Neurons enhance blood–brain barrier function via upregulating claudin-5 and VE-cadherin expression due to glial cell line-derived neurotrophic factor secretion
eLife assessment
This revised study presents valuable evidence that a combination of endothelial cells, astrocytes, and neuroblastoma cells of human origin can integrate to form an in vitro brain blood barrier, that recapitulates key aspects of its natural counterpart, especially at short times. Convincingly, the mechanism by which neuroblastoma-secreted GDNF increases Claudin-5 and VE-cadherin is described. To substantiate the role of GDNF in vivo, authors demonstrated that knock-down of this neurotrophic factor, increased the permeability of the brain blood barrier in mice. This in vitro system can be used to study the permeability of the human brain blood barrier to different drugs.
https://doi.org/10.7554/eLife.96161.3.sa0Valuable: Findings that have theoretical or practical implications for a subfield
- Landmark
- Fundamental
- Important
- Valuable
- Useful
Convincing: Appropriate and validated methodology in line with current state-of-the-art
- Exceptional
- Compelling
- Convincing
- Solid
- Incomplete
- Inadequate
During the peer-review process the editor and reviewers write an eLife Assessment that summarises the significance of the findings reported in the article (on a scale ranging from landmark to useful) and the strength of the evidence (on a scale ranging from exceptional to inadequate). Learn more about eLife Assessments
Abstract
Blood–brain barrier (BBB) prevents neurotoxins from entering central nervous system. We aimed to establish and characterize an in vitro triple co-culture BBB model consisting of brain endothelial cells hCMEC/D3, astrocytoma U251 cells, and neuroblastoma SH-SY5Y cells. Co-culture of SH-SY5Y and U251 cells markedly enhanced claudin-5 and VE-cadherin expression in hCMEC/D3 cells, accompanied by increased transendothelial electrical resistance and decreased permeability. Conditioned medium (CM) from SH-SY5Y cells (S-CM), U251 cells (U-CM), and co-culture of SH-SY5Y and U251 cells (US-CM) also promoted claudin-5 and VE-cadherin expression. Glial cell line-derived neurotrophic factor (GDNF) levels in S-CM and US-CM were significantly higher than CMs from hCMEC/D3 and U-CM. Both GDNF and US-CM upregulated claudin-5 and VE-cadherin expression, which were attenuated by anti-GDNF antibody and GDNF signaling inhibitors. GDNF increased claudin-5 expression via the PI3K/AKT/FOXO1 and MAPK/ERK pathways. Meanwhile, GDNF promoted VE-cadherin expression by activating PI3K/AKT/ETS1 and MAPK/ERK/ETS1 signaling. The roles of GDNF in BBB integrity were validated using brain-specific Gdnf silencing mice. The developed triple co-culture BBB model was successfully applied to predict BBB permeability. In conclusion, neurons enhance BBB integrity by upregulating claudin-5 and VE-cadherin expression through GDNF secretion and established triple co-culture BBB model may be used to predict drugs’ BBB permeability.
Introduction
As a dynamic interface between the blood circulatory system and the central nervous system (CNS), the blood–brain barrier (BBB) maintains homeostasis and normal function of CNS by strictly regulating material exchange between the blood and brain (Palmiotti et al., 2014). The maintenance of BBB is mainly attributed to the expression of tight junctions (TJ) and adherent junctions (AJ) between adjacent brain endothelial cells and a variety of drug transporters. However, as a double-edged sword that protects CNS function, BBB also restricts the transport of some drugs from blood to brain, leading to poor CNS therapeutic effects and even CNS treatment failure (Banks, 2016).
Several in silico, in vitro, in situ, and in vivo methods have been developed to assess the permeability of drugs across BBB, but each method has its limitations (Hanafy et al., 2021). The in situ brain perfusion (ISBP) is considered the ‘gold standard’ for assessing BBB permeability but there exist limits related to animal ethics. Moreover, ISBP is not suitable for human or high-throughput workflows. Thus, a suitable, accurate, and high-throughput in vitro BBB model is required to predict the permeability of drug candidates through BBB.
