Drosophila epidermal cells are intrinsically mechanosensitive and modulate nociceptive behavioral outputs
Figures
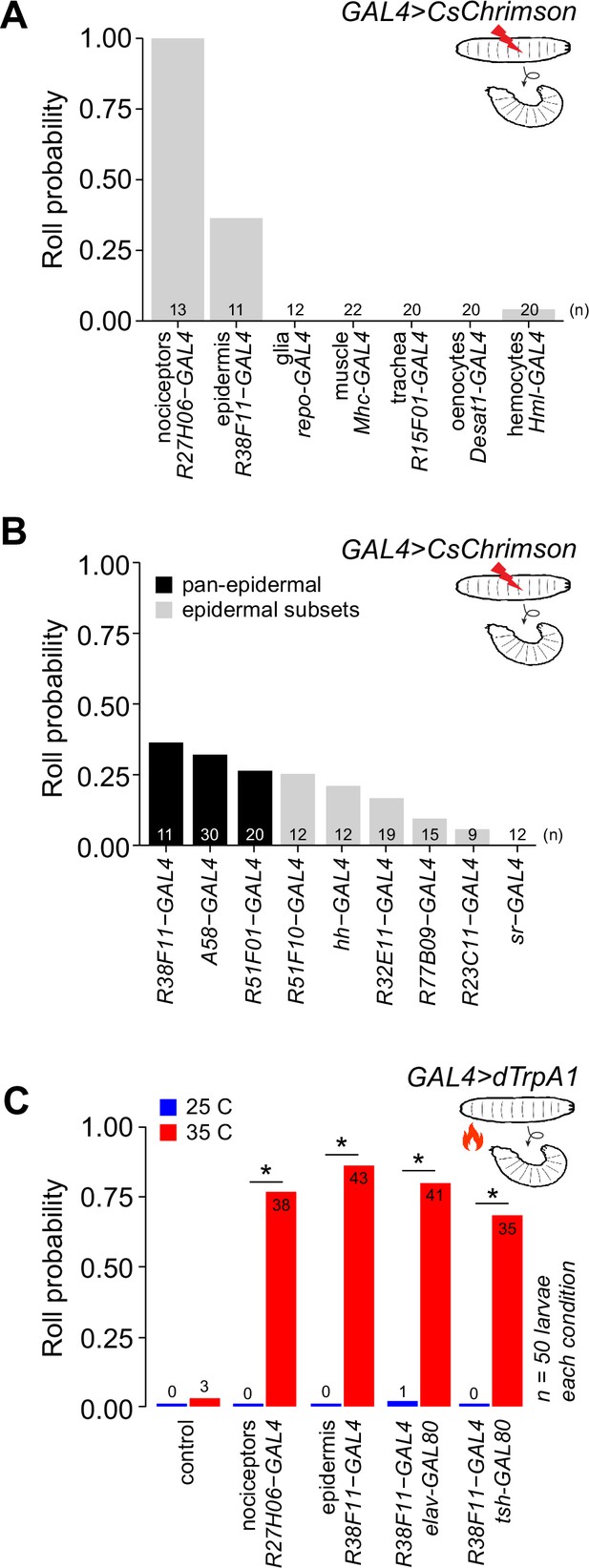
Stimulation of epidermal cells elicits nociceptive behaviors.
(A) Fraction of larvae that exhibited optogenetic-induced rolling (roll probability) using the indicated GAL4 lines to drive UAS-CsChrimson expression. All experimental genotypes, except for larvae expressing UAS-CsChrimson in class IV dendrite arborization (C4da) neurons, included elav-GAL80 to suppress neuronal GAL4 activity. Genotypes: GAL4, UAS-CsChrimson, elav-GAL80/+. (B) Roll probability of larvae following optogenetic stimulation using the indicated GAL4 lines in combination with elav-GAL80 (or tsh-GAL80+cha-GAL80 in the case of A58-GAL4) to drive UAS-CsChrimson expression in epidermal cells. All epidermal drivers except for sr-GAL4, which is expressed in apodemes but no other epidermal cells, elicited rolling responses. Genotypes: GAL4, UAS-CsChrimson, elav-GAL80/+. (C) Roll probability of larvae following thermogenetic stimulation using the indicated GAL4 line to express the warmth (35°C)-activated UAS-TrpA1. The number of rolling larvae (out of 50) is indicated for each group. Genotypes: GAL4, UAS-TrpA1, GAL80 (as indicated)/+. Control, UAS-TrpA1/+. 50 larvae were tested for each genotype/stimulus combination; the number of rolling larvae is indicated on each bar. Asterisk (*) indicates p<0.05 in this and subsequent figures. Raw data for all figures is provided in Source data 1 and details of statistical analyses, including tests performed, p-values, and q-values are provided in Supplementary file 1.
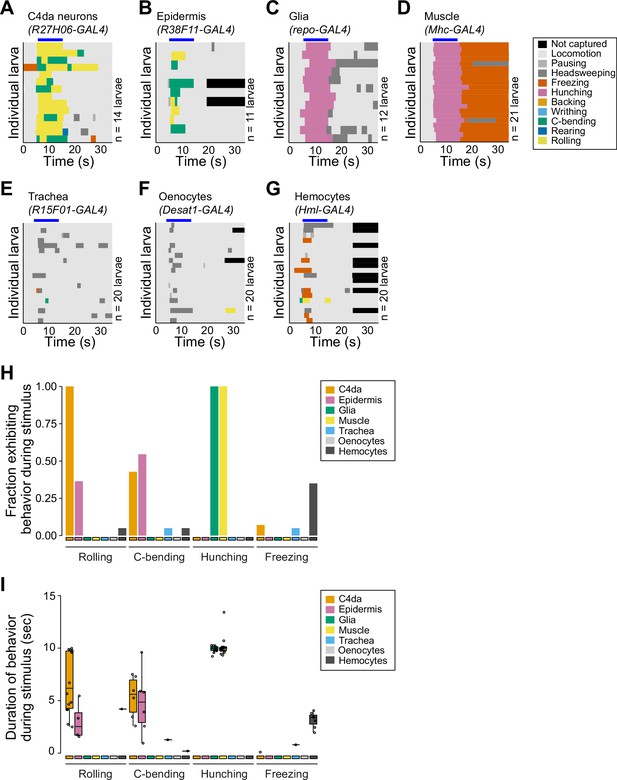
Related to Figure 1A.
Detailed behavior analyses of larvae following optogenetic stimulation using the indicated GAL4 lines to express UAS-CsChrimson. (A–G) Behavior ethograms show behaviors of individual larvae (rows) prior to, during, and after application of the light stimulus (indicated by the blue bar above the ethogram). (H) Fraction of larvae exhibiting indicated behaviors and (I) duration of indicated behaviors during light stimulus. The number of larvae tested is indicated for each driver.
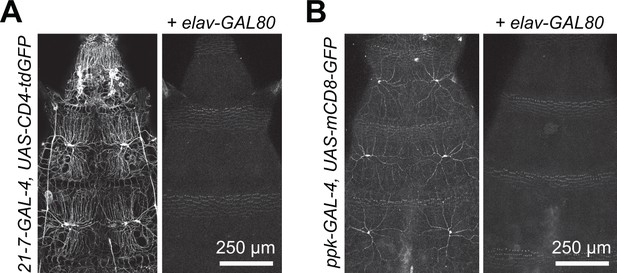
Related to Figure 1A.
Efficacy of GAL80 transgenes. Maximum intensity projections show body wall expression patterns of (A) the pan-da neuron driver GAL421-7 and (B) the nociceptor-specific driver ppk-GAL4 with or without elav-GAL80 to silence neuronal GAL4 expression. Note that elav-GAL80 completely suppresses GFP reporter expression from each of the drivers. Genotypes: (A) GAL421-7 UAS-CD4-tdGFP/+ and GAL421-7 UAS-CD4-tdGFP/+, elav-GAL80/+, (B) ppk-GAL4, UAS-CD4-tdGFP/+ and ppk-GAL4, UAS-CD4-tdGFP/+, elav-GAL80/+.
