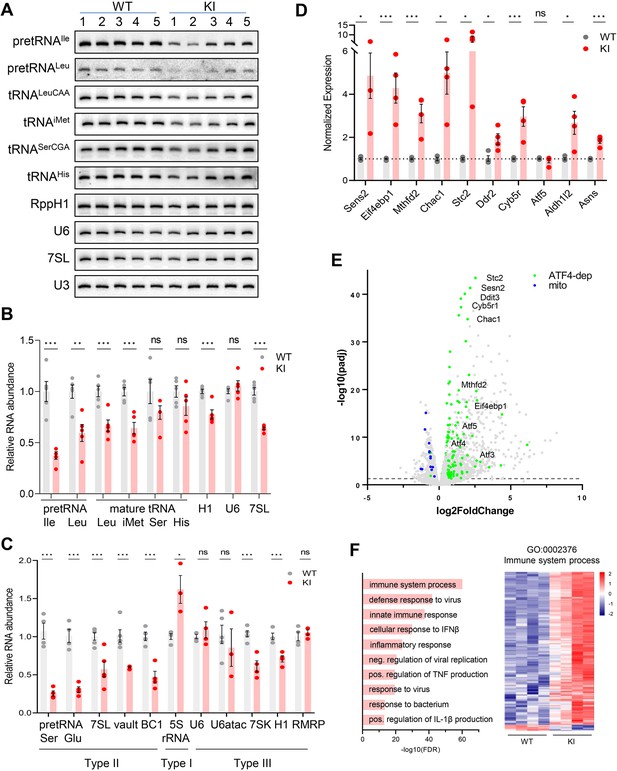
Transcriptome analysis of Polr3a-tamKI cerebra at P75.
(A) Northern blot of Pol III transcripts. Precursor tRNAs (Ile-TAT, Leu-CAA), mature tRNAs (Leu-CAA, iMet, Ser-CGA, His), RPPH1, U6 and 7SL RNAs and the loading control U3 snRNA were detected by hybridization using transcript-specific oligonucleotide probes. All blots represent sequential probing of a single gel. The cropped images frame the relevant regions. (B) Quantitation of Pol III transcripts in panel A (mean ± SEM, n=5 biological replicates). (C) RT-qPCR of Pol III transcript abundance (mean ± SEM, n=3–6 biological replicates). (D) RT-qPCR of selected ATF4-regulated ISR genes (mean ± SEM, n=3–5 biological replicates). (E) Global gene expression quantified by RNA-seq. The volcano plot shows gene expression changes (KI/WT, n=4 biological replicates). Differentially expressed (DE) ATF4-regulated genes (green) (p-adj <0.05, log2FC >|0.58|) and mitochondrially encoded transcripts (blue). ATF4-regulated genes assayed in panel D and Ddit3/Chop are labeled. (F) Top ten GO bioprocesses among up-regulated DE genes are shown with a heatmap of Z-scores for DE genes annotated to the GO:0002376 immune system process. For all graphs: WT, gray bars and circles; KI, red bars and circles. ns, not significant; * p≤0.05; ** p≤0.01; *** p≤0.005.
-
Figure 3—source data 1
PDF file containing original northern blots for Figure 3A, indicating the relevant bands and conditions.
- https://cdn.elifesciences.org/articles/95314/elife-95314-fig3-data1-v1.pdf
-
Figure 3—source data 2
Original files for northern analysis displayed in Figure 3A.