BBB is formed by neurovascular units (NVU), composed of neural (neurons, microglia, and astrocytes) and vascular components (vascular endothelial cells, pericytes, and vascular smooth muscle cells) (Potjewyd et al., 2018). The constant crosstalk and interactions among these cells contribute to the structural and signaling-based regulation of transcellular and paracellular transport, control of BBB permeability, and regulation of cerebral circulation (Arvanitis et al., 2020; Muoio et al., 2014; Potjewyd et al., 2018). Although brain microvascular endothelial cells (BMECs) are commonly utilized as an in vitro BBB model to assess drug permeability across BBB, in vitro mono-culture of BMECs tends to lose some unique characteristics without the support of other cell types. To overcome the drawbacks of mono-culture BBB models, a range of multicellular co-culture BBB models co-cultured with other NVU elements (such as astrocytes or pericytes) have been developed. Astrocytes are the most abundant glial cell type in brain (Clasadonte and Prevot, 2018). Their terminal feet cover >80% of the surface of capillaries to form interdigitating coverage without slits (Mathiisen et al., 2010). In addition, astrocytes also contain several proteins related to the tight binding of the basement membrane. The in vitro multicellular co-culture BBB models composed of astrocytes, pericytes, and endothelial cells are considered to mimic the vascular structure and offer paracellular tightness in BBB (Ito et al., 2019; Nakagawa et al., 2009; Watanabe et al., 2021).
Neurons also play a crucial role in the NVU and may be involved in the regulation of BBB function. The maturation of BBB in mice considerably overlaps with the establishment of neuronal activity (Biswas et al., 2020). A study revealed that the conditioned medium (CM) collected from primary rat neurons also attenuated cell death caused by glucose–oxygen–serum deprivation (Lin et al., 2016). Furthermore, the primary rat neurons were reported to affect the differentiation and formation of BMECs (Savettieri et al., 2000; Schiera et al., 2003; Xue et al., 2013). Recent research reported that, compared with the double co-culture of hCMEC/D3 and 1321N1 (human astrocytoma cells), the triple co-culture of hCMEC/D3, 1321N1, and human neuroblastoma SH-SY5Y cells exhibited a higher transendothelial electrical resistance (TEER) (Barberio et al., 2022). A similar report showed that co-cultivation of RBE4.B (rat brain capillary endothelial cells) and neurons resulted in the lower permeability of [3H] sucrose than RBE4.B cells grown alone (Schiera et al., 2005). All these studies indicate that neurons, as the elements of the NVU, may be tightly connected to the formation and maintenance of BBB functions.
The aims of the study were: (1) to establish and characterize an in vitro triple co-culture BBB model consisting of human brain endothelial cells (hCMEC/D3), human astrocytoma cells (U251), and human neuroblastoma cells (SH-SY5Y); (2) to investigate whether neurons were involved in the formation and maintenance of BBB integrity and explore their underlying mechanisms. The integrity of BBB was evaluated by quantifying the leakage of both fluorescein and FITC-Dextran 3–5 kDa (FITC-Dex); (3) to validate the in vitro results through in vivo experiments. Finally, the in vivo/in vitro correlation assay was analyzed to prove that the triple co-culture BBB model could better predict the BBB penetration of CNS drugs compared with the mono-culture BBB model.