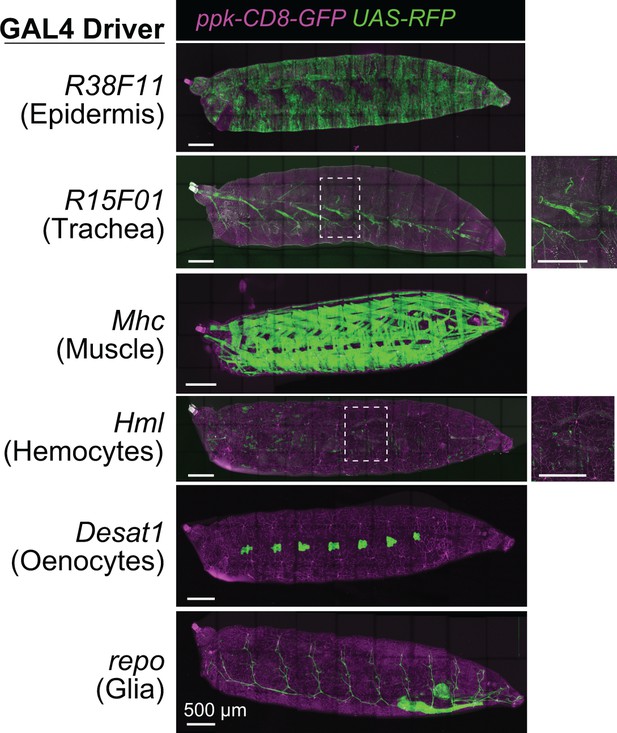
Related to Figure 1A and C.
Expression patterns of body wall GAL4 drivers. Maximum intensity projections of tiled confocal stacks show larval expression of membrane-targeted RFP (UAS-mCD2-Cherry) by the indicated drivers in larvae additionally expressing the class IV dendrite arborization (C4da) neuron-specific marker ppk-CD8-GFP. Scale bars, 500 µm. Genotypes: ppk-mCD8-GFP/+; UAS-mCD2-Cherry/+; GAL4/+.
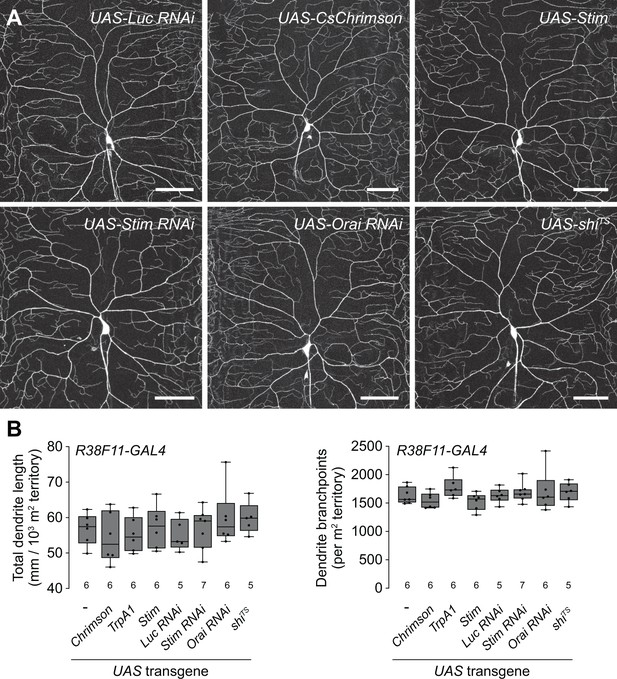
Related to Figure 1A.
Epidermal manipulations have no effect on class IV dendrite arborization (C4da) neuron dendrite morphogenesis. (A) Maximum intensity projections show dendrites of representative C4da neurons labeled with ppk-CD4-tdGFP in the indicated treatment groups. (B) Morphometric analysis of C4da dendrites. Box plots depict the total dendrite length (left) and the number of dendrite branch points (right) normalized to larval segmental area in larvae containing the pan-epidermal driver R38F11-GAL4 and the indicated UAS-transgenes at 120 hr AEL. Prior to imaging, ppk-CD4-tdGFP/+, R38F11-GAL4/UAS-shiTS larvae were incubated for 10 min at 30°C, as in Figure 6N. N>5 neurons for each genotype, points represent measurements from individual neurons, boxes display the first and third quartiles, hatches mark medians, and whiskers mark maximum and minimum values. ANOVA with post hoc Tukey’s test revealed no significant difference between control and treatment groups for both metrics. The number of larvae tested is indicated for each genotype. Genotypes: ppk-CD4-tdGFP/+; R38F11-GAL4/+ without (-) or with a single copy of the indicated UAS-transgenes.
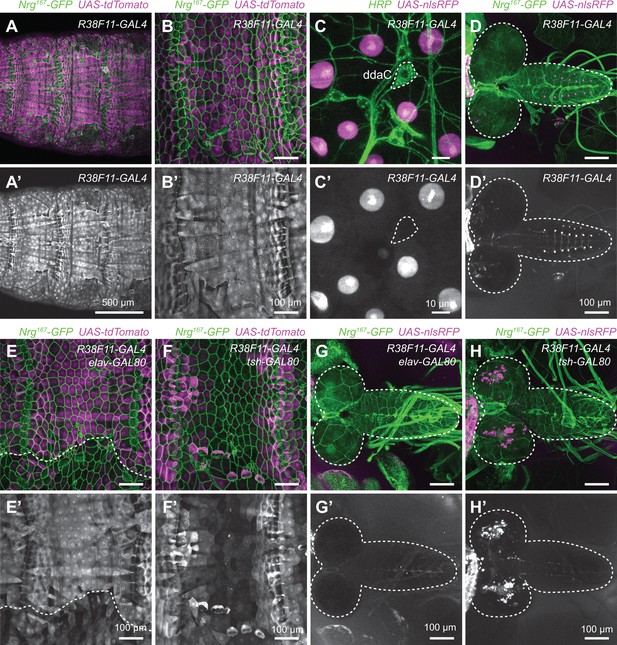
Related to Figure 1A and C.
Expression analysis of the pan-epidermal R38F11-GAL4 driver. (A–B) Maximum intensity projections of confocal stacks show larval expression of a red fluorescent protein (UAS-tdTomato) under control of R38F11-GAL4 in larvae additionally expressing Nrg167-GFP, an exon trap line that labels epidermal and glial membranes (Morin et al., 2001; Yamamoto et al., 2006). (A) Low-magnification view showing stereotyped expression in dorsal epidermis across multiple larval segments. Larvae are oriented dorsal-up. R38F11-GAL4 is likewise expressed throughout the ventral and lateral epidermis. Compared to other epidermal drivers, R38F11-GAL4 exhibited more uniform expression from segment to segment and among epidermal subpopulations within a given segment. (B) R38F11-GAL4 expression in the dorsal epidermis of a single abdominal segment, A2. (C) High-resolution images of R38F11-GAL4 expression visualized using a nuclear localized red fluorescent protein (UAS-NLS-RFP). Images depict expression in larval skin territory containing dorsal cluster of somatosensory neurons (SSNs) (visualized by HRP immunoreactivity), which includes the nociceptive neuron ddaC (outlined by dashed lines). Note that although R38F11-GAL4 is expressed in epidermal cells, expression is undetectable in da neurons. The R38F11-GAL4 expression domain likewise excludes SSNs throughout the body wall. (D) R38F11-GAL4 expression of UAS-NLS-RFP in the larval central nervous system (CNS). R38F11-GAL4 is expressed in a single motor neuron in each segment of the ventral ganglion, ~10 VNC interneurons and <50 additional neurons in each brain hemisphere. (E–H) GAL80-mediated refinement of R38F11-GAL4 expression. (E) Tsh-GAL80-induced variegation in epidermal R38F11-GAL4 expression and (F) suppressed VNC expression while expanding brain expression domains of R38F11-GAL4. (G) Elav-GAL80 likewise induced epidermal variegation in R38F11-GAL4 expression but (H) completely attenuated CNS expression. Dashed lines in (D, G, and H) outline the larval brain and ventral ganglion. Genotypes: (A–B) NrgG00305/+; UAS-tdTomato/+; GAL4GMR38F11/+, (C) GAL4GMR38F11/+, UAS-NLS-RedStinger/+, (E–F) NrgG00305/+; UAS-tdTomato/tsh-GAL80; GAL4GMR38F11/+, (G–H) NrgG00305/+; UAS-NLS-RedStinger/+; GAL4GMR38F11/elav-GAL80.
