Coordination of cell cycle and morphogenesis during organ formation
Figures
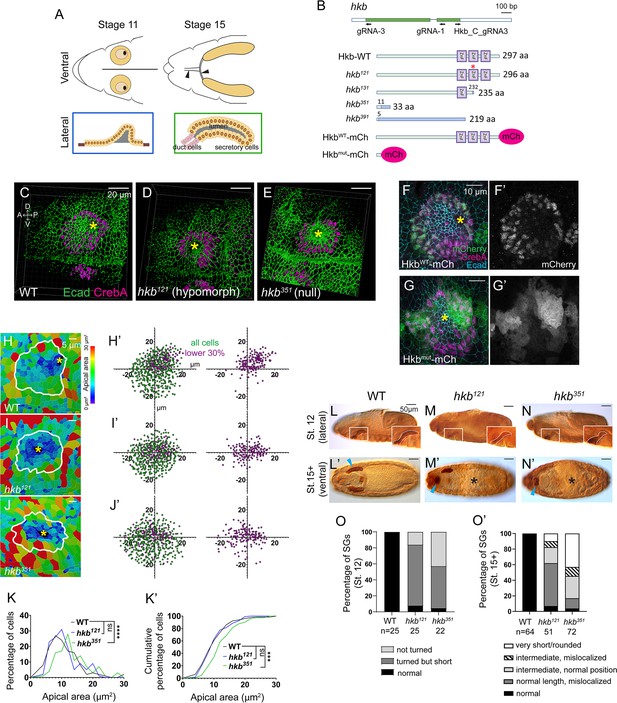
Generation of new huckebein (hkb) alleles and fluorescent knock-in lines reveals its function and tissue-specific localization during salivary gland (SG) morphogenesis.
(A) Top cartoon diagram: Anterior region of stage 11 and 15 Drosophila embryos. SGs shown in yellow. Brown area, clustered apical constriction of SG cells during invagination. Black, invagination pit. Arrowheads, salivary duct. Bottom cartoon diagram: Lateral view of SG at stages 11 and 15. Orange circles, SG cell nuclei. Gray, SG lumen. (B) Top cartoon diagram: hkb transcript displayed with coding exon in green and gRNAs used for hkb alleles and mCh knock-in lines. Bottom: Hkb protein structures produced in each allele and knock-in lines. In hkb121, two amino acids (N237 and E238) are substituted with K237 (asterisk). In hkb351 and hkb391, a short N-terminal region of the Hkb protein (green) is followed by random amino acid sequences (blue) due to a frameshift. hkb mRNA with the premature stop codon in hkb351 might be degraded by nonsense-mediated mRNA decay. ZnF, zinc finger domain. (C–E) Three-dimensional (3D) reconstruction of the confocal images of invaginating SGs immunolabeled with E-Cadherin (Ecad) and CrebA. Asterisks, invagination pit. (F-G’) Confocal images of stage 11 SGs immunostained for mCh (green), CrebA (magenta), and Ecad (cyan). (H–J) Heat maps showing apical area distribution in invaginating SGs. White lines, SG boundary determined by SG-specific CrebA signals. Yellow asterisks, invagination pits. (H’-J’) Scatter plots showing the relative position of cells in the SG placode. X and Y axes represent the distance along the A/P and D/V axes, respectively, from the center of the placode. Cells in the bottom 30% of the apical area (magenta) and all cells (green) are plotted (n=5 SGs from different embryos (WT, 572 cells; hkb121, 505 cells; hkb351, 498 cells)). (K, K’) Quantification of the apical area distribution in the SG cells of H-J. Mann-Whitney U test (percentage of cells) and Kolmogorov-Smirnov test (cumulative percentage of cells). **p<0.01; ***p<0.001; ****p<0.0001. (L-N’) Embryos immunostained for CrebA, with magnified views (insets) showing SGs outlined by white dashed lines. Cyan arrowheads, late-stage SGs. Asterisks, defective gut morphology. (O, O’) Quantification of the phenotypic distribution of SGs. n, number of embryos. Statistical significance was determined using two-way ANOVA (p<0.05).
© 2024, Matthew et al. Figure 1 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.

Indels of the CRISPR/Cas9 huckebein (hkb) alleles and predicted protein sequences for each allele.
(A) The nucleotide sequence of the hkb-RA transcript. The open reading frame (ORF) is marked in blue. Three zinc-finger (ZnF) domains are highlighted in green (ZnF1), yellow (ZnF2), and magenta (ZnF3), respectively. Two gRNA sequences used to create hkb alleles are indicated in bold and underlined. (B) CRISPR/Cas9-induced indels in hkb mutants. Nucleotides deleted and inserted in each mutant are indicated by dashed lines and red letters, respectively. (C) Sequences of the wild-type (WT) Hkb protein and predicted proteins in each hkb mutant.
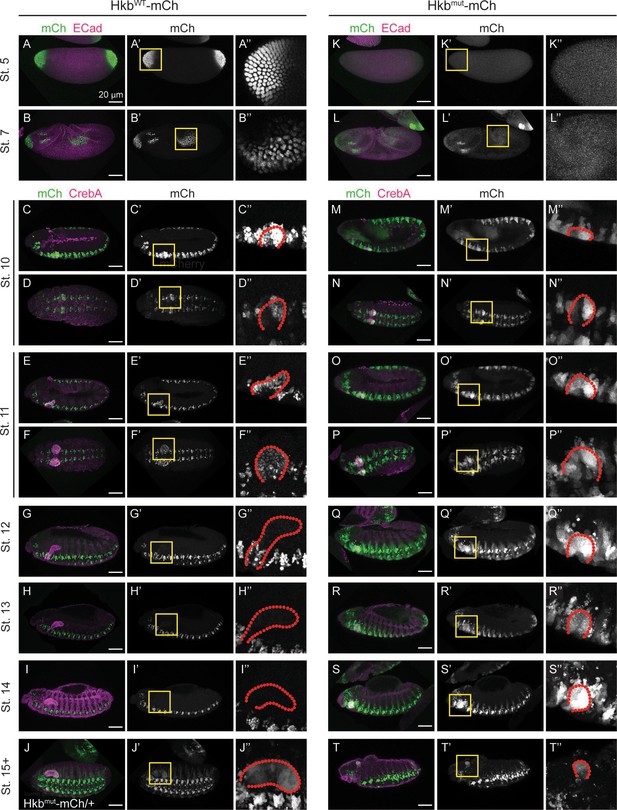
Hkbwt-mCh and Hkbmut-mCh signals during embryonic development.