- https://cdn.elifesciences.org/articles/95314/elife-95314-fig3-data2-v1.zip
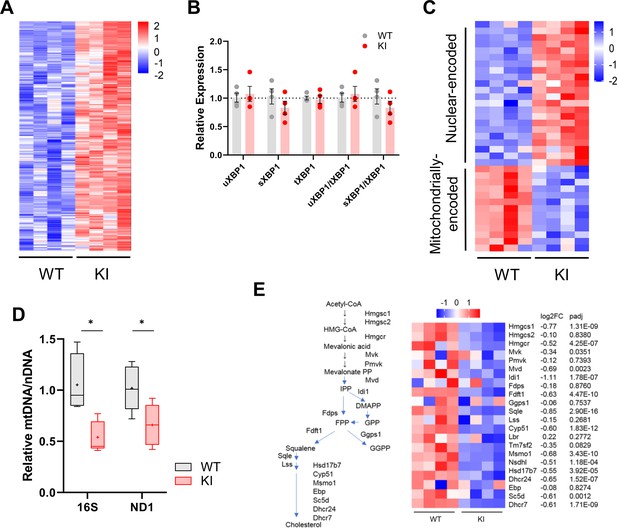
Gene expression in WT and Polr3a-tamKI (KI) cerebra at P75.
(A) Heatmap represents Z scores of DESeq2 normalized read counts for ATF4-dependent ISR genes activated in cerebra at P75 (p-adj <0.05). (B) RT-qPCR analysis of XBP1 splicing. The relative expression of unspliced Xbp1 (XBP1u, constitutively expressed) and spliced Xbp1 (XBP1s, spliced in response to ER stress) is normalized to total Xbp1 mRNA (Yoon et al., 2019). The fold change in expression was calculated by the ΔΔCt method and expressed relative to the WT value. Data are presented as mean ± SEM, n=4 biological replicates. (C) A heatmap of mitochondrial and nuclear-encoded mitochondrial proteins showing differential expression (DE) of their genes in KI and WT cerebra at P75. DE genes (p-adj <0.05, log2FC|0.58|) were queried against the Mitocarta 3.0 gene list of mitochondrially located proteins (Rath et al., 2021; 34/1140 genes). The heatmap represents Z scores of DESeq2 normalized read counts for DE nuclear-encoded and mitochondrially encoded protein-coding genes (p-adj <0.05, log2FC >|0.58|). (D) Relative mitochondrial genome abundance in KI and WT cerebra at P75. PCR analysis of mitochondrial 16 S rDNA and Nd1 gene abundance was normalized to a nuclear gene (HK2). Fold change was calculated by the ΔΔCt method (Quiros et al., 2017) and expressed relative to the WT value. Box plot and whiskers (min to max) are shown for 16 S rDNA and Nd1 for WT (gray) and Polr3a-tamKI (red), three to four biological replicates repeated three times. Median is represented by +, * p≤0.05 (Student’s standard t-test). (E) Sterol biosynthesis is down-regulated in KI cerebra. Scheme of sterol biosynthesis (adapted from Rye et al., 2018) and gene expression changes of enzymes annotated to the GO term sterol biosynthetic process pathway for cholesterol synthesis (GO:0016126). The heatmap shows Z-scores of DESeq2 normalized read counts adjacent to the log2FoldChange and p-adj values from Supplementary file 1.
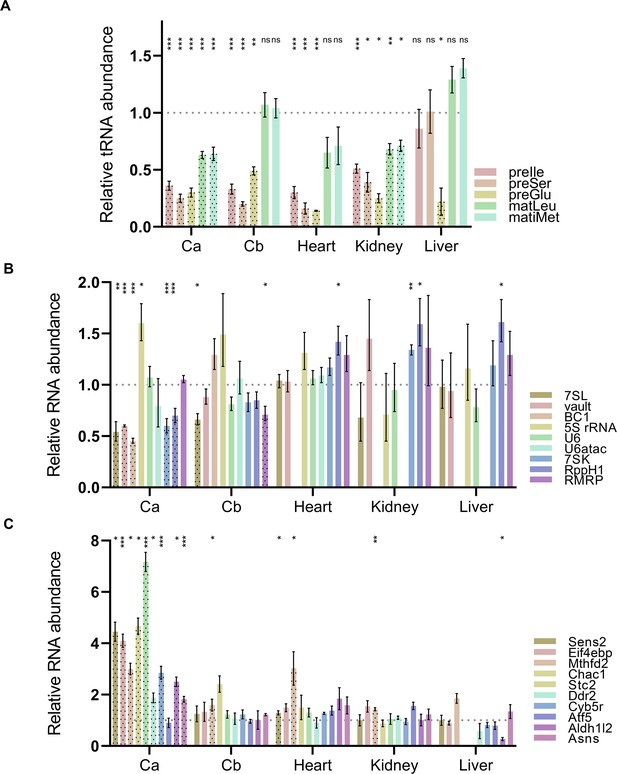
Pol III and Pol II transcript levels in various tissues from WT and Polr3a-tamKI (KI) mice at P75.