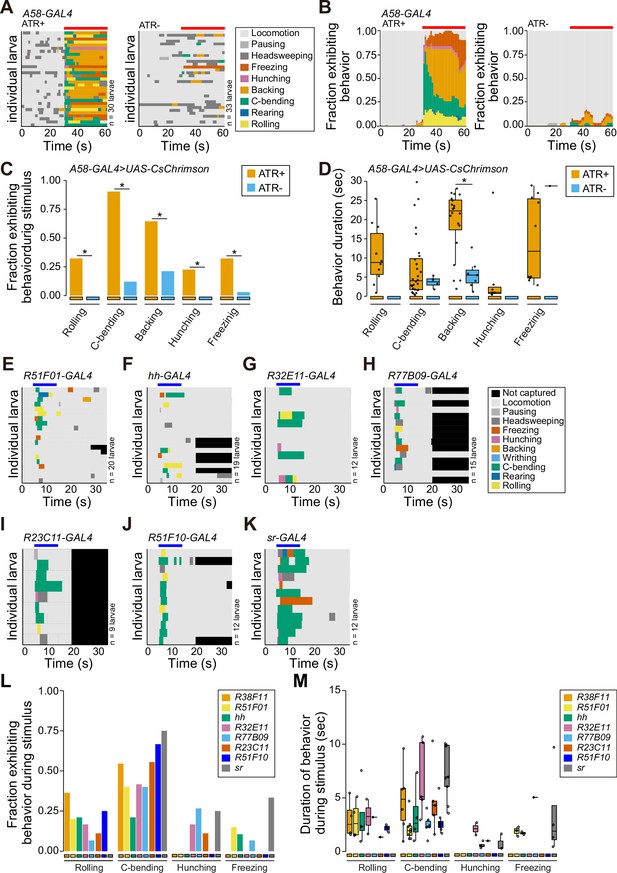
Related to Figure 1B.
(A–B) Behavior ethograms of larvae following pan-epidermal stimulation (denoted with a red bar) mediated by A58-GAL4. (C) Fraction of larvae exhibiting indicated behaviors and (I) duration of indicated behaviors during optogenetic stimulation (genotype: A58-GAL4, UAS-CsChrimson, tsh-GAL80, cha-GAL80/+). (E–K) Behavioral ethograms of larvae following optogenetic stimulation (denoted with a blue bar) with a panel of epidermal GAL4 drivers. (L) Fraction of larvae exhibiting indicated behaviors and (M) duration of indicated behaviors during light stimulus. The number of larvae tested is indicated next to each ethogram. Genotype: GAL4, UAS-CsChrimson, elav-GAL80/+.
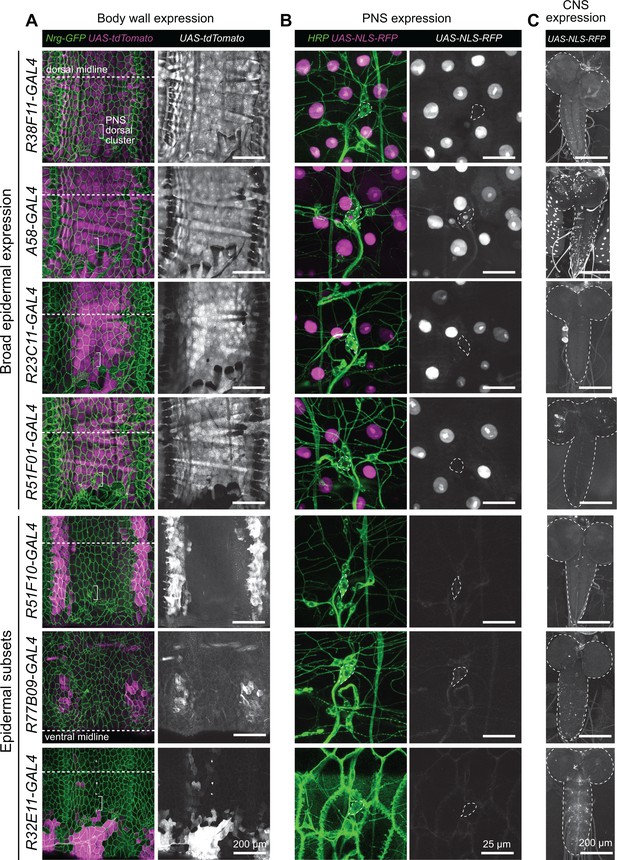
Related to Figure 1B.
Expression patterns of epidermal GAL4 drivers in the larval body wall and central nervous system (CNS). (A) Maximum intensity projections of confocal stacks show larval expression of a red fluorescent protein (UAS-tdTomato) under control of the indicated epidermal drivers in larvae additionally expressing Nrg167-GFP, an exon trap line that labels epidermal and glial membranes (Morin et al., 2001; Yamamoto et al., 2006). White brackets mark the location of the dorsal cluster of somatosensory neurons (SSNs), which are visualized at high resolution in (B). (B) Maximum intensity projections of confocal stacks show larval expression of a nuclear-localized form of RFP (UAS-RedStinger) under control of the indicated GAL4 drivers in larval fillets stained with fluorescently conjugated anti-HRP antibody to label sensory neurons. The nociceptive class IV dendrite arborization (C4da) neuron soma is outlined with a white hatched line in composite images and the outline is superimposed on images depicting UAS-RedStinger signal. Sensory neuron expression is undetectable for all epidermal drivers except for A58-GAL4, for which stochastic sensory neuron expression has been previously described (Jiang et al., 2014). (C) Maximum intensity projections show CNS expression epidermal drivers used in this study. Aside from A58-GAL4, each driver exhibits sparse nervous system expression. Genotypes: (A) NrgG00305/+; UAS-tdTomato/+; GAL4/+, (B–C) UAS-NLS-RFP/+; GAL4/+.
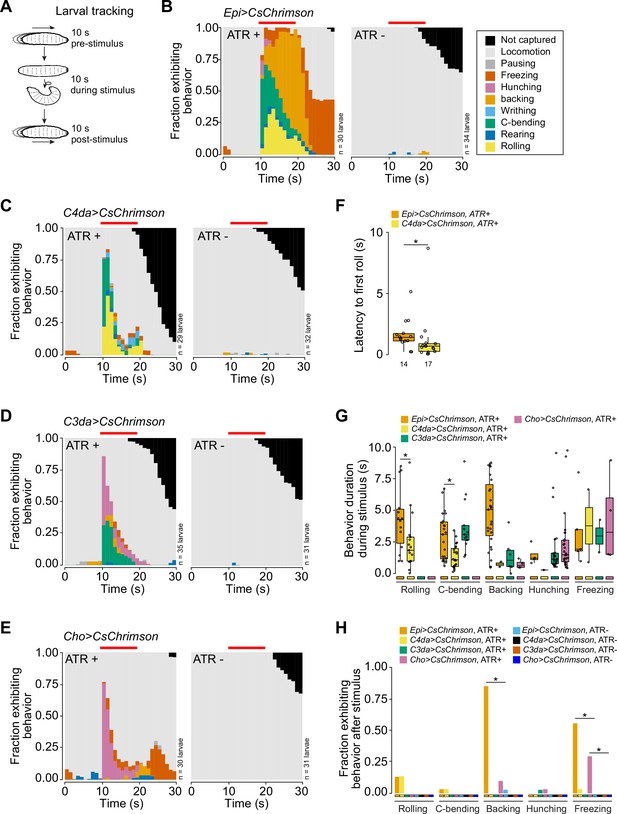
Stimulation of epidermal cells evokes multimodal behavioral responses.
(A) Larval behaviors were scored for 10 s before, during, and after optogenetic stimulation. (B–E) Fraction of larvae exhibiting indicated behaviors over time in 1 s bins expressing CsChrimson in (B) epidermal cells, (C) class IV dendrite arborization (C4da) neurons, (D) C3da neurons, and (E) Cho neurons in the presence and absence of all-trans retinal (ATR). Red line indicates the presence of light stimulation. (F) The latency to the first roll of the larvae that rolled from Epi>Chrimson ATR+ and C4da>Chrimson ATR+ treatment groups (n=14, 17, respectively). In this and subsequent box plots, points represent measurements from individual neurons, boxes display the first and third quartiles, hatches mark medians, and whiskers mark 1.5 times the interquartile range. (G) The duration of indicated behaviors of the larvae that displayed those behaviors during optogenetic stimulation. (H) The fraction of larvae that exhibited indicated behaviors following removal of the light stimulus of all larvae from panels (B–E). The number of larvae tested for each genotype/condition is indicated (B–E). Genotypes: GAL4, UAS-CsChrimson/+.

Ethograms and kinetic analysis of behavioral responses to epidermal stimulation.