(A-T’’) Hkbwt-mCh (A-I’’) and Hkbmut-mCh (J-T’’) expression patterns in embryos at different stages. Yellow-boxed regions in A’-T’ are shown in higher magnification in A’’-T’’. Salivary glands (SGs) are outlined by red dotted lines in M’’-T’’. (A-A’’) At stage 5, Hkbwt-mCh signals localize to the nuclei of the cells at the anterior and posterior poles of the embryo. (B-B’’) At stage 7, Hkbwt-mCh signals are seen in the nuclei of the anterior region and in the posterior midgut of the embryo. (C-D’’) At stage 10, strong nuclear Hkbwt-mCh signals are seen along the CNS and in the posterior region of the SG of the embryo. mCherry signals localize to the nuclei of the cells. (C-C’’) Lateral view. (D-D’’) Dorsal view. (E-F’’) At stage 11, strong nuclear Hkbwt-mCh signals are seen in all SG cells. (G-G’’) At stage 12, Hkbwt-mCh signals are significantly reduced in SG nuclei, while mCh signals are consistently high in CNS cells. (H-H’’) At stage 13, very low levels of Hkbwt-mCh signals are seen in SG nuclei. (I-I’’) At stage 14, Hkbwt-mCh signals of SG nuclei are reduced to almost undetectable levels. (J-T’’) Hkbmut-Ch signals are seen throughout the cell. (J-J’’) mCh signals in Hkbmut-mCh/+heterozygous embryo at stage 16. mCh signals persist in the SG. (M-T’’) mCh signals in Hkbmut-mCh homozygous embryos. mCh signals persist in the SG throughout embryogenesis and are seen throughout the cell.
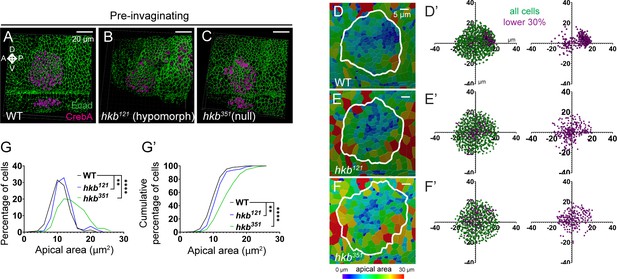
Quantification of apical areas in the pre-invaginating salivary gland (SG) of wild-type (WT) and huckebein (hkb) mutant embryos.
(A–C) Three-dimensional reconstruction of confocal images of pre-invaginating SGs immunolabeled with E-Cadherin (Ecad) and CrebA. (D–F) Heat maps of apical area distribution in pre-invaginating SGs. (D’-F’) Scatter plots showing the relative position of total SG cells (green) and cells with small apical areas (magenta). The X and Y axes represent the distance along the A/P and D/V axes, respectively, from the center of the placode. Cells in the bottom 30% of the apical area (magenta) and all cells (green) are plotted. n=7 SGs (WT, 932 cells; hkb121, 727 cells; hkb351, 864 cells) (G, G’) Quantification of apical area distribution in SG cells in D’-F’ before invagination. Mann-Whitney U test (percentage of cells) and Kolmogorov-Smirnov test (cumulative percentage of cells). **p<0.01; ****p<0.0001.
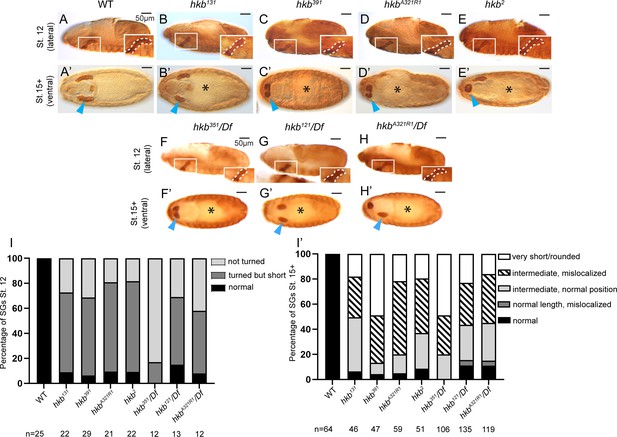
Salivary gland (SG) phenotypes in different huckebein (hkb) alleles.
(A-H’) Stage 12 (A-E; lateral) and stage 15+ (A’-H’; ventral) embryos immunostained for CrebA. The insets show a magnified view of the region in the white rectangles, with the SG outlined by white dashed lines. Cyan arrowheads, SGs in stage 15+ embryos. Asterisks, defective gut morphology. (I, I’) Quantification of the SG phenotypic distribution at stage 12 (I) and stage 15 (I’). The number of embryos used is indicated below each genotype. Statistical significance was calculated by two-way ANOVA. p<0.05.