(A) The relative abundance of precursor and mature tRNAs in total RNA from cerebella (Cb), heart, kidney, and liver. Cerebral (Ca) data (from Figure 3B and C) are included for ease of comparison. Precursor-tRNAIle-TAT (preIle) and mature tRNAs Leu-CAA and iMet (matLeu and matiMet, respectively) levels were determined by Northern blotting of five to six biological replicates and normalized to a U3 snRNA loading control. RT-qPCR of four to six biological replicates was used to determine relative pre-tRNA levels for pre-tRNASer Ts12 and pre-tRNAGlu Te5/7/8 (preSer and preGlu, respectively). The fold change in expression was calculated by the ΔΔCt method (Taylor et al., 2019) and expressed relative to the WT value. Data represent the mean ± SEM. The results for preIle, preSer, preGlu, matLeu, and matiMet are plotted from left to right for each tissue. Only KI values are shown for simplicity. tRNAs with significant differences between WT and Polr3a-tamKI samples are highlighted with black dots. ns, not significant; * p≤0.05; ** p≤0.01; *** p≤0.005 (Student’s standard t-test). (B) RT-qPCR analysis of non-tRNA Pol III transcripts across WT and KI cerebella (Cb), heart, kidney, and liver are shown. Cerebral (Ca) values (from Figure 3C) are included for ease of comparison. Only KI values are shown for simplicity. Pol III transcripts with significant differences between WT and KI are highlighted with black dots. Data were calculated, normalized and plotted as in panel A, n=3–6 biological replicates. (C) RT-qPCR analysis of a set of ATF4-regulated ISR genes across WT and KI cerebella (Cb), heart, kidney, and liver are shown. Cerebral (Ca) values (from Figure 3D) are included for ease of comparison. Data were calculated, normalized, and plotted as in Figure 3D, n=3–5 biological replicates.
-
Figure 3—figure supplement 2—source data 1
PDF file containing original northern blots for Figure 3—figure supplement 2A.
- https://cdn.elifesciences.org/articles/95314/elife-95314-fig3-figsupp2-data1-v1.pdf
-
Figure 3—figure supplement 2—source data 2
Original files for northern analysis displayed in Figure 3—figure supplement 2A.
- https://cdn.elifesciences.org/articles/95314/elife-95314-fig3-figsupp2-data2-v1.zip
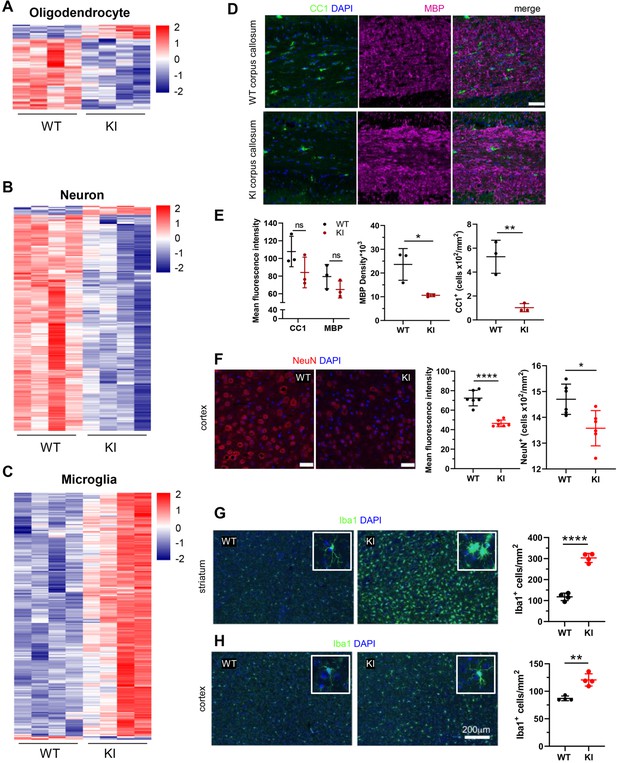
Cell-type-specific changes in Polr3a-tamKI cerebra at P75.