Results
Establishment and characterization of the in vitro triple co-culture BBB model
Four types of BBB in vitro models were established to compare the contributions of U251 and SH-SY5Y cells to hCMEC/D3 cells (Figure 1A). TEER values were measured during the co-culture (Figure 1B). TEER values of the four in vitro BBB models gradually increased until day 6. On day 7, the TEER values showed a decreasing trend. Thus, 6-day co-culture period was used for subsequent experiments. The highest TEER values were observed in the triple co-culture BBB model, followed by double co-culture with U251 cells and double co-culture with SH-SY5Y cells BBB models, hCMEC/D3 cells mono-culture BBB model showed the lowest TEER values. We also measured the TEER values of U251 monolayer cells, and the results showed that U251 cells themselves also contributed to the physical barrier of the model (Figure 1C). The apparent permeability coefficient (Papp) values of fluorescein and FITC-Dex were measured to characterize the integrity of four BBB models (Figure 1D, E). Consistent with the TEER values, the triple co-culture BBB model showed the lowest permeability of fluorescein and FITC-Dex, followed by double co-culture with U251 cells and double co-culture with SH-SY5Y cells BBB models. It was noticed that monolayer of U251 cells itself also worked as a barrier, preventing the leakage of permeability markers, which may explain why the permeability of FITC-Dex in double co-culture model with U251 cells is lower than that in double co-culture model with SH-SY5Y cells (Figure 1F). Co-culture with SH-SY5Y, U251, and U251 + SH-SY5Y cells also enhanced the proliferation of hCMEC/D3 cells. Moreover, the promoting effect of SH-SY5Y cells was stronger than that of U251 cells (Figure 1G–I). Furthermore, hCMEC/D3 cells were incubated with basic fibroblast growth factor (bFGF), which promotes cell proliferation without affecting both claudin-5 and VE-cadherin expression (Figure 2F). The results showed that incubation with bFGF increased cell proliferation and reduced permeabilities of fluorescein and FITC-Dex across hCMEC/D3 cell monolayer. However, the permeability reduction was less than that by double co-culture with U251 cells or triple co-culture. These results inferred that contribution of cell proliferation to the barrier function of hCMEC/D3 was minor (Figure 1—figure supplement 1).
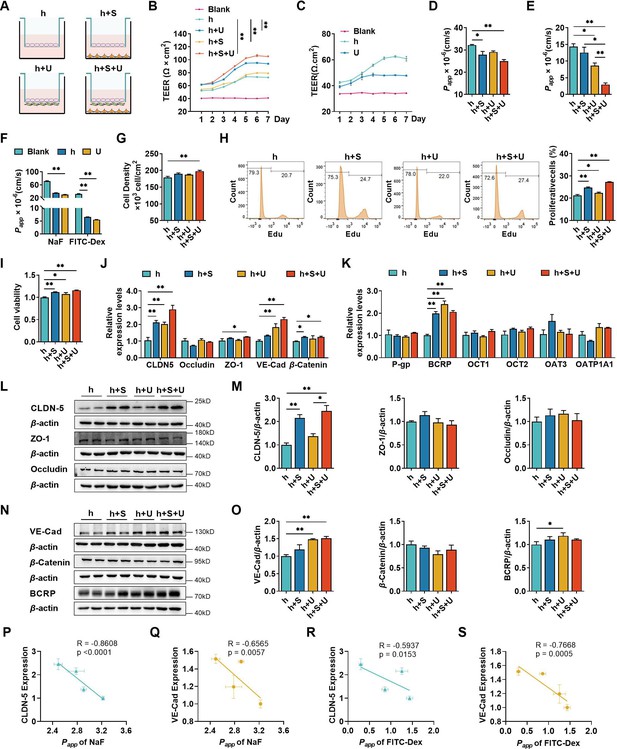
The effects of co-culture with U251 and/or SH-SY5Y cells on the integrity of hCMEC/D3 and blood–brain barrier (BBB) function.