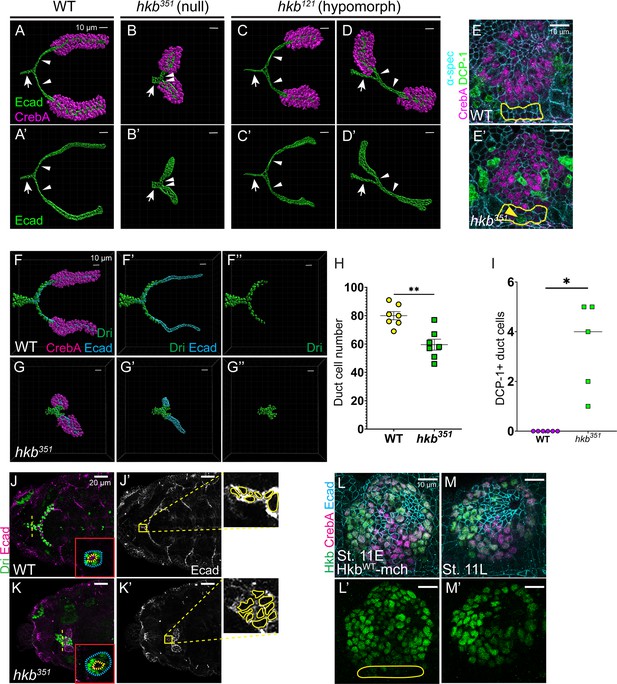
huckebein (hkb) mutants show short salivary duct phenotypes with reduced cell numbers and cell intercalation defects.
(A-D’) Three-Dimensional (3D) reconstruction of the confocal images of stage 15+ salivary glands (SGs) immunolabeled with E-Cadherin (Ecad) (green) and CrebA (magenta). Arrows, common duct. Arrowheads, individual duct. (E-E’) Confocal images of stage 11 SGs immunolabeled with α-spectrin (cyan; lateral membrane), CrebA (magenta), and death caspase-1 (DCP-1) (green). Yellow dotted lines, SG duct cells. Yellow arrowhead, DCP-1-positive duct cell. (F-G’’) 3D reconstruction of the confocal images of stage 15+ SGs immunolabeled with dead ringer (Dri) (green), CrebA (magenta), and Ecad (cyan). (H) Quantification of ductal cell number. n=7. Statistical significance was calculated by Student’s t-test. **p<0.01. (I) Quantification of DCP-1-positive duct cells. n=6 (wild-type, WT); 5 (hkb351). Statistical significance was calculated by Student’s t-test. *p<0.05. (J-K’) Confocal images of SGs immunolabeled with Ecad (magenta) and Dri (green). Z-sections along the yellow dashed lines in H and I are shown as insets. Yellow-boxed regions in H’ and I’ are shown at higher magnification, with the boundary of each salivary duct cell marked with yellow lines. (L-M’) Confocal images of SGs immunolabeled with mCh (green), CrebA (magenta), and Ecad (Cyan). Yellow line, duct cells.
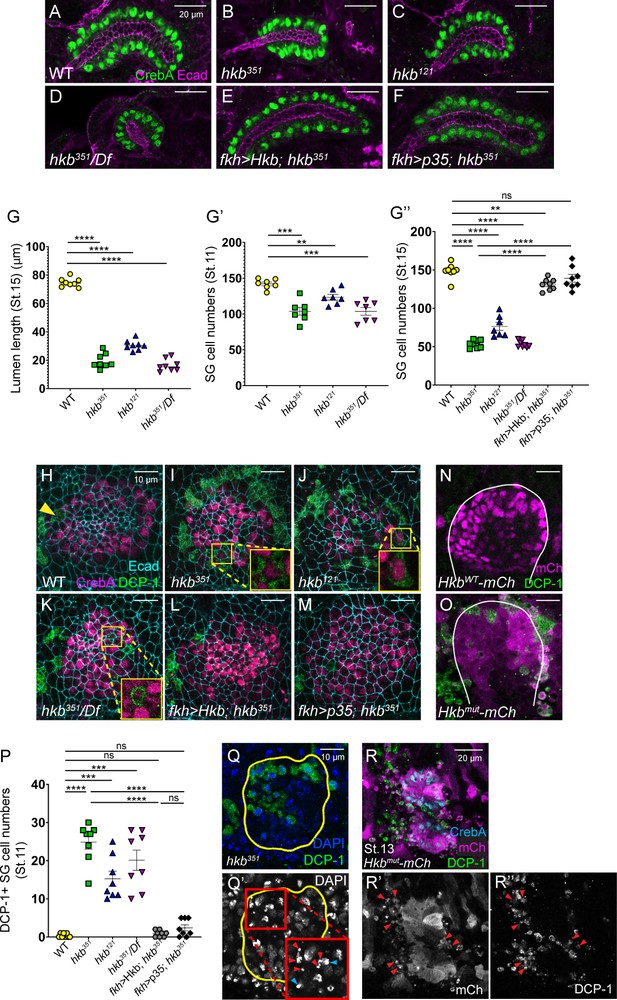
Huckebein (Hkb) is required to prevent apoptosis in salivary gland (SG) cells to ensure proper organ size.