(A–E) Behavior ethograms depict behaviors of individual larvae displayed in 1 s bins, scored for 10 s before, during, and after optical stimulus. Plots depict responses of larvae which contain a single copy of UAS-CsChrimson together with the indicated GAL4 driver in the absence and presence of ATR (A–D), or responses of effector-only (UAS-CsChrimson/+, ATR+) controls (E). (F) Plot depicts the fraction of larvae exhibiting indicated behaviors during light stimulus. (G) Latency to the first bend, hunch, back and freeze behaviors following optogenetic stimulation. The number of larvae tested for each genotype/condition is indicated. Genotypes: GAL4, UAS-CsChrimson/+.
Behavioral response of representative larva to optogenetic epidermal stimulation.
Movies were captured under infrared light and annotated with behaviors that were scored post hoc. Nociceptive behaviors (indicated in red) precede non-nociceptive behaviors (blue). Genotype: R38F11-GAL4, UAS-CsChrimson/+.
Behavioral response of representative larva to optogenetic nociceptor stimulation.
Genotype: ppk-GAL4, UAS-CsChrimson/+.
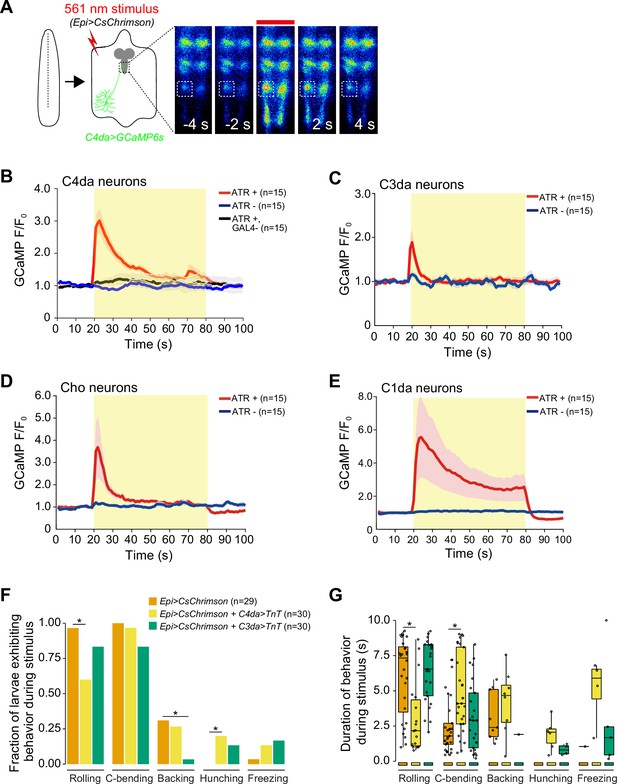
Optogenetic epidermal stimulation elicits somatosensory neuron activation.
(A) Optogenetic activation of CsChrimson-expressing epidermal cells in the body wall triggers calcium transients in the axon terminal of GCaMP6s-expressing nociceptive somatosensory neurons (SSNs). Images show responses from one representative animal. Plots depict mean GCaMP6s fluorescence intensity of the axon terminals of (B) class IV dendrite arborization (C4da), (C) C3da, (D) Cho, and (E) C1da neurons following optogenetic activation (light stimulus, yellow box) of epidermal cells over time. Solid lines depict mean GCaMP6s fluorescence across replicates (n=15 larval fillet preparations), shading indicates SEM, red traces are GAL4+ ATR+ , blue traces are GAL4+ ATR-, black trace is GAL4- ATR+. (F) The fraction of larvae exhibiting indicated behaviors during optogenetic epidermal stimulation in combination with SSN silencing via tetanus toxin (TnT) expression. We note that although baseline rolling probability is elevated in all genetic backgrounds containing the AOP-LexA-TnT insertion, silencing C4da and C3da neurons significantly attenuates responses to epidermal stimulation. (G) The duration of the behavioral responses during optogenetic epidermal stimulation with neuronal TnT expression. The number of larvae tested for each genotype/condition is indicated. Genotypes: (A–B) R27H06-LexA (C4da neurons), AOP-GCaMP6s, UAS-CsChrimson/+; R38F11-GAL4/+ or R27H06-LexA (C4da neurons), AOP-GCaMP6s, UAS-CsChrimson/+ (GAL4-ATR- effector-only control); (C) AOP-GCaMP6s, UAS-CsChrimson/+; R38F11-GAL4/NompC-LexA (C3da neurons); (D) UAS-GCaMP6s, AOP-CsChrimson, R61D08-GAL4 (Cho neurons)/R38F11-LexA; (E) UAS-GCaMP6s, AOP-CsChrimson, R11F05-GAL4 (C1da neurons)/R38F11-LexA; (F–G) R38F11-GAL4, UAS-CsChrimson, AOP-LexA-TnT/+ (Epi>CsChrimson); R38F11-GAL4, UAS-CsChrimson, AOP-LexA-TnT/ppk-LexA (Epi>CsChrimson+C4da>TnT); R38F11-GAL4, UAS-CsChrimson, AOP-LexA-TnT/NompC-LexA (Epi>CsChrimson+C3da>TnT).
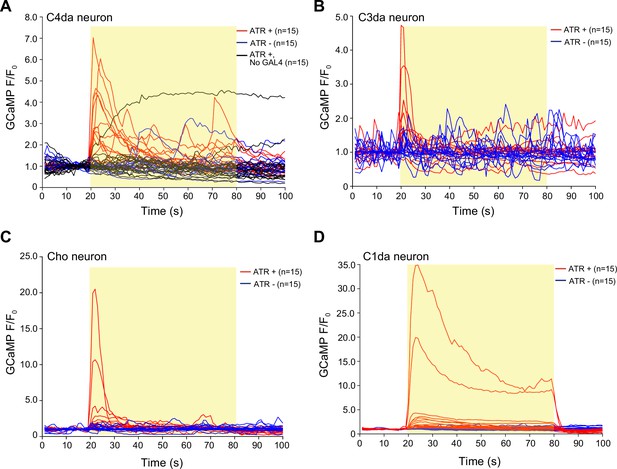
Related to Figure 3A–D.
GCaMP6s responses in axon terminals of (B) class IV dendrite arborization (C4da), (C) C3da, (D) Cho, and (E) C1da neurons following optogenetic activation (light stimulus, yellow box) of epidermal cells over time. Each trace represents the GCaMP6s fluorescence of an individual larval fillet. Red traces are GAL4+ ATR+ , blue traces are GAL4+ ATR-, black trace is GAL4- ATR+. The number of larvae tested for each genotype/condition is indicated. Genotypes: (A) R27H06-LexA, AOP-GCaMP6s, UAS-CsChrimson/+; R38F11-GAL4/+ or R27H06-LexA, AOP-GCaMP6s, UAS-CsChrimson/+ (GAL4-ATR- effector-only control); (B) AOP-GCaMP6s, UAS-CsChrimson/+; R38F11-GAL4/NompC-LexA; (C) UAS-GCaMP6s, AOP-CsChrimson, R61D08-GAL4/R38F11-LexA; (D) UAS-GCaMP6s, AOP-CsChrimson, R11F05-GAL4/R38F11-LexA.
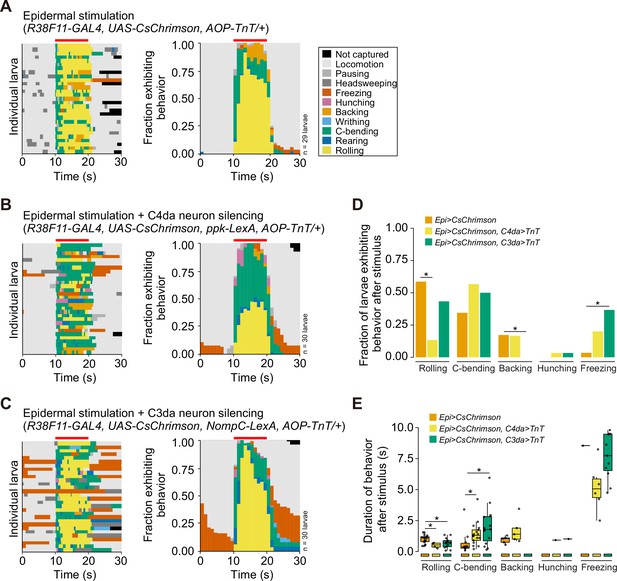
Related to Figure 3E and F.