(A) Four different types of BBB models were prepared from hCMEC/D3 cells (h), SH-SY5Y cells (S), and U251 cells (U). (B) The transendothelial electrical resistance (TEER) of four models, and the TEER values in day 6 were compared. Blank: no cells. Four biological replicates per group. (C) The TEER of hCMEC/D3 and U251 cells monolayer. Four biological replicates per group. (D, E) The apparent permeability coefficient (Papp, ×10−6 cm/s) of fluorescein (NaF) and FITC-Dextran 3–5 kDa (FITC-Dex) of four BBB models. Four biological replicates per group. (F) The Papp (×10−6 cm/s) of NaF and FITC-Dex across the blank inserts, and hCMEC/D3 or U251 mono-culture models. Four biological replicates per group. The cell density (G), EdU incorporation (H) of hCMEC/D3 cells after mono/co-culturing. Three biological replicates per group. (I) Cell viability of hCMEC/D3 cells after mono/co-culturing. Four biological replicates per group. (J, K) The mRNA levels of tight junction proteins, adherent junction proteins, and transporters. Four biological replicates per group. The protein expression levels of claudin-5 (CLDN-5), ZO-1, occluding (L, M), VE-cadherin (VE-Cad), β-catenin, and BCRP (N, O) in hCMEC/D3 cells. Four biological replicates per group. The correlations between the Papp (×10−5 cm/s) of NaF and claudin-5 expression (P), or VE-cadherin expression (Q). The correlation between Papp (×10−5 cm/s) of FITC-Dex and claudin-5 expression (R), or VE-cadherin expression (S). The above data are shown as the mean ± SEM. For J and K, two technical replicates per biological replicate. One technical replicate per biological replicate for the rest. *p < 0.05; **p < 0.01 by one-way ANOVA test followed by Fisher’s LSD test, Welch’s ANOVA test, or Kruskal–Wallis test. The simple linear regression analysis was used to examine the presence of a linear relationship between two variables.
-
Figure 1—source data 1
The western blot raw images in Figure 1.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig1-data1-v1.zip
-
Figure 1—source data 2
The labeled western blot images in Figure 1.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig1-data2-v1.zip
-
Figure 1—source data 3
Excel file containing summary data and data analysis of Figure 1.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig1-data3-v1.xlsx
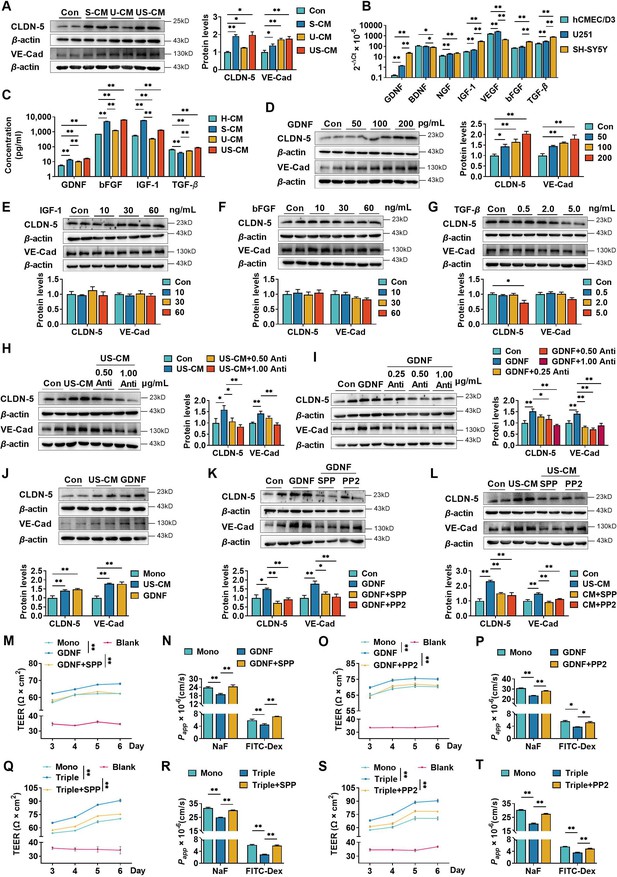
Neurons and astrocytes upregulated claudin-5 and VE-cadherin expression in hCMEC/D3 cells due to glial cell line-derived neurotrophic factor (GDNF) secretion.