(A–F) Stage 15+ SGs immunolabeled for E-Cadherin (Ecad) (magenta) and CrebA (green). (G) Quantification of the stage 15+ SG lumen length. n=9 (WT), 12 (hkb351), 11 (hkb121), and 8 (hkb351/Df). One-way ANOVA. ****p<0.0001. (G’-G’’) Quantification of the number of SG secretory cells at stage 11 (G’; n=7 for all genotypes) and stage 15 (G’’; n=8 for all genotypes except hkb121 (n=7)). One-way ANOVA. **p<0.01; ***p<0.001; ****p<0.0001. (H–M) Pre-invaginating SGs immunolabeled for death caspase-1 (DCP-1) (green), CrebA (magenta), and Ecad (cyan). Insets, magnified views. (N, O) HkbWT-mCh and Hkbmut-mCh SGs immunostained for mCh (magenta) and DCP-1 (green). White lines, SG boundary determined by SG-specific CrebA signals. (P) Quantification of the DCP-1 positive SG cells. n=8 for all genotypes. One-way ANOVA. ***p<0.001; ****p<0.0001. (Q, Q’) Stage 11 hkb null mutant SG stained for DAPI (blue) and DCP-1 (green). Insets, higher magnification of the red-boxed region. Red arrowheads, fragmented nuclei. Cyan arrowheads, pyknotic nuclei. Yellow lines, SG boundary determined by SG-specific CrebA signals. (R-R’’) SG of a stage 13 Hkbmut-mCh embryo immunostained for mCh (cyan), CrebA (magenta), and DCP-1 (green). Arrowheads, mCh- and DCP-1-positive cells near the SG.
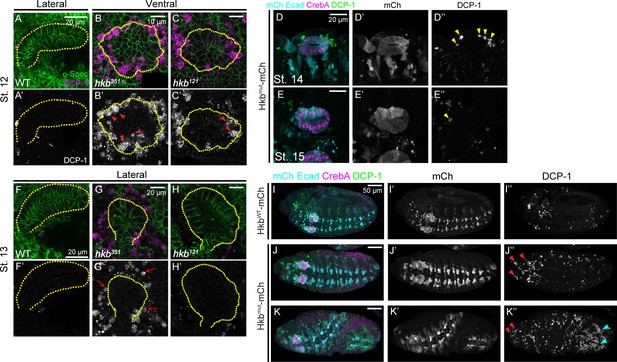
Death caspase-1 (DCP-1) signals in late-stage salivary glands (SGs) and other regions of huckebein (hkb) mutant embryos.
(A-C’) Confocal images of SGs in stage 12wild-type (WT) and hkb mutant embryos immunostained for α-Spec (green) and DCP-1 (magenta). Yellow dotted lines, SG boundary. Arrowheads in B’ and C’, DCP-1 signals in the cells within the SG placode. (D-E’’) Confocal images of SGs in stage 14 and 15 Hkbmut-mCh embryos stained for mCh and E-Cadherin (Ecad) (cyan), CrebA (magenta), and DCP-1 (green). Arrows in J’’, DCP-1-positive cells near the SG at stage 14. (F-H’) Confocal images of SGs in stage 13 WT and hkb mutants immunostained for α-Spec (green) and DCP-1 (magenta). Yellow dotted lines, SG boundary. Arrows in G’, DCP-1 signals near the SG placode. (I-K’’) Confocal images of stage 11 embryos immunostained for Ecad and mCh (cyan), CrebA (magenta), and DCP-1 (green). Hkbmut-mCh embryos show increased DCP-1 signals in other regions of the embryo compared to HkbWT-mCh embryos, with a greater increase in the head region (red arrowheads) and posterior region (cyan arrowheads in I’’).
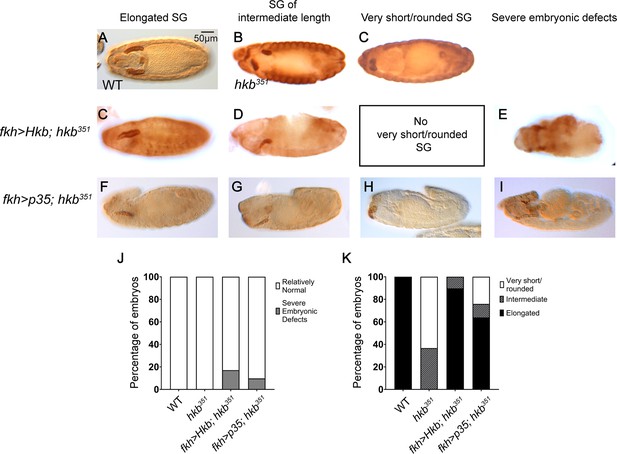
Overexpression of Huckebein (Hkb) or p35 rescues hkb mutant salivary gland (SG) phenotypes.
(A–I) Embryos stained for CrebA. Wild-type (WT) (A), hkb mutant (B, C), overexpression of Hkb (C–E) or p35 (F–I) in the SG using the fkh-Gal4 driver in the hkb mutant background results in a range of the phenotypes consisting mostly of fully elongated SGs as in WT (C, F), some SGs of intermediate length (D, G), and a few SGs with a very short/rounded phenotype as in hkb null mutants (H). Some embryos also show severe morphological defects (E, I). (J–K) Quantification of embryonic (J) and SG (K) morphology. n=25 (WT), 30 (hkb351), 29 (fkh >Hkb; hkb351), 92 (fkh >p35; hkb351).

huckebein (hkb) mutant salivary glands (SGs) have a smaller apical domain size with Rab11 mislocalized to the basolateral domain.
(A–D) Confocal images of SGs in stage 15 embryos immunostained for E-Cadherin (Ecad). Apical domains in the yellow-boxed regions are shown at higher magnification. (E) Quantification of the apical area of individual SG cells. n=8 SGs, 40 cells (WT, hkb351, hkb121); n=7 SGs, 35 cells (hkb351/Df). One-Way ANOVA with multiple comparisons. *p<0.05, ***p<0.001. (F, G) Confocal images of SGs in stage 15 embryos immunostained for Rab11. Magenta-boxed regions are shown at higher magnification. Yellow dotted lines, basal boundary of the SG. Red arrowheads and small punctate Rab11 signals are often observed in the basolateral region of SG cells in hkb mutants.