(A–C) Oligodendrocyte (A), neuron (B), and microglia (C) cell type-specific DE genes. Heatmaps show Z-scores of DE genes identified in the top 500 mouse cell-type-specific genes defined in McKenzie et al., 2018. (D–E) Immunostaining of oligodendrocytes at the midline of the corpus callosum. Scale bar, 40 μm. Mean fluorescence intensities of MBP and CC1, MBP staining density and CC1 cell counts represent mean ± SD, n=3. (F) NeuN immunostaining of neurons in the motor cortex. Scale bar, 40 μm. Mean fluorescence intensities and cell counts represent mean ± SD of bilaterally symmetric regions (n=3). (G–H) Iba1 staining of microglia in the striatum (G) and cerebral cortex (H). Insets show larger magnifications of individual microglia. Note the striking shortening of processes and enlargement of the cell body in the striatum. Scale bar, 200 μm. Cell counts represent mean ± SD of bilaterally symmetric regions, (n=2). For all graphs: WT, black; KI, red; ns, not significant; * p≤0.05; ** p≤0.01; *** p≤0.005; **** p<0.0001.
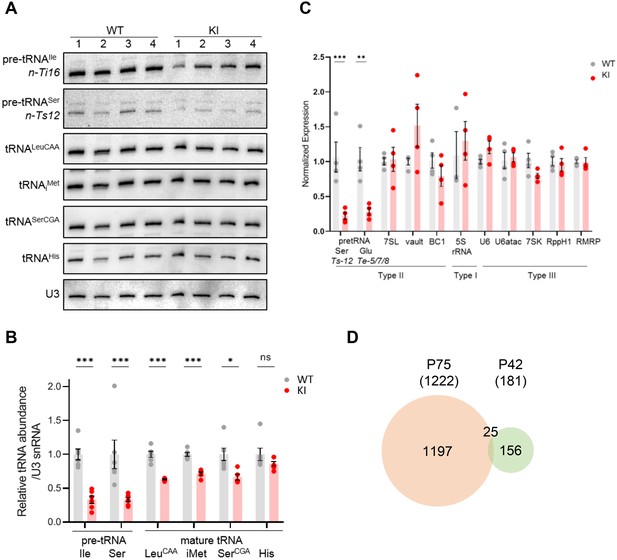
Transcriptome analysis of Polr3a-tamKI cerebra at P42.
(A) Northern blots of Pol III transcripts. Precursor tRNAs (Ile-TAT, Ser-CGA), mature tRNAs (Leu-CAA, iMet, Ser-CGA, His) and U3 snRNA were detected as in Figure 3A. (B) Quantitation of Pol III transcripts in panel A. Mean ± SEM, n=5 biological replicates. (C) RT-qPCR of Pol III transcript abundance. Mean ± SEM, n=3–5 biological replicates. (D) Venn diagram of the overlap between P75 and P42 DE genes (p-adj <0.05, log2FC >|0.58|).
-
Figure 5—source data 1
PDF file containing original northern blots for Figure 5A, indicating the relevant bands and conditions.
- https://cdn.elifesciences.org/articles/95314/elife-95314-fig5-data1-v1.pdf
-
Figure 5—source data 2
Original files for northern analysis displayed in Figure 5A.