(A–C) Behavior ethograms (left) and fraction of larvae (right) exhibiting indicated behaviors to optogenetic epidermal stimulation in combination with (A) an AOP-TNT transgene (control lacking LexA driver), (B) class IV dendrite arborization (C4da) neuron silencing via tetanus toxin (TnT), and (C) C3da neuron silencing via TnT. (D) Fraction of larvae exhibiting each behavior and (E) the duration of behavior responses after removal of the light stimulus. The number of larvae tested for each genotype is indicated. Genotypes are indicated in (A–C).
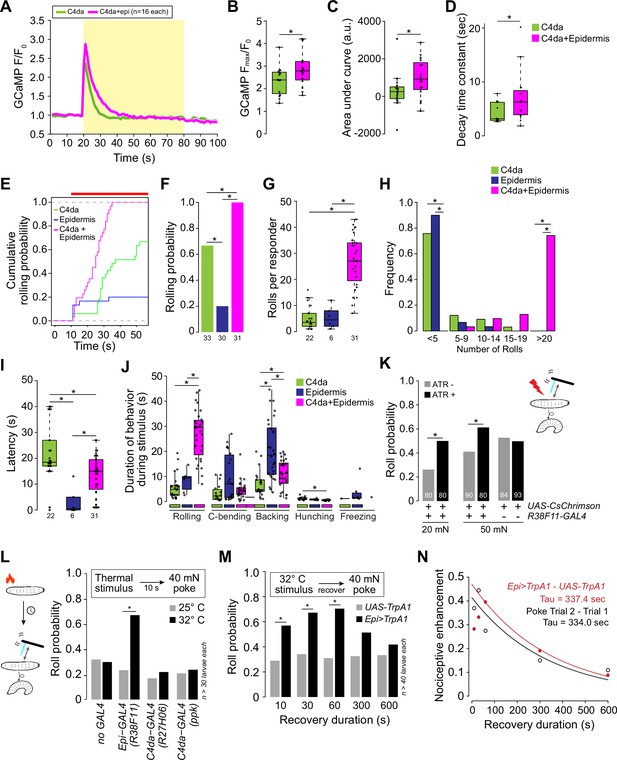
Epidermal stimulation augments nociceptive responses.
(A) Mean GCaMP6s responses (F/F0) in class IV dendrite arborization (C4da) axons during optogenetic stimulation (yellow box) of C4da neurons alone (green) or of C4da neurons and epidermal cells (magenta), shading indicates SEM. (B) Simultaneous epidermal stimulation increased the peak calcium response (Fmax/F0), (C) total calcium influx (area under the curve), and (D) duration of C4da neuron calcium responses compared to stimulation of C4da neurons alone. Genotypes: ppk-LexA, AOP-GCAMP6s/+; R27H06-GAL4/UAS-CsChrimson (C4da) and ppk-LexA, AOP-GCAMP6s/+; R27H06-GAL4/R38F11-GAL4, UAS-CsChrimson (C4da+epi). (E–J) Characterization of the behavioral responses to low-intensity optogenetic stimulation of C4da neurons, epidermal cells, or simultaneous C4da neurons and epidermal cells. (E) Cumulative and (F) total roll probability during optogenetic stimulation (indicated by the red bar). n=33 (C4da>CsChrimson), 30 (Epi>CsChrimson), and 31 (C4da+Epi>CsChrimson) larvae. (G, H) Number and frequency distribution of rolls, (I) latency to the first roll observed for larvae of the indicated genotypes, and (J) the duration of the indicated behaviors during light stimulus. Genotypes: UAS-CsChrimson/+; R27H06-GAL4/+ (C4da), UAS-CsChrimson/+; R38F11-GAL4/+ (Epidermis), UAS-CsChrimson/+; R27H06-GAL4/R38F11-GAL4 (C4da +Epidermis). (K) Roll probability of larvae to a 20 mN or 50 mN von Frey mechanical stimulus and epidermal optogenetic activation (a light stimulus, 1.16 μW/mm2 that was insufficient on its own to induce nocifensive rolling). Larvae were reared in the presence or absence of ATR, as indicated. Genotypes: UAS-CsChrimson/+; R38F11-GAL4/+. (L–N) Prior epidermal but not nociceptor stimulus potentiates mechanical nociceptive responses. (L) Roll probability of control larvae (UAS-TrpA1/+) or larvae expressing TrpA1 in the epidermis (Epi-GAL4: R38F11-GAL4) or C4da neurons (UAS-TrpA1/+; C4da-GAL4 #1: R27H06-GAL4, UAS-TrpA1/+; C4da-GAL4 #2: ppk-GAL4, UAS-TrpA1/+), or control larvae (no GAL4: UAS-TrpA1/+;) in response to 40 mN mechanical stimulus 10 s following 10 s of a thermal stimulus (25°C or 32°C). To control for effects of genetic background, we confirmed that each of the experimental genotypes exhibited mechanically induced nociceptive sensitization (Figure 4—figure supplement 1C). (M) Roll probability of control larvae (UAS-TrpA1/+) or larvae expressing TrpA1 in the epidermis (Epi>TrpA1: R38F11-GAL4, UAS-TrpA1/+) in response to a 40 mN mechanical stimulus delivered at the indicated time interval following a 32°C thermal stimulus. (N) Nociceptive enhancement (difference in the roll probability to the first and second stimulus) is plotted against the recovery duration and results were fit to an exponential curve to derive the decay time constant. The red line indicates nociceptive enhancement of a mechanical stimulus by a prior epidermal thermogenetic stimulus; the black line indicates nociceptive enhancement by a prior mechanical stimulus. The number of larvae tested for each genotype/condition is indicated.
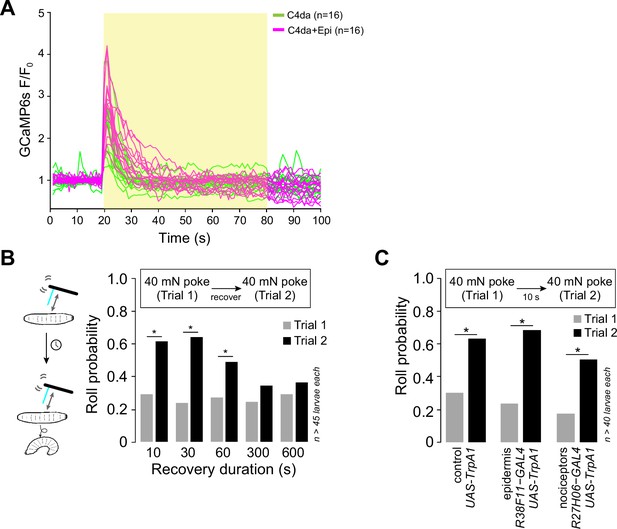
Epidermal stimulation induces persistent sensitization of nociceptors.
(A) GCaMP6s responses (F/F0) of individual replicates during optogenetic stimulation (yellow box) of class IV dendrite arborization (C4da) neurons alone (green) or C4da neurons and epidermal cells (magenta), yellow box indicates presence of light stimulus. Genotypes: ppk-LexA, AOP-GCAMP6s/+; R27H06-GAL4/UAS-CsChrimson (C4da) and ppk-LexA, AOP-GCAMP6s/+; R27H06-GAL4/R38F11-GAL4, UAS-CsChrimson (C4da+epi). (B) Roll probability of larvae in response to two successive 40 mN mechanical stimuli spaced by the indicated amount of time (recovery duration). Genotype: R38F11-GAL4/+. (C) Roll probability of larvae expressing UAS-TrpA1 in nociceptors exhibited levels of mechanically induced nociceptive potentiation comparable to other experimental genotypes used in the study (control: UAS-TrpA1/+; epidermis: R38F11-GAL4, UAS-TrpA1/+; nociceptors: R27H06-GAL4, UAS-TrpA1/+). The number of larvae tested for each genotype/condition is indicated.
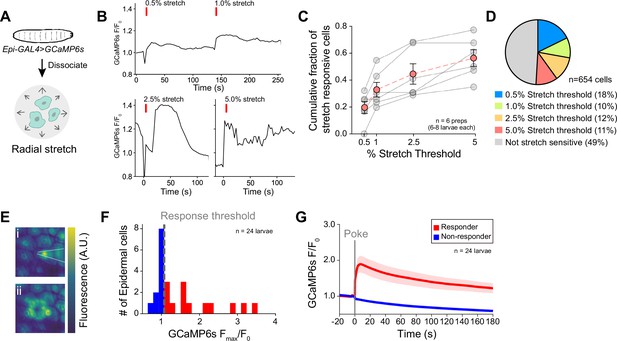
Epidermal cells are intrinsically mechanosensitive.