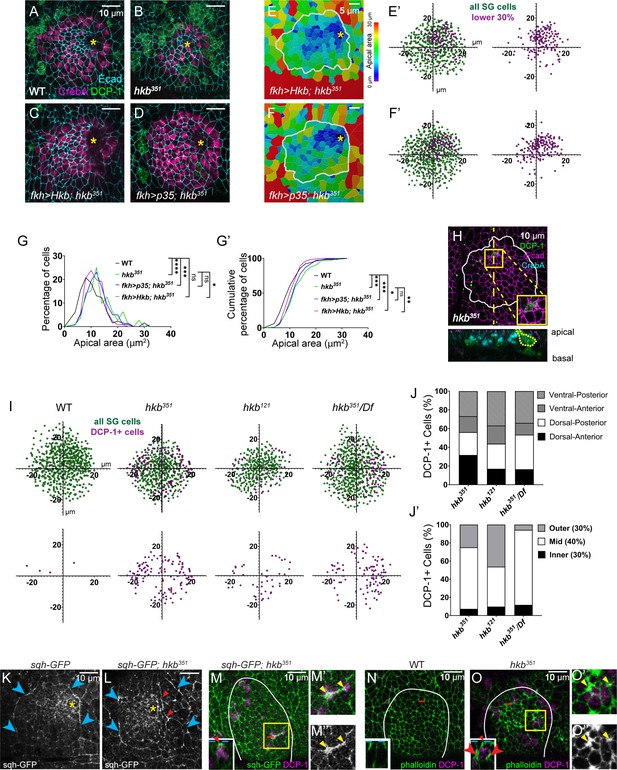
Aberrant cell death results in centered invagination in the huckebein (hkb) mutant salivary gland (SG).
(A–D) Stage 11 SGs immunolabeled with death caspase-1 (DCP-1) (green), CrebA (magenta), and E-Cadherin (Ecad) (cyan). (E–F) Heat maps depicting apical area distribution in invaginating SGs. (E’-F’) Scatter plots showing the relative position of cells in the SG placode. X and Y axes represent the distance along the A/P and D/V axes, respectively, from the center of the placode. Cells in the bottom 30% of the apical area (magenta) and all cells (green) are plotted (n=5 SGs from five different embryos). (G-G’) Quantification of apical area distribution in invaginating SG cells. Mann-Whitney U test (percentage of cells) and Kolmogorov-Smirnov test (cumulative percentage of cells). n=5 SGs for all genotypes (WT (555 cells), hkb351 (488 cells), fkh >Hkb; hkb351 (526 cells)) except for fkh >p35; hkb351 (6 SGs, 632 cells) *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.(H) hkb mutant SG immunostained for DCP-1 (green), Ecad (magenta), and CrebA (cyan). Insets, magnified view of the yellow boxed regions. Z-section along the dotted line displayed below. (I) Scatter plots showing the relative position of total SG cells (green) and DCP-1-positive cells (magenta) in stage 11 SGs. (J,J’) Quantification of the number of DCP-1-positive SG cells in different regions of the placode. (K,L) Stage 11 SGs immunostained for GFP (for sqh-GFP). Cyan arrowheads, supracellular myosin cable. Red arrowheads, short cables inside the SG placode. Yellow asterisk, invagination pit. (M-M’’) Stage 11 SGs immunostained for GFP (for sqh-GFP; green) and DCP-1 (magenta). (N-O’’) Stage 11 SGs stained for F-actin (green) and DCP-1 (magenta). Insets in L-N, z-sections along red dotted lines. Dotted cyan lines, apical boundary of cells. Red arrowheads/arrows indicate increased myosin (M) and F-actin (O) signals in the apical and lateral regions of the DCP-1-positive cell, respectively. (M’, M’’, O’, O’’) Magnified views of yellow boxed regions in M and O. Yellow arrowheads, increased myosin/F-actin signals at the apical-junctional region of DCP-1-positive cells. White lines, SG boundary determined by SG-specific CrebA signals.
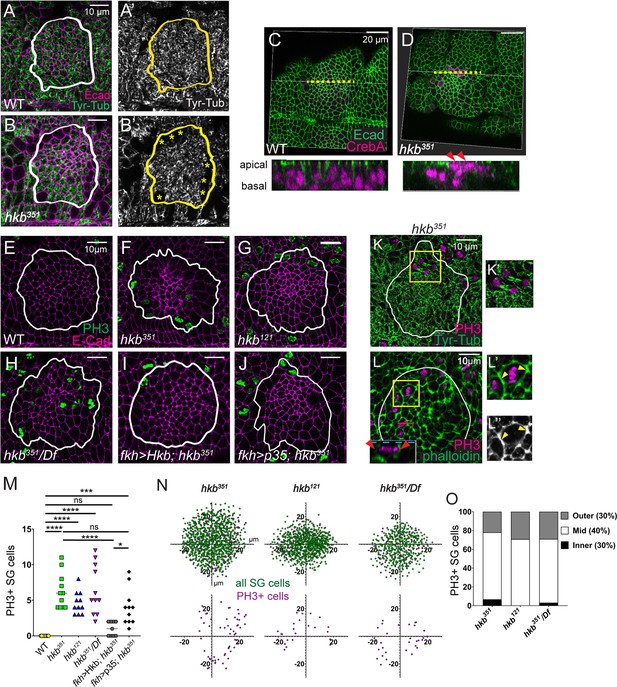
Salivary gland (SG) cells undergo abnormal cell division in huckebein (hkb) mutants.