- https://cdn.elifesciences.org/articles/95314/elife-95314-fig5-data2-v1.zip
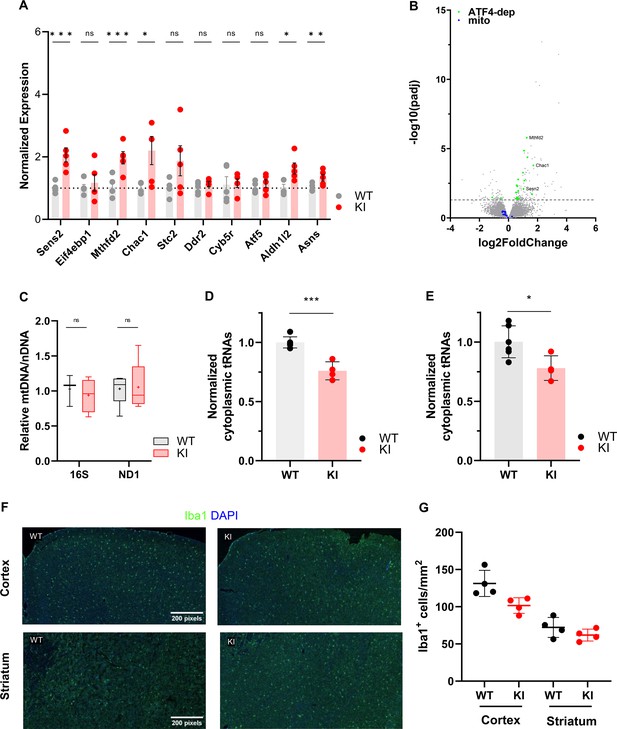
Gene expression and Iba1 staining in adolescent WT and Polr3a-tamKI (KI) cerebra.
(A) RT-qPCR analysis of a set of ATF4-regulated ISR genes in WT and KI cerebra at P42. Data were calculated, normalized and plotted as in Figure 3D, n=3–5 biological replicates. ns, not significant; * p≤0.05; ** p≤0.01; *** p≤0.005 (Student’s standard t-test). (B). Fold change in global gene expression in WT and KI cerebra at P42. A volcano plot shows RNA-seq expression changes (KI/WT, n=4 biological replicates). Differentially expressed (DE) ATF4-regulated ISR genes are highlighted in green (p-adj <0.05, log2FC >|0.58|). Mitochondrial-encoded mRNAs are shown in blue for comparison with Figure 3E. Representatives of the ATF4-regulated ISR gene set assayed in panel A are labeled. (C). Relative mitochondrial genome abundance in KI and WT cerebra at P42. PCR analysis of mitochondrial 16 S rDNA and Nd1 gene abundance was calculated, normalized and expressed as described in Supplemental Fig. S4D. Box plot and whiskers (min to max) are shown for 16 S rDNA and Nd1 for WT and KI cerebra at P42, 4–5 biological replicates, replicated three times. Median is represented by +, ns, not significant (Student’s standard t-test). (D–E). Total tRNA reads in WT and KI cerebra at P42 were normalized to the sum of endogenous mitochondrially-encoded tRNA reads (D) or exogenous spike-in reads (E) and expressed relative to the mean WT value. Data represent the mean ± SD, KI n=4 and WT n=6 biological replicates, p=0.0003 and 0.024 (Student’s standard t-test) for panels D and E, respectively. (F). Iba1 staining of microglia in the cerebral cortex and striatum at P44. Scale bar, 200 μm, applies to all panels. (G). Iba1 + cell counts in the cerebral cortex and striatum represent the mean ± SD of bilaterally symmetric regions (n=2).