(A) Schematic of preparation to measure radial stretch evoked calcium responses of dissociated epidermal cells. (B) Representative calcium responses of a dissociated epidermal cell to 0.5% and 1% radial stretch (successive stimuli), 2.5% radial stretch, and 5% radial stretch. (C) Dose-response curve displaying the fraction of epidermal cells activated by increasing magnitudes of stretch. Red trace displays the mean ± SEM across six independent dissociated cell preparations, obtained from a minimum of six larvae. Gray traces display fraction responding in each dissociated cell preparation replicate. (D) Subsets of epidermal cells display varying stretch thresholds, n=6 dissociated cell preparations, for a total of 654 epidermal cells. (E) Representative mechanically induced epidermal calcium responses in the larval body wall. Images show GCaMP6s fluorescence intensity 100 ms prior to (i) and 20 s following (ii) a 25 μm membrane displacement (poke). (F) Distribution of the peak calcium response (Fmax/F0) to a 25 μm membrane displacement (poke) of 24 cells from 24 independent larval fillets. Cells were classified as responders (>10% increase in normalized GCaMP6s fluorescence). (G) Mean calcium responses (F/F0) of poke responders and non-responders (n=12 cells each). Solid lines depict mean normalized GCaMP6s fluorescence and shading indicates SEM. Sample number is indicated for each genotype/condition. Genotype: R38F11-GAL4, UAS-GCaMP6s.
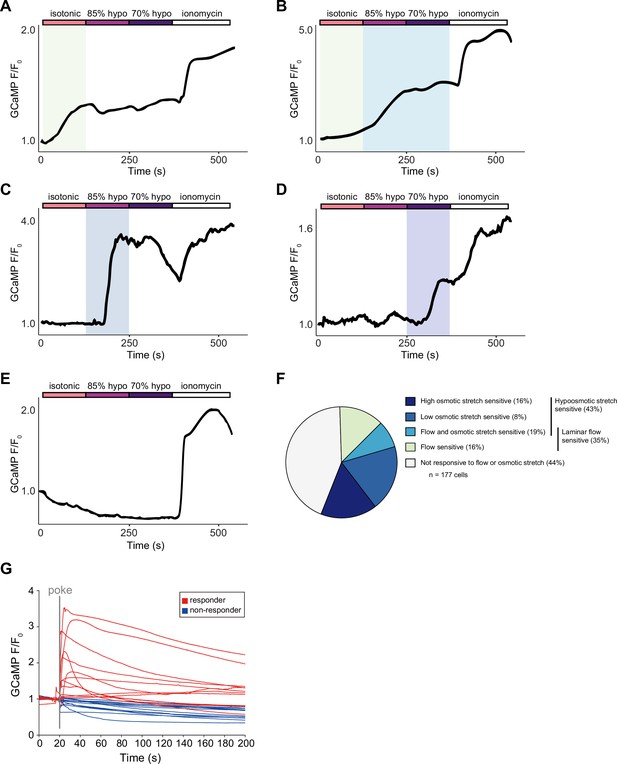
Subsets of epidermal cells display calcium responses to diverse mechanical stimuli.
Representative calcium traces of epidermal cells that respond to: (A) laminar flow via perfusion, (B) laminar flow and osmotic stretch, (C) 15% hypo-osmotic stretch, and (D) 30% hypo-osmotic stretch, or (E) do not respond to either flow or stretch. (F) Proportion of epidermal cells that are sensitive to mechanical stimuli. n=3 distinct cell preparations, 6 larvae per preparation, for a total of 177 cells. (G) Related to Figure 5G. GCaMP6s responses (F/F0) of individual epidermal cells to a 25 μm membrane displacement. Genotype: R38F11-GAL4, UAS-GCaMP6s.
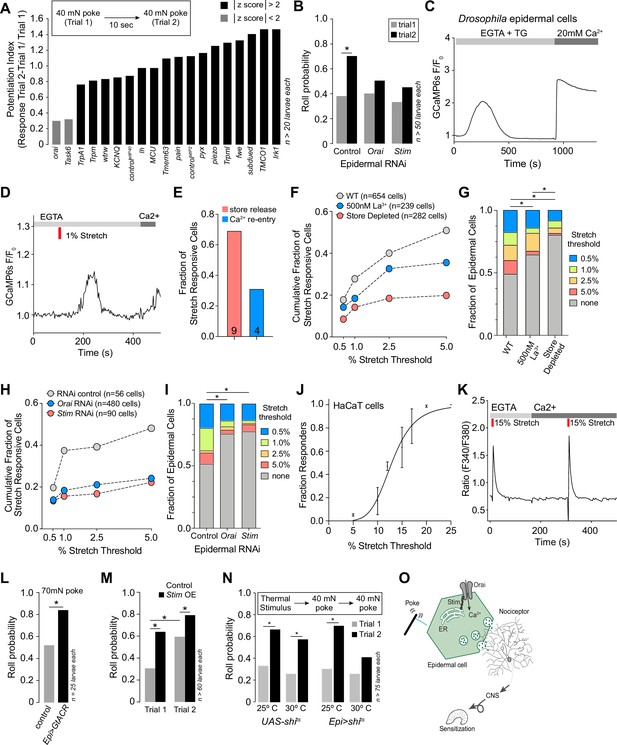
Ca2+ release-activated Ca2+ (CRAC) channels are required for epidermal mechanosensory responses and epidermal nociceptive potentiation.
(A) RNAi screen for epidermal ion channels required for mechanically induced nociceptive potentiation. Bars depict nociceptive potentiation index (difference in the larval roll probability to the first and second mechanical stimuli divided by roll probability to the first mechanical stimulus). Candidate channels were chosen for further analysis if they had a z-score greater than 2 (absolute value). (B) The CRAC channels Orai and Stim are required in epidermal cells for mechanically evoked nociceptive potentiation. Roll probability of larvae of the indicated genotypes (control RNAi, R38F11-GAL4, UAS-RFP-RNAi/+; Stim RNAi, R38F11-GAL4, UAS-Stim-RNAi/+; Orai RNAi, R38F11-GAL4, UAS-Orai-RNAi/+) to a 40 mN mechanical stimulus followed by a second 40 mN mechanical stimulus 10 s later. (C) Drosophila epidermal cells display classical store-operated calcium entry (SOCE). Treatment with the drug thapsigargin (TG) in the absence of extracellular calcium promoted depletion of intracellular calcium stores and calcium influx, following extracellular calcium re-addition. (D) Like TG, 1% stretch in the absence of extracellular calcium-induced depletion of intracellular calcium stores and calcium influx, following extracellular calcium re-entry. (E) 69% of stretch-responsive cells displayed greater calcium influx during intracellular calcium stores release than during the calcium re-entry phase. (F–G) The Orai blocker, lanthanum chloride (500 nM), or the depletion of intracellular stores by TG (1 µM) reduces the fraction of stretch-sensitive epidermal cells. (H–I) The fraction of stretch-sensitive epidermal cells is significantly decreased in cells isolated from larvae expressing Stim RNAi, or Orai RNAi, as compared to control RNAi. (J) Stretch stimuli evoke dose-dependent calcium signals in the human keratinocyte HaCaT cell line. (K) Representative stretch-evoked SOCE calcium response in HaCaT cells. Stretch induces calcium release from stores in the absence of extracellular calcium and a greater calcium influx in the presence of extracellular calcium. (L) Epidermal hyperpolarization enhances mechanical nocifensive responses. Roll probability of larvae expressing GtACR in epidermal cells (R38F11-GAL4, UAS-GtACR/+) or control larvae (R38F11-GAL4/+) to a single 70 mN mechanical stimulus. (M) Epidermal Stim overexpression enhances mechanical nocifensive responses. Roll probability of Stim-overexpressing larvae (R38F11-GAL4, UAS-Stim/+) and control larvae (R38F11-GAL4/+) to two successive 40 mN mechanical stimuli delivered 10 s apart. (N) Epidermal potentiation of mechanical nociceptive responses requires exocytosis. Roll probability of control larvae (UAS-shits/+) or larvae expressing temperature-sensitive dominant-negative shi in epidermal cells (R38F11-GAL4, UAS-shits/+) in response to two successive mechanical stimuli that followed 10 min of conditioning at the permissive (25°C) or non-permissive (30°C) temperature. (O) Model of epidermal-neuronal signaling. Mechanically evoked Stim/Orai calcium signaling in epidermal cells drives calcium influx and vesicle release that drives nociceptor activation and mechanical sensitization via activation of class IV dendrite arborization (C4da) nociceptors. Sample number is indicated for each genotype/condition.