(A-B’) Stage 11 SGs immunolabeled with tyrosinated α-tubulin (Tyr-Tub) (green) and E-Cadherin (Ecad) (magenta). Asterisks, cells without Tyr-Tub signals. (C, D) 3D reconstructions of confocal images of pre-invaginating SGs immunolabeled with Ecad (green) and CrebA (magenta). Z-sections along yellow dotted lines are shown below. Arrowheads, apically localizing nuclei in hkb mutant SG. (E–J) Stage 11 SGs immunolabeled with phosphorylated histone H3 (PH3) (green) and Ecad (magenta). (K, K’) Stage 11 SGs immunolabeled with Tyr-Tub (green) and PH3 (magenta). Higher magnification of yellow boxed region shown in K’. (L-L’’) Stage 11 SGs immunolabeled with PH3 (magenta) and phalloidin (green). Inset; lateral section (XZ; red line). Higher magnification of yellow boxed region shown in L’ and L’’. (M) Quantification of the number of PH3-positive SG cells. n=11 for all genotypes except fkh >Hkb; hkb351(n=9). One-way ANOVA. *p<0.05; ***p<0.001; ****p<0.0001. (N) Scatter plots showing the position of total SG cells (green) and PH3-positive cells (magenta) in the SG placode. X and Y axes represent the distance along the A/P and D/V axes, respectively, from the center of the placode (n=5 SGs from five different embryos). (O) Quantification of the number of PH-positive SG cells in different regions of the placode. White and yellow lines, SG boundary determined by SG-specific CrebA signals.
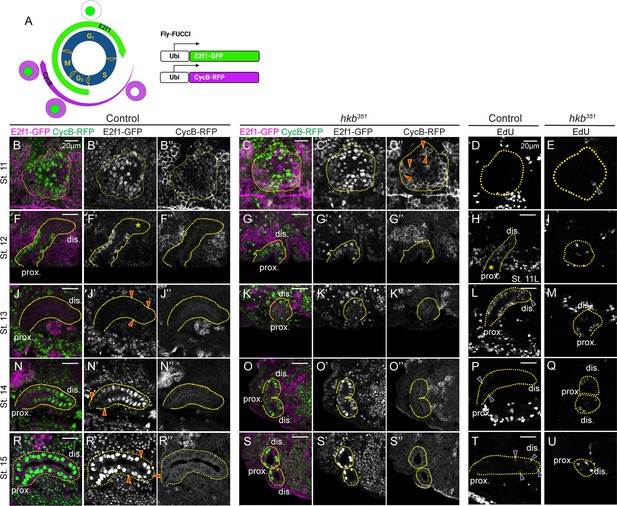
The distal-to-proximal progression of the endoreplication is disrupted in the huckebein (hkb) mutant salivary gland (SG).
(A) Schematic of the Fly-FUCCI system. (B-C’’, F-G’’, J-K’’, N-O’’, R-S’’) SGs immunolabeled with GFP (for E2f1-GFP; green) and cytoplasmic cyclin B (CycB) (for CycB-RFP; magenta) at different stages. Red arrowheads in C’’, CycB signals distributed throughout the dividing cells. Asterisk in F’, absence of E2F transcription factor 1 (E2f1) signals in distal cells of stage 12 SG. Red arrowheads in J’, weak E2f1 signals in the proximal SG cells. Red arrowheads in N’ and R’, little to no E2f1 signal in proximal (N’) and distal (R’) SG cells. (D–E, H–I, L–M, P–Q, T–U) 5-Ethynyl-2'-deoxyuridine (EdU)-labeled SGs at different developmental stages. Blue arrowhead in E, EdU-positive single SG cell. Asterisks in H, absence of EdU signals in proximal cells of late stage 11 SG. Blue arrowheads in L, P, and T, weak EdU signals in distal (L, T) and proximal (P) SG cells. Yellow dotted lines, SG boundary determined by SG-specific CrebA signals. Due to the proximity of the two SGs in hkb mutants, one or two SGs are shown depending on the orientation of the embryo. dis. denotes distal; prox. denotes proximal.
© 2024, Matthew et al. Figure 5 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
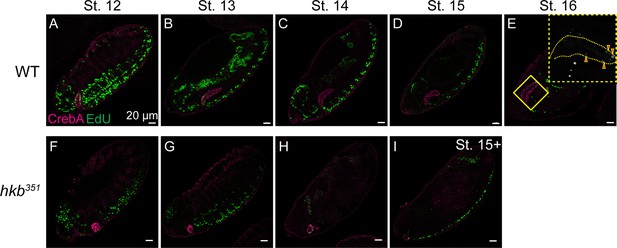
5-Ethynyl-2'-deoxyuridine (EdU) signals during mid to late embryogenesis in wild-type (WT) and huckebein (hkb) mutant embryos.
(A–I) Confocal images of WT (A–E) and hkb mutant (F–I) embryos pulsed and labeled for EdU (green) and immunolabeled for CrebA (magenta). Insets, higher magnification images for the regions highlighted with the yellow boxes. Arrowheads, cells with EdU signals.
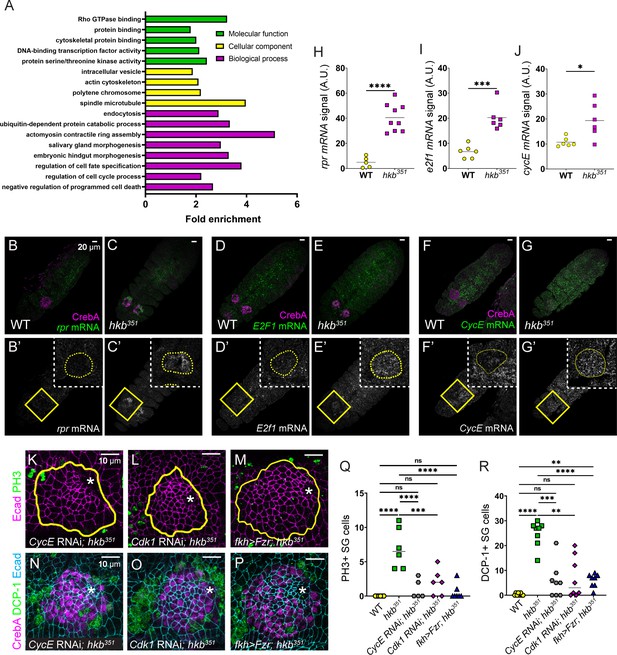
Huckebein (Hkb) regulates key cell cycle and cell death genes transcriptionally.