(A) Effects of conditioned medium (CM) on claudin-5 and VE-cadherin expression. Con: the normal medium; S-CM: the CM from SH-SY5Y cells; U-CM: the CM from U251 cells; US-CM: the CM from SH-SY5Y cells co-culture with U251 cells. (B) The mRNA expression levels of neurotrophic factors in hCMEC/D3, U251, and SH-SY5Y cells. (C) Concentrations of GDNF, basic fibroblast growth factor (bFGF), insulin-like growth factor-1 (IGF-1), and transforming growth factor-β (TGF-β) in the CMs. H-CM: the CM from hCMEC/D3 cells. Effects of GDNF (D), IGF-1 (E), bFGF (F), and TGF-β (G) on the expression of claudin-5 and VE-cadherin. The dosages have been marked in the figure. Effects of anti-GDNF antibody on the upregulation of claudin-5 and VE-cadherin expression induced by US-CM (H) or 200 pg/ml GDNF (I). (J) Effects of 200 pg/ml GDNF and US-CM on claudin-5 and VE-cadherin expression in primary rat brain microvascular endothelial cells. Effects of 3 μM RET tyrosine kinase inhibitor SSP-86 (SPP), and 5 μM Src family kinases inhibitor PP2 on the upregulation of claudin-5 and VE-cadherin induced by 200 pg/mL GDNF (K) and US-CM (L). Effects of SPP on the transendothelial electrical resistance (TEER) on day 6 (M), the permeability of NaF, and FITC-Dex (N) of the hCMEC/D3 mono-culture blood–brain barrier (BBB) model treating 200 pg/ml GDNF. Effects of PP2 on the TEER on day 6 (O), the permeability of NaF, and FITC-Dex (P) of the hCMEC/D3 mono-culture BBB model treating 200 pg/ml GDNF. Effects of SPP on the TEER on day 6 (Q), the permeability of NaF, and FITC-Dex (R) of the triple co-culture BBB model. Effects of PP2 on the TEER on day 6 (S), the permeability of NaF, and FITC-Dex (T) of the triple co-culture BBB model. The above data are shown as the mean ± SEM. Four biological replicates per group. For B and C, two technical replicates per biological replicate. One technical replicate per biological replicate for the rest. *p < 0.05; **p < 0.01 by one-way ANOVA test followed by Fisher’s LSD test, Welch’s ANOVA test, or Kruskal–Wallis test.
-
Figure 2—source data 1
The western blot raw images in Figure 2.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig2-data1-v1.zip
-
Figure 2—source data 2
The labeled western blot images in Figure 2.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig2-data2-v1.zip
-
Figure 2—source data 3
Excel file containing summary data and data analysis of Figure 2.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig2-data3-v1.xlsx
The paracellular barrier of BBB is also associated with TJs, AJs, and transporters (Abbott, 2013). The mRNA levels of TJs (claudin-5, ZO-1, and occludin), AJs (VE-cadherin and β-catenin), and transporters (P-gp, BCRP, OCT-1, OCT-2, OAT-3, and OATP1A1) in hCMEC/D3 cells from the four BBB models were analyzed using quantitative real-time PCR (qPCR) (Figure 1J, K). Compared with hCMEC/D3 cell mono-culture model, double co-culture with SH-SY5Y, double co-culture with U251, and triple co-culture BBB models showed markedly increases in claudin-5, VE-cadherin, β-catenin, and BCRP mRNA expression. Expression of corresponding proteins was measured using western blot (Figure 1L–O). Notably increased claudin-5 expression was detected in double co-culture with SH-SY5Y cells and triple co-culture BBB models, while VE-cadherin expression was markedly increased in double co-culture with U251 cells and triple co-culture BBB models. Expression levels of other TJ proteins (ZO-1 and occludin) and AJ protein (β-catenin) were unaltered. The expression of BCRP was slightly affected by co-cultivation with U251 cells. Significant negative correlations were found between Papp values of fluorescein and the expression of claudin-5 or VE-cadherin. Papp values of FITC-Dex were also negatively correlated to the expression levels of claudin-5 or VE-cadherin (Figure 1P–S). These results indicate that the decreased permeability of fluorescein and FITC-Dex mainly results from the upregulated expression of both claudin-5 and VE-cadherin.