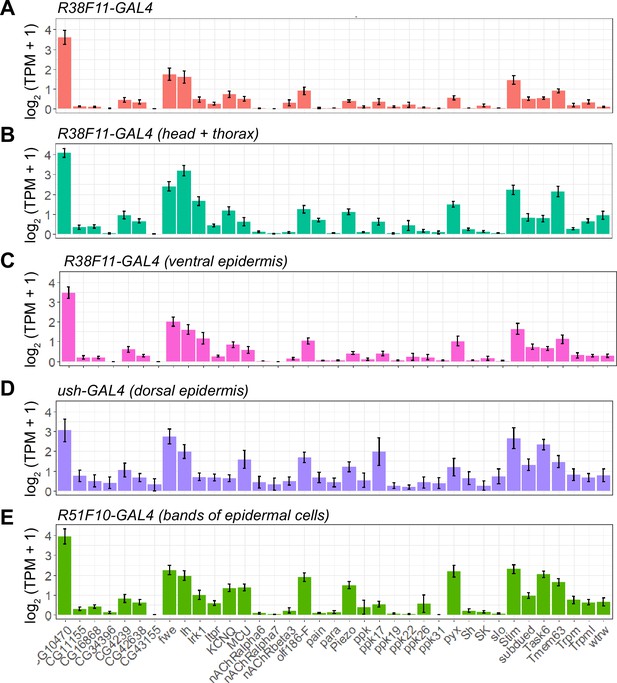
Related to Figure 6A.
Ion channel expression in epidermal cells. Five populations of UAS-GFP-expressing epidermal cells were selected for RNA-seq analysis. GFP-positive epidermal cells were dissociated into single-cell suspensions, manually picked, and subjected to RNA-seq analysis. Plots depict expression levels (mean ± standard deviation) of the indicated ion channels (log2(TPM+1)) from n>6 independent libraries for each sample type. (A) Expression in epidermal cells collected from R38F11-GAL4 which labels all epidermal cells. (B) Anatomically defined subsets of epidermal cells were profiled by removing posterior segments from R38F11-GAL4, UAS-GFP larvae (R38F11-GAL4 head and thorax), or (C) dissecting the ventral epidermis (R38F11-GAL4 ventral epidermis). (D) Expression in the dorsal epidermis (ush-GAL4) and/or (E) bands of epidermal cells (R51F10-GAL4).
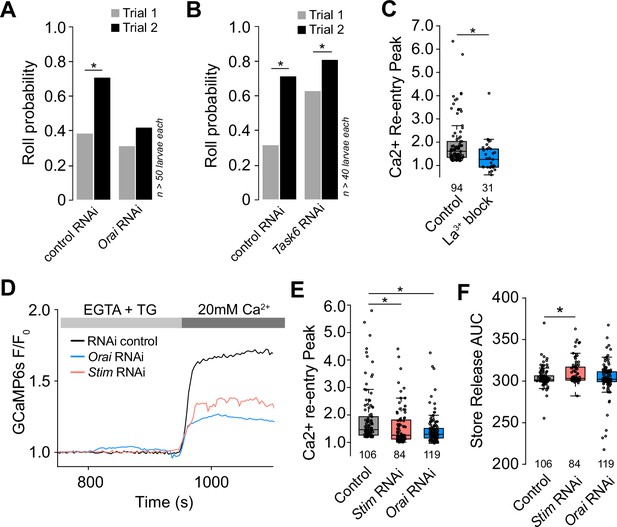
Mechanically evoked responses of epidermal cells involve SOCE.
(A–B) Epidermal knockdown of ion channels affects larval mechanical nociceptive responses. Roll probability in response to the first and second mechanical stimulus for larvae expressing RNAi transgenes to (A) Orai or attP40 RNAi control transgene and (B) Task6 or attP2 RNAi control transgene. (C) Epidermal cell store-operated calcium entry (SOCE) is attenuated by pre-treatment with the Orai blocker, lanthanum chloride (100 nM). (D) Representative SOCE after treatment with thapsigargin (TG) (1 μm) in epidermal cells isolated from larvae treated with control RNAi, Stim RNAi (red), or Orai RNAi, (blue). (E) Peak calcium response following SOCE in epidermal cells isolated from larvae treated with control RNAi, Stim RNAi (red), or Orai RNAi (blue). (F) Stim RNAi decreases calcium store content of epidermal cells. Calcium store content was measured as the area under the curve of the cytosolic calcium response to TG (1 μm) in the absence of extracellular calcium. Sample number is indicated for each genotype/condition.
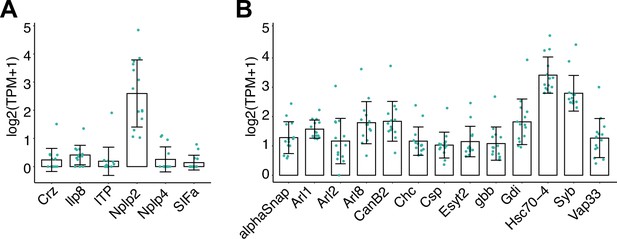
Epidermal expression of neuropeptide genes and neuropeptide release machinery.
Plots depict expression levels (log2(TPM+1) mean ± standard deviation) of (A) neuropeptide genes and (B) genes involved in neuropeptide release. n = 14 independent libraries of epidermal cells labeled by R38F11-GAL4.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | Stim | GenBank | FLYB:FBgn 0045073 | |
| Gene (D. melanogaster) | Orai | GenBank | FLYB:FBgn 0041585 | |
| Genetic reagent (D. melanogaster) | A58-GAL4 | Maintained in the Parrish Lab | Flybase_FBal0181674 | GAL4 driver (epidermis) |
| Genetic reagent (D. melanogaster) | Desat1-GAL4 | Bloomington Drosophila Stock Center | BDSC_65405 | GAL4 driver (oenocytes) |
| Genetic reagent (D. melanogaster) | GMR15F01-GAL4 | Bloomington Drosophila Stock Center | BDSC_45071 | GAL4 driver (trachea) |
| Genetic reagent (D. melanogaster) | GMR23C11-GAL4 | Bloomington Drosophila Stock Center | BDSC_45123 | GAL4 driver (epidermis) |
| Genetic reagent (D. melanogaster) | GMR27H06-GAL4 | Bloomington Drosophila Stock Center | BDSC_49440 | GAL4 driver (C4da neurons) |
| Genetic reagent (D. melanogaster) | GMR32E11-GAL4 | Bloomington Drosophila Stock Center | BDSC_48109 | GAL4 driver (epidermis) |
| Genetic reagent (D. melanogaster) | GMR38F11-GAL4 | Bloomington Drosophila Stock Center | BDSC_50014 | GAL4 driver (epidermis) |
| Genetic reagent (D. melanogaster) | GMR51F01-GAL4 | Bloomington Drosophila Stock Center | BDSC_38787 | GAL4 driver (epidermis) |
| Genetic reagent (D. melanogaster) | GMR51F10-GAL4 | Bloomington Drosophila Stock Center | BDSC_38793 | GAL4 driver (epidermis) |
| Genetic reagent (D. melanogaster) | GMR61D08-GAL4 | Bloomington Drosophila Stock Center | BDSC_39272 | GAL4 driver (Cho neurons) |
| Genetic reagent (D. melanogaster) | GMR77B09-GAL4 | Bloomington Drosophila Stock Center | Flybase_FBti0138381 | GAL4 driver (epidermis) |
| Genetic reagent (D. melanogaster) | hh-GAL4 | Bloomington Drosophila Stock Center | DGGR_118117 | GAL4 driver (epidermis) |
| Genetic reagent (D. melanogaster) | Hml-GAL4 | Bloomington Drosophila Stock Center | BDSC_6395 | GAL4 driver (hemocytes) |
| Genetic reagent (D. melanogaster) | MHC-GAL4 | Bloomington Drosophila Stock Center | BDSC_55132 | GAL4 driver (muscle) |
| Genetic reagent (D. melanogaster) | NompC-GAL4 | Bloomington Drosophila Stock Center | BDSC_36361 | GAL4 driver (C3da neurons) |
| Genetic reagent (D. melanogaster) | Piezo-GAL4 | Bloomington Drosophila Stock Center | BDSC_78335 | GAL4 driver (piezo-expressing cells) |
| Genetic reagent (D. melanogaster) | ppk-GAL4 | Bloomington Drosophila Stock Center | BDSC_32079 | GAL4 driver (C4da neurons) |
| Genetic reagent (D. melanogaster) | 21-7-GAL4 | Maintained in the Parrish Lab | Flybase_FBti0131369 | GAL4 driver (MD neurons) |
| Genetic reagent (D. melanogaster) | repo-GAL4 | Bloomington Drosophila Stock Center | BDSC_7415 | GAL4 driver (glia) |
| Genetic reagent (D. melanogaster) | sr-GAL4 | Bloomington Drosophila Stock Center | BDSC_26663 | GAL4 driver (apodemes) |
| Genetic reagent (D. melanogaster) | ppk-LexA | Maintained in the Parrish Lab | Flybase_FBtp0125814 | LEXA driver (C4da neurons) |
| Genetic reagent (D. melanogaster) | NompC-LexA | Bloomington Drosophila Stock Center | BDSC_52241 | LEXA driver (C3da neurons) |
| Genetic reagent (D. melanogaster) | elav-GAL80 | Maintained in the Parrish Lab | Flybase_FBtp0079702 | GAL80 (pan-neuronal) |
| Genetic reagent (D. melanogaster) | tsh-GAL80 | Bloomington Drosophila Stock Center | Flybase_FBti0114123 | GAL80 (VNC) |
| Genetic reagent (D. melanogaster) | ush-GAL4 | Bloomington Drosophila Stock Center | BDSC_36524 | GAL4 driver (epidermis) |
| Genetic reagent (D. melanogaster) | Nrg167GFP | Bloomington Drosophila Stock Center | BDSC_6844 | Reporter (epidermal cell junctions) |
| Genetic reagent (D. melanogaster) | UAS-tdTomato | Bloomington Drosophila Stock Center | BDSC_36328 | Reporter (RFP) |
| Genetic reagent (D. melanogaster) | UAS-RedStinger | Bloomington Drosophila Stock Center | BDSC_8546 | Reporter (NLS-RFP) |