(A) GO enrichment analysis of ModENCODE Hkb ChIP-Seq data. (B-G’’) Fluorescence in situ hybridization images of stage 11 embryos showing mRNA levels of reaper (rpr) (B-C’), E2F transcription factor 1 (E2f1) (D-E’), and cyclin E (CycE) (F-G’). Yellow-boxed regions are shown at a higher magnification in insets. (H–J) Quantification of the signal intensity of the mRNA levels shown in B’-G’. n=5 (wild-type, WT) and 9 salivary glands (SGs) (hkb351) (H); 6 SGs for both genotypes (I, J). Student’s t-test with Welch’s correction. *p<0.05; ***p<0.001; ****p<0.0001. (K–P) Cdk1 and CycE knockdowns and Fzr overexpression in hkb mutant SGs immunostained for E-Cadherin (Ecad) (magenta) and phosphorylated histone H3 (PH3) (green) (K–M) and Ecad (magenta), death caspase-1 (DCP-1) (green), and CrebA (cyan) (N–P). Asterisks, invagination pit. (Q, R) Quantification of the number of PH3-positive (Q) and DCP-1-positive (R) SG cells. n=6 (Q) and 8 (R) SGs from different embryos for all genotypes. One-way ANOVA. **p<0.01; ***p<0.001; ****p<0.0001. Yellow lines and yellow dotted lines, SG boundary determined by SG-specific CrebA signals.

Expression levels of key cell cycle and cell death genes in wild-type (WT) and huckebein (hkb) mutant embryos.
(A-L’) Confocal images of embryos from fluorescence in situ hybridization experiment showing mRNA levels of reaper (rpr) at stage 12–13 (A-D’), E2F transcription factor 1 (E2f1) at stage 12 (E-F’), cyclin E(CycE) at stage 12 (G-H’), cyclin A (CycA) at stage 11 (I-J’), and string (stg) at stage 11 of embryogenesis (K-L’). (A-D’) At stage 12, compared to the ubiquitously low level of rpr in wild-type (WT) salivary glands (SGs), a few SG cells maintain high levels of rpr (yellow arrowheads). At stage 13, rpr transcript levels are very low in the SG of both genotypes. (E-F’) In WT SGs, E2F1 mRNA shows basal levels comparable to neighboring cells. In huckebein (hkb) mutant SGs, E2F1 transcript levels are slightly higher than in neighboring cells. (G-H’) In WT SGs, CycE levels are not significantly higher than the neighboring cells at stage 12, and in hkb mutant SGs, CycE mRNA levels are slightly elevated and comparable to neighboring cells. (I-J’) In WT SGs, CycA shows placode-wide underexpression. In hkb mutants, CycA mRNA levels are significantly elevated in some SG cells (red arrowheads). (K-L’) Compared to most ventral epidermal tissues, stg is downregulated in both WT and hkb mutant SGs at stage 11. (M) Quantification of stg levels in WT and hkb mutant SGs. n=5 (WT), 6 (hkb351). Statistical analysis was performed using Student’s t-test with Welch’s correction. Red boxes indicate the SG regions at higher magnification shown as insets in A’-L’. Yellow dotted lines indicate the SG boundary.
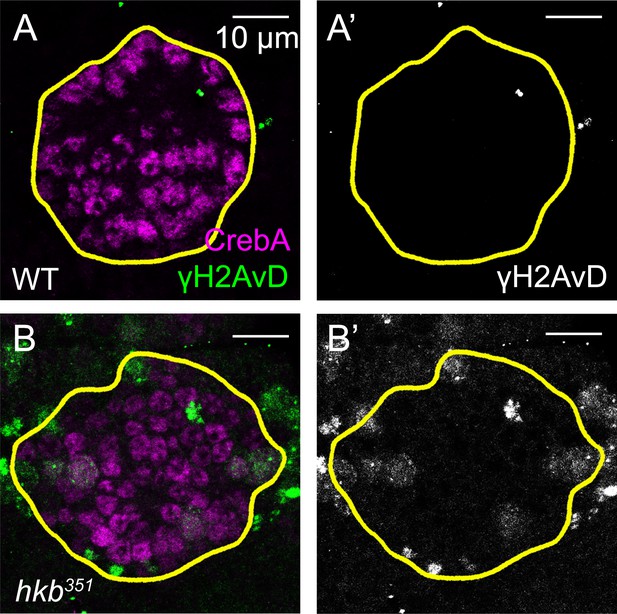
γ-H2Av signals are increased in huckebein (hkb) mutant salivary glands (SGs) at stage 11.
(A-B’) Confocal images of SGs in wild-type (WT) (A-A’) and hkb mutant (B-B’) embryos immunostained for CrebA (magenta) and γ-H2Av (green). hkb mutant SGs show an increase in γ-H2Av signals in the entire placode compared to WT. Yellow lines, SG boundary.

Genetic inhibition of the function of key cell cycle genes is sufficient to suppress cell death and cell division in huckebein (hkb) mutants.