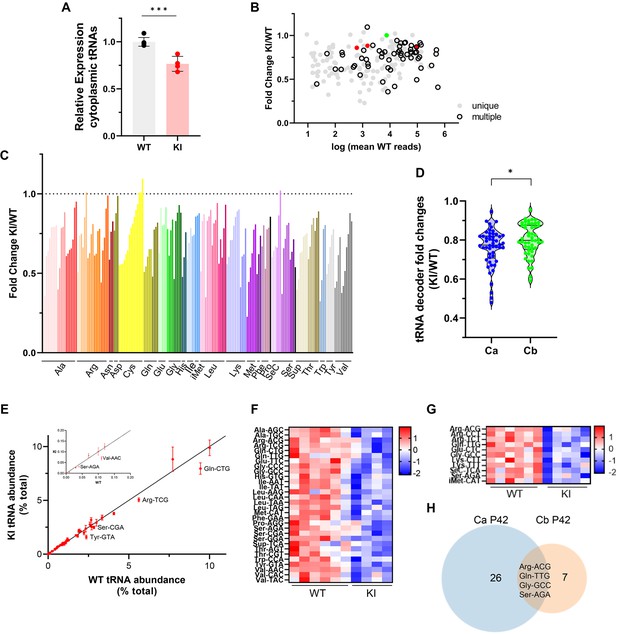
Cytosolic tRNA abundance in Polr3a-tamKI cerebra at P42.
(A) Total cytoplasmic tRNA reads in WT and KI cerebra, normalized to the sum of spike-in and mitochondrially-encoded tRNA reads are expressed relative to the mean WT value. Mean ± SD, KI n=4, WT n=6 biological replicates, p=0.0003. (B) tRNA fold change (KI/WT) is plotted against Log mean WT reads. Symbols show tRNAs encoded by unique loci (gray), identical tRNAs encoded by multiple loci (hollow black), iMet tRNAs (red) and n-TRtct5 (green). (C) tRNA fold change (KI/WT) for all decoder families. Individual tRNAs are ordered from most to least fold change and grouped by codon recognition (tRNA decoder family). The amino acid for each tRNA decoder family is indicated. (D) Violin plots of tRNA decoder fold changes (KI/WT) for cerebra (Ca) and cerebella (Cb) at P42. tRNA reads were summed for each tRNA decoder family and normalized to spike-in and mitochondrial tRNA reads, p=0.0250. For Ca, n=4 (KI) and n=6 (WT) biological replicates and for Cb, n=5 (KI) and n=6 (WT) biological replicates. (E) The cytoplasmic tRNA profile for KI cerebra is plotted against the WT profile. tRNA decoder reads are expressed as a percentage of their respective total cytoplasmic decoder pool, mean ± SEM. tRNA decoders that are significantly lower in the KI compared to WT (p≤0.05) fall below the regression line and are labeled. The inset shows tRNA decoders representing <0.2% of the total tRNA pool. (F) DE tRNA decoders in cerebra (Ca). (G) DE tRNA decoders in cerebella (Cb). Heatmaps represent Z scores of normalized read counts for significant DE genes (p-adj <0.05). (H) Venn diagram of the overlap between DE tRNA decoders in Ca and Cb (p-adj <0.05).
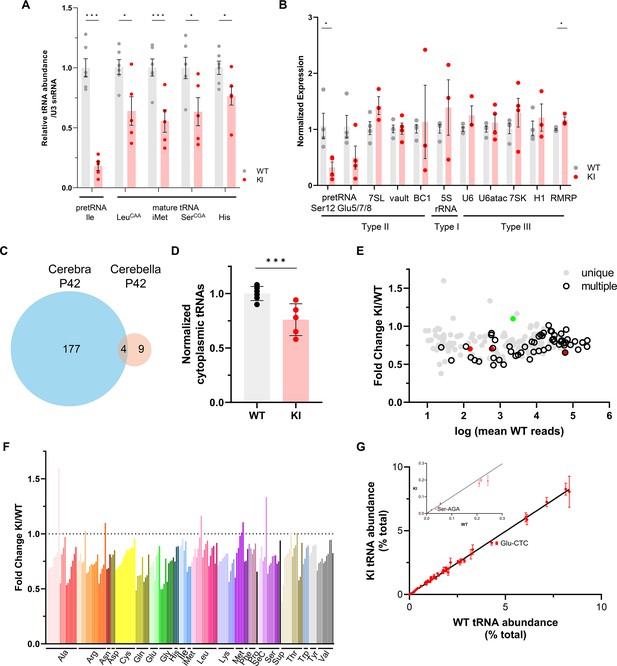
Gene expression in WT and Polr3a-tamKI (KI) cerebella at P42.