| Genetic reagent (D. melanogaster) | UAS0GCaMP6s | Bloomington Drosophila Stock Center | BDSC_42749 | Reporter (calcium indicator) |
| Genetic reagent (D. melanogaster) | UAS-GCaMP6s | Bloomington Drosophila Stock Center | BDSC_42746 | Reporter (calcium indicator) |
| Genetic reagent (D. melanogaster) | AOP-GCaMP6s | Bloomington Drosophila Stock Center | BDSC_44273 | Reporter (calcium indicator) |
| Genetic reagent (D. melanogaster) | AOP-TNT | Maintained in the Parrish Lab | Flybase_FBtp0144631 | Neuronal silencing (tetanus toxin) |
| Genetic reagent (D. melanogaster) | UAS-shi-ts | Bloomington Drosophila Stock Center | BDSC_44222 | Inducible inhibition of endocytosis |
| Genetic reagent (D. melanogaster) | UAS-CsChrimson | Bloomington Drosophila Stock Center | BDSC_55135 | Inducible cation channel |
| Genetic reagent (D. melanogaster) | UAS-CsChrimson | Bloomington Drosophila Stock Center | BDSC_55136 | Inducible cation channel |
| Genetic reagent (D. melanogaster) | UAS-TRPA1 | Bloomington Drosophila Stock Center | BDSC_26263 | Inducible cation channel |
| Genetic reagent (D. melanogaster) | UAS-GtACR | Bloomington Drosophila Stock Center | BDSC_92983 | Inducible anion channel |
| Genetic reagent (D. melanogaster) | UAS-luciferase RNAi | Bloomington Drosophila Stock Center | BDSC_31603 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-RFP-RNAi | Bloomington Drosophila Stock Center | BDSC_67852 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-fwe-RNAi | Bloomington Drosophila Stock Center | BDSC_27323 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-Ih-RNAi | Bloomington Drosophila Stock Center | BDSC_29574 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-Ih-RNAi | Bloomington Drosophila Stock Center | BDSC_58089 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-Irk1-RNAi | Bloomington Drosophila Stock Center | BDSC_42644 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-KCNQ-RNAi | Bloomington Drosophila Stock Center | BDSC_80446 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-MCU-RNAi | Bloomington Drosophila Stock Center | BDSC_67897 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-orai-RNAi | Bloomington Drosophila Stock Center | BDSC_53333 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-orai-RNAi | Vienna Drosophila Resource Center | VDRC_12221 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-pain-RNAi | Bloomington Drosophila Stock Center | BDSC_51835 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-piezo-RNAi | Vienna Drosophila Resource Center | FlyBase_FBst0457216 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-piezo-RNAi | Kyoto Drosophila Stock Center | Flybase_FBtp0071516 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-pyx-RNAi | Bloomington Drosophila Stock Center | BDSC_31297 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-stim-RNAi | Bloomington Drosophila Stock Center | BDSC_41759 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-stim-RNAi | Vienna Drosophila Resource Center | FlyBase_FBst0478081 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-subdued-RNAi | Vienna Drosophila Resource Center | FlyBase_FBst0480747 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-Task6-RNAi | Bloomington Drosophila Stock Center | BDSC_28016 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-Task6-RNAi | Vienna Drosophila Resource Center | FlyBase_FBst0471360 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-Tmem63-RNAi | Vienna Drosophila Resource Center | FlyBase_FBst0470667 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-TMCO1-RNAi | Bloomington Drosophila Stock Center | BDSC_42896 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-TMCO1-RNAi | Bloomington Drosophila Stock Center | BDSC_55909 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-TrpA1-RNAi | Bloomington Drosophila Stock Center | BDSC_66905 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-Trpm-RNAi | Bloomington Drosophila Stock Center | BDSC_31291 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-Trpml-RNAi | Bloomington Drosophila Stock Center | BDSC_31294 | RNAi transgene |
| Genetic reagent (D. melanogaster) | UAS-wtrw-RNAi | Bloomington Drosophila Stock Center | BDSC_51563 | RNAi transgene |
| Cell line (Homo sapiens) | Immortalized Human Keratinocytes (HaCaT cells) | Cytion | Cat. #: 300493 | |
| Peptide, recombinant protein | Collagenase type I | Thermo Fisher | Cat. #: 17-100-017 | |
| Chemical compound, drug | Fura-2AM | Thermo Fisher | Cat. #: F1221 | |
| Chemical compound, drug | Pluronic F-127 | Thermo Fisher | Cat. #: P3000MP | |
| Chemical compound, drug | All-trans retinal | Millipore Sigma | Cat. #: R2500 | |
| Chemical compound, drug | Ionomycin | Thermo Fisher | Cat. #: I24222 | |
| Chemical compound, drug | Lanthanum chloride | Millipore Sigma | Cat. #: 211605 | 500 nM |
| Software, algorithm | SPSS | SPSS | RRID:SCR_002865 | |
| Software, algorithm | MATLAB | Mathworks | RRID:SCR_001622 | |
| Software, algorithm | ImageJ | ImageJ | RRID:SCR_003070 | |
| Software, algorithm | CASAVA | Illumina, http://www.illumina.com/software/genome_analyzer_software.ilmn | ||
| Software, algorithm | FasQC | Babraham bioinformatics, https://www.bioinformatics.babraham.ac.uk/ | ||
| Software, algorithm | Cutadapt v2.4 | Martin, 2011 | ||
| Software, algorithm | Kallisto version 0.46.0 | Bray et al., 2016 |
Additional files
-
Supplementary file 1
Details of statistical analyses performed for this study, including comparison groups, statistical tests, and results.
- https://cdn.elifesciences.org/articles/95379/elife-95379-supp1-v1.pdf
-
Supplementary file 2
Larval behaviors scored in this study, including a description and scoring criteria for each behavior.
- https://cdn.elifesciences.org/articles/95379/elife-95379-supp2-v1.pdf
-
Supplementary file 3
Recipes for solutions used in imaging and physiology experiments.
- https://cdn.elifesciences.org/articles/95379/elife-95379-supp3-v1.pdf
-
Source data 1
Raw data for each experiment sorted by figure panel.
- https://cdn.elifesciences.org/articles/95379/elife-95379-data1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/95379/elife-95379-mdarchecklist1-v1.pdf