(A–G) Salivary glands (SGs) in stage 11 embryos of wild-type (WT) and mutants for hkb and several key cell cycle genes, immunostained for E-Cadherin (Ecad) (magenta) and phosphorylated histone H3 (PH3) (green). Single mutants for cyclin A (CycA), cyclin D (CycD), cyclin E (CycE), and Cdk1 have relatively normal SG morphology, with few SG cells expressing PH3 (green) in CycD and CycE mutants. Cdk2 mutant embryos have severe morphological defects, including significantly fewer SG cells. (H-I’) SGs in stage 11 embryos double mutant for Cdk2 and hkb (H) and CycA and hkb (I-I’). Cdk2 hkb double mutant embryos show severe morphological defects similar to Cdk2 single mutant embryos, with significantly reduced SG cell numbers. CycA hkb double mutants show the same range of phenotypes as hkb mutants, including the centralized invagination phenotype and small SG at later embryonic stages. (J–O) Confocal images of SGs at stage 11 embryos double mutant for CycE and hkb (J, L), Cdk1 and hkb (K, M), or overexpression of p35 and fizzy-related (Fzr) in the hkb mutant SG using the fkh-Gal4 driver (N, O). (J, K, N) Embryos immunostained for PH3 (green), Ecad (J, K; magenta), or crumbs (Crb) (N; magenta). (L, M, O) Embryos are immunostained for CrebA (magenta), death caspase-1 (DCP-1) (green), and Ecad (L, M; cyan) or Crumbs (O; cyan). (P–Q) Quantification of the number of PH3-positive (P) and DCP-1-positive SG cells (Q) at stage 11. (P) n=12 SGs for all genotypes except for fkh >p35, Fzr; hkb351 (7 SGs). (Q) n=8 SGs for all genotypes except for fkh >p35, Fzr; hkb351 (6 SGs). Statistical significance was calculated using a one-way ANOVA test with multiple comparisons. ****p<0.0001.

Inhibition of cell division or cell death partially rescues the salivary gland (SG) phenotypes of huckebein (hkb) mutants.
(A–Z) Embryos stained for CrebA. Overexpression of Hkb (D–F) or p35 (G–J) as well as knockdown of cyclin E (CycE) (K–N), Cdk1 (O–R), overexpression of fizzy-related (Fzr) (S–V), or overexpression of both Fzr and p35 (W–Z) in hkb mutant SGs using the fkh-Gal4 driver results in a range of rescue effects, including fully elongated SGs as in wild-type (WT) (A, D, G, K, O, S, W), SGs of intermediate length (B, E, H, L, P, T, X), SGs with a very short/rounded phenotype as in hkb null mutants (C, I, M, Q, U, Y). Some embryos also show severe morphological defects (F, J, N, R, V, Z). (AA) Quantification of embryonic morphology. (AB) Quantification of SG morphology. Embryos with normal morphology (AB) as shown in A-C, D-E, G-I, K-M, O-Q, S-U, and W-Y were used for quantification. Note that the quantification for WT, hkb351, fkh >Hkb; hkb351, and fkh >p35; hkb351 is the same as shown in Figure 2—figure supplement 2. n=25 (WT), 30 (hkb351), 29 (fkh >Hkb; hkb351), 92 (fkh >p35; hkb351), 68 (fkh >Cdk1 RNAi; hkb351), 35 (fkh >CycERNAi; hkb351), and 60 (fkh >Fzr; hkb351), and 86 (fkh >p35, Fzr; hkb351) embryos. Statistical significance was calculated by two-way ANOVA.
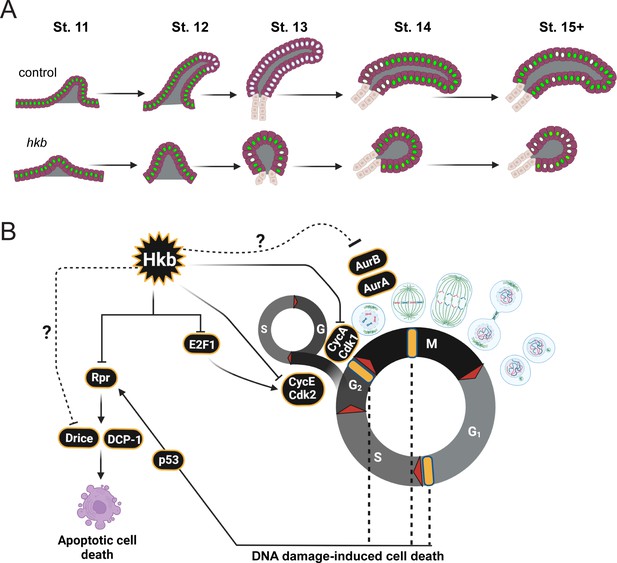
A model of endoreplication regulation by Huckebein (Hkb) during salivary gland (SG) morphogenesis.
(A) Cartoon diagram showing the cell cycle status and progression of endoreplication during SG morphogenesis. Cells with green nuclei represent cells with E2F transcription factor (E2f1) signals (G phase). Cells with white nuclei represent cells with 5-Ethynyl-2'-deoxyuridine (EdU signals S phase). (B) Summary of the role of Hkb in cell cycle regulation in the SG. Hkb helps to inhibit cell division and cell death and promotes endoreplication progression through transcriptional inhibition of key cell cycle and pro-apoptotic genes. Cell death in hkb mutant SG may also be caused by DNA damage-induced cell death. Yellow bars represent cell cycle checkpoints.
© 2024, Matthew et al. Figure 7 was created using BioRender, and is published under a CC BY-NC-ND 4.0 license. Further reproductions must adhere to the terms of this license.
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/95830/elife-95830-mdarchecklist1-v2.pdf
-
Supplementary file 1
Fly lines used in the study.
- https://cdn.elifesciences.org/articles/95830/elife-95830-supp1-v2.docx
-
Supplementary file 2
Antibodies used in the study.
- https://cdn.elifesciences.org/articles/95830/elife-95830-supp2-v2.docx
-
Supplementary file 3
A full list of Hkb target genes, GO clusters, and the list of genes in each cluster.
- https://cdn.elifesciences.org/articles/95830/elife-95830-supp3-v2.xlsx