Neurons and astrocytes upregulated the expression of claudin-5 and VE-cadherin by glial cell line-derived neurotrophic factor secretion
The hCMEC/D3 cells did not direct contact with U251 or SH-SY5Y cells in the double co-culture and triple co-culture BBB models, indicating that cell–cell interaction between U251, SH-SY5Y, and hCMEC/D3 cells relied on secreted active factors. To test this hypothesis, the effects of CM from SH-SY5Y cells (S-CM), U251 cells (U-CM), and co-culture of SH-SY5Y and U251 cells (US-CM) on the expression of claudin-5 and VE-cadherin in hCMEC/D3 cells were analyzed (Figure 2A). Both S-CM, U-CM, and US-CM markedly increased the expression of claudin-5 and VE-cadherin. US-CM showed the strongest induction effects on claudin-5 and VE-cadherin.
To investigate which cytokines were involved in the promotion of hCMEC/D3 cell integrity by U251 and SH-SY5Y cells, the mRNA expression levels of various cytokines in these three types of cells were compared. The results showed that U251 or SH-SY5Y cells exhibited significantly higher expression levels of glial cell line-derived neurotrophic factor (GDNF), nerve growth factor, insulin-like growth factor-1 (IGF-1), vascular endothelial growth factor, bFGF, and transforming growth factor-β (TGF-β) compared to hCMEC/D3 cells (Figure 2B). Furthermore, the mRNA expression of GDNF, IGF-1, TGF-β, and bFGF in SH-SY5Y cells was higher than those in U251 cells.
In these cytokines, GDNF (Dong and Ubogu, 2018; Igarashi et al., 1999; Shimizu et al., 2012), bFGF (Shimizu et al., 2011; Wang et al., 2016), IGF-1 (Ko et al., 2009; Nowrangi et al., 2019), and TGF-β (Fu et al., 2021) have been reported to promote BBB integrity. Thus, the concentrations of GDNF, bFGF, IGF-1, and TGF-β in the CMs were measured (Figure 2C). The results showed that levels of GDNF, bFGF, and IGF-1 in S-CM and US-CM were significantly higher than CMs from hCMEC/D3 cells (H-CM) and U-CM, but levels of TGF-β in S-CM and U-CM were lower than those in H-CM. Interestingly, the level of IGF-1 in US-CM was remarkably lower than that in S-CM, indicating that U251 cells suppressed IGF-1 secretion from SH-SY5Y. The effects of GDNF, bFGF, IGF-1, and TGF-β on the expression of claudin-5 and VE-cadherin were investigated (Figure 2D–G). Among the four tested neurotrophic factors, only GDNF induced the expression of claudin-5 and VE-cadherin in a concentration-dependent manner (Figure 2D). In contrast, a high level (5 ng/ml) of TGF-β slightly downregulated claudin-5 expression (Figure 2G). These results demonstrate that upregulation of claudin-5 and VE-cadherin expression by US-CM are attributed to secreted GDNF.
To provide additional verification of the deduction, anti-GDNF antibody was used to neutralize exogenous and endogenous GDNF in culture medium. Consistent with our expectation, the anti-GDNF antibody concentration-dependent reversed the US-CM-induced claudin-5 and VE-cadherin expression (Figure 2H). Furthermore, GDNF-induced upregulation of claudin-5 and VE-cadherin expression was also reversed by the anti-GDNF antibody (Figure 2I). The induction effects of US-CM and GDNF on the claudin-5 and VE-cadherin expression in hCMEC/D3 cells were also confirmed in primary rat BMECs (Figure 2J).
GDNF forms a heterohexameric complex with two GFRα1 molecules and two RET receptors to activate the GDNF–GFRα1–RET signaling (Fielder et al., 2018). The RET receptor tyrosine kinase inhibitor SPP-86 (Bhallamudi et al., 2021) and Src-type kinase inhibitor PP2 (Morita et al., 2006) were used to further investigate whether GDNF upregulated the expression of claudin-5 and VE-cadherin in hCMEC/D3 cells by activating the GDNF–GFRα1–RET signaling pathway. Both SPP-86 and PP2 markedly attenuated claudin-5 and VE-cadherin expression induced by GDNF (Figure 2K) and US-CM (Figure 2L).