(A–B) The relative abundance of Pol III transcripts in total RNA from WT and KI cerebella (Cb) at P42 determined by northern analysis (A) and RT-qPCR (B). (A) Precursor (pre-tRNAIle-TAT) and mature tRNAs (Leu-CAA, iMet, Ser-CGA, His) detected by northern blotting were quantified as in Figure 3A. Data represent the mean ± SEM, n=6 WT and n=5 KI biological replicates. (B) Pol III transcripts detected by RT-qPCR in KI and WT Cb. Data were normalized as in Figure 3C and represent the mean ± SEM, n=3–6 biological replicates; ns, not significant; * p≤0.05; ** p≤0.01; *** p≤0.005 (Student’s standard t-test). (C) Venn diagram shows limited overlap between significant DE genes from cerebra (Ca) and cerebella (Cb) at P42 (p-adj <0.05, log2F|0.58|), KI n=4 and WT n=4 biological replicates for both Ca and Cb. (D) Total cytoplasmic tRNA reads in WT and KI cerebella at P42 were normalized to the sum of endogenous mitochondrially-encoded tRNA reads and exogenous spike-in reads and expressed relative to the mean WT value. Data represent the mean ± SD, KI n=5 and WT n=5 biological replicates, p=0.0006 (Student’s standard t-test). (E) Fold change for KI/WT cytoplasmic tRNAs is plotted against log mean WT reads. The symbols show tRNAs encoded by unique loci (gray), identical tRNAs encoded by multiple loci (hollow black), iMet tRNAs (red) and n-TRtct5 (green). (F) Fold change for cytoplasmic tRNAs (KI/WT) is plotted for all decoder families. Individual tRNAs are ordered from most to least fold change and grouped by codon recognition (tRNA decoder) family. The amino acid that is charged by each tRNA decoder family is indicated. (G) The cytoplasmic tRNA profile for KI P42 cerebella is plotted against the WT profile. tRNA decoder reads from each KI and WT replicate were expressed as a percentage of their respective total cytoplasmic decoder pool. Data represent the mean ± SEM. tRNA decoders that are significantly lower in KI compared to WT (p≤0.05, Student’s standard t-test) fall below the regression line and are labeled. The inset shows tRNA decoders with <0.3% of the tRNA pool.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | C57BL/6J-Polr3atm1Iwil | Merheb et al., 2021 | PMCID:PMC8501794 | |
| Genetic reagent (M. musculus) | B6.Cg-Tg(CAG-cre/Esr1*5Amc/J) | Jackson Laboratory | #004682, RRID:IMSR_JAX:004682 | |
| Genetic reagent (M. musculus) | B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | Jackson Laboratory | #007676, RRID:IMSR_JAX:007676 | |
| Antibody | Anti-MBP, rat monoclonal | Abcam | #ab7349, RRID:AB_305869 | 1:100 |
| Antibody | Anti-APC (CC1), mouse monoclonal | Millipore | #OP80, RRID:AB_2057371 | 1:20 |
| Antibody | Anti-NeuN, mouse monoclonal | Abcam | #ab104224, RRID:AB_10711040 | 1:100 |
| Antibody | Anti-Iba1, rabbit polyclonal | FUJIFILM Wako | #019–19741, RRID:AB_839504 | 1:250 |
| Antibody | Anti-mouse IgG, Alexa Fluor 488 goat polyclonal | Invitrogen | #A28175, RRID:AB_2536161 | 1:1000 |