The contributions of GDNF-induced claudin-5 and VE-cadherin expression to TEER and permeability were investigated using hCMEC/D3 cells mono-culture BBB model. GDNF significantly increased TEER values (Figure 2M, O) and decreased the permeability of fluorescein and FITC-Dex (Figure 2N, P), which were almost abolished by SPP-86 or PP2. Furthermore, treatment with SPP-86 or PP2 completely reversed the increased TEER values (Figure 2Q, S) and decreased permeability of fluorescein and FITC-Dex (Figure 2R, T) in the triple co-culture BBB model. These results indicate that neurons but also astrocytes upregulate claudin-5 and VE-cadherin expression in hCMEC/D3 cells by secreting GDNF. Subsequently, GDNF induces claudin-5 and VE-cadherin expression by activating GDNF–GFRα1–RET signaling.
GDNF-induced claudin-5 and VE-cadherin expression of hCMEC/D3 by activating the PI3K/AKT and MAPK/ERK pathways
GDNF exerts its biological activities by activating several signaling pathways, including the phosphatidylinositol-3-kinase (PI3K)/protein kinase B (AKT), mitogen-activated protein kinase (MAPK)/extracellular regulated kinase (ERK), MAPK/c-Jun N-terminal kinase (JNK), and MAPK/ p38 pathways (Fielder et al., 2018). The effects of the PI3K/AKT, MAPK/ERK, MAPK/JNK, and MAPK/p38 pathway inhibitors LY294002 (Figure 3A), U0126 (Figure 3B), SP600125 (Figure 3C), and SB203580 (Figure 3D), respectively, on GDNF-induced claudin-5 and VE-cadherin expression in hCMEC/D3 cells were investigated. GDNF increased claudin-5 and VE-cadherin expression, accompanied by the phosphorylation of AKT (p-AKT) and ERK (p-ERK). However, it did not stimulate the phosphorylation of JNK and p38. SPP-86, LY29002, and U0126 significantly suppressed GDNF-induced claudin-5 and VE-cadherin expression, while SP600125 and SB203580 had almost no effect on GDNF-induced claudin-5 and VE-cadherin expression. GDNF-induced phosphorylation of AKT and ERK was also markedly attenuated by the anti-GDNF antibody (Figure 3E). Similarly, US-CM remarkably upregulated the expression of claudin-5, VE-cadherin, p-AKT, and p-ERK, which were also markedly reversed by SPP-86, LY29002, U0126, or anti-GDNF antibody (Figure 3F–J). These findings indicate that GDNF induces the expression of claudin-5 and VE-cadherin in hCMEC/D3 cells by activating both the PI3K/AKT and MAPK/ERK pathways.
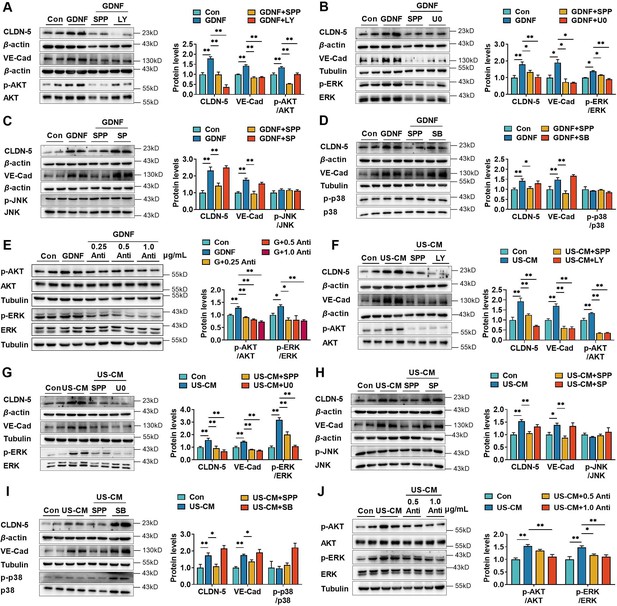
Glial cell line-derived neurotrophic factor (GDNF)-induced claudin-5 and VE-cadherin expression in hCMEC/D3 cells by activating the PI3K/AKT and MAPK/ERK signaling.