| Antibody | Anti-mouse IgG2b Alexa-Fluor 568 goat polyclonal | Invitrogen | #A21144, RRID: AB_2535780 | 1:1000 |
| Antibody | Anti-rat IgG Alexa Fluor 633 goat polyclonal | Invitrogen | #A21094, RRID: AB_2535749 | 1:1000 |
| Antibody | Anti-rabbit IgG Alexa Fluor 488 goat polyclonal | Invitrogen | #A11008 RRID: AB_143165 | 1:1000 |
| Commercial assay or kit | Adult brain dissociation kit, mouse | Miltenyi | #130-107-677 | |
| Commercial assay or kit | Lightcycler 480 SYBR Green I Master mix | Roche LifeScience | #04707516001 | |
| Chemical compound, drug | ProLong diamond antifade with DAPI | Invitrogen | #36962 | |
| Chemical compound, drug | Paraformaldehyde 32% | Electron Microscopy Sciences | #15,714 S | |
| Chemical compound, drug | Tamoxifen | Millipore Sigma | #T5648 | |
| Chemical compound, drug | Corn oil | Millipore Sigma | #C8267 | |
| Chemical compound, drug | SuperScript III | Thermo Fisher | #18080051 | |
| Chemical compound, drug | SuperScript IV | Thermo Fisher | #18091050 | |
| Chemical compound, drug | TRIzol Reagent | Thermo Fisher | #15596018 | |
| Chemical compound, drug | RNaseOUT | Invitrogen | #10777019 | |
| Chemical compound, drug | T4 polynucleotide kinase | New England Biolabs | #M0201 | |
| Software, algorithm | CaseViewer v2.4 software | 3DHistech | RRID:SCR_017654 | |
| Software, algorithm | Viewer software | Biobserve | RRID:SCR_014337 | |
| Software, algorithm | Volocity v5.3 | Perkin Elmer | RRID:SCR_002668 | |
| Software, algorithm | Prism v9.0 | GraphPad Software | RRID:SCR_002798 | |
| Software, algorithm | ImageQuant v5.2 | GE Healthcare | RRID:SCR_014246 | |
| Software, algorithm | ImageJ v1.53 | Github | RRID:SCR_003070 | |
| Software, algorithm | R studio v1.3.10.93 | Posit | RRID:SCR_000432 | |
| Software, algorithm | DEseq2 | Bioconductor | RRID:SCR_015687 |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/95314/elife-95314-mdarchecklist1-v1.docx
-
Supplementary file 1
RNAseq analysis of WT and Polr3a-tamKI cerebra at P75.
- https://cdn.elifesciences.org/articles/95314/elife-95314-supp1-v1.xlsx
-
Supplementary file 2
Mouse cell type-specific DE genes in Polr3a-tamKI cerebra at P75.
- https://cdn.elifesciences.org/articles/95314/elife-95314-supp2-v1.xlsx
-
Supplementary file 3
RNAseq analysis of WT and Polr3a-tamKI cerebra at P42.
- https://cdn.elifesciences.org/articles/95314/elife-95314-supp3-v1.xlsx
-
Supplementary file 4
RNAseq of WT and Polr3a-tamKI cerebella at P42.
- https://cdn.elifesciences.org/articles/95314/elife-95314-supp4-v1.xlsx
-
Supplementary file 5
tRNAseq P42_Ca. tRNAseq results and analysis of P42 cerebra.
- https://cdn.elifesciences.org/articles/95314/elife-95314-supp5-v1.xlsx
-
Supplementary file 6
tRNAseq analysis of WT and Polr3a-tamKI cerebella at P42.
- https://cdn.elifesciences.org/articles/95314/elife-95314-supp6-v1.xlsx
-
Supplementary file 7
Oligonucleotide sequences.
- https://cdn.elifesciences.org/articles/95314/elife-95314-supp7-v1.xlsx