(A) Effects of 3 μM LY294002 (LY) on the levels of claudin-5, VE-cadherin, and p-AKT/AKT in hCMEC/D3 cells stimulated by 200 pg/ml GDNF. (B) Effects of 2 μM U0126 (U0) on the levels of claudin-5, VE-cadherin, and p-ERK/ERK in hCMEC/D3 cells stimulated by 200 pg/ml GDNF. (C) Effects of 5 μM SP600125 (SP) on the levels of claudin-5, VE-cadherin, and p-JNK/JNK in hCMEC/D3 cells stimulated by 200 pg/ml GDNF. (D) Effects of 2 μM SB203580 (SB) on the levels of claudin-5, VE-cadherin, and p-p38/p38 in hCMEC/D3 cells stimulated by 200 pg/ml GDNF. (E) Effects of anti-GDNF antibody on the GDNF-induced p-AKT/AKT and p-ERK/ERK ratios. (F) Effects of 3 μM LY on the levels of claudin-5, VE-cadherin, and p-AKT/AKT in hCMEC/D3 cells stimulated by US-CM. (G) Effects of 2 μM U0 on the levels of claudin-5, VE-cadherin, and p-ERK/ERK in hCMEC/D3 cells stimulated by US-CM. (H) Effects of 5 μM SP on the levels of claudin-5, VE-cadherin, and p-JNK/JNK in hCMEC/D3 cells stimulated by US-CM. (I) Effects of 2 μM SB on the levels of claudin-5, VE-cadherin, and p-p38/p38 in hCMEC/D3 cells stimulated by US-CM. (J) Effects of anti-GDNF antibody on the US-CM-induced p-AKT/AKT and p-ERK/ERK ratios. The above data are shown as the mean ± SEM. Four biological replicates per group. One technical replicate for each biological replicate. *p < 0.05; **p < 0.01 by one-way ANOVA test followed by Fisher’s LSD test or Welch’s ANOVA test.
-
Figure 3—source data 1
The western blot raw images in Figure 3.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig3-data1-v1.zip
-
Figure 3—source data 2
The labeled western blot images in Figure 3.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig3-data2-v1.zip
-
Figure 3—source data 3
Excel file containing summary data and data analysis of Figure 3.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig3-data3-v1.xlsx
GDNF upregulated the claudin-5 expression in hCMEC/D3 cells by activating the PI3K/AKT/FOXO1 pathway
Claudin-5 is negatively regulated by the transcriptional repressor forkhead box O1 (FOXO1) (Beard et al., 2020). FOXO1 is also an important target of PI3K/AKT signaling. FOXO1 phosphorylation results in FOXO1 accumulation in the cytoplasm (Zhang et al., 2011) and lowers its level in the nucleus. Here, we investigated whether GDNF-induced claudin-5 expression is involved in FOXO1 nuclear exclusion. As shown in Figure 4A, both GDNF and US-CM significantly enhanced FOXO1 phosphorylation (p-FOXO1). Similarly, GDNF and US-CM increased the levels of phosphorylated and unphosphorylated FOXO1 in the cytoplasm and decreased the levels of nuclear FOXO1 (Figure 4B). Whether FOXO1 was involved in the GDNF-induced regulation of claudin-5 and VE-cadherin expression was investigated in hCMEC/D3 cells transfected with FOXO1 small interfering RNA (siRNA). Silencing FOXO1 significantly decreased FOXO1 levels in both whole-cell lysates and nucleus of hCMEC/D3 cells (Figure 4C), demonstrating FOXO1 silencing efficacy. Consistent with our expectation, silencing FOXO1 upregulated the expression of claudin-5 rather than VE-cadherin expression (Figure 4D). In contrast, high levels of FOXO1 were observed in both whole-cell lysates and nucleus of hCMEC/D3 cells that were transfected with plasmids containing FOXO1. Meanwhile, FOXO1 overexpression resulted in a decrease in both basal and GDNF-induced claudin-5 expression (Figure 4E), consistent with the known role of FOXO1 on claudin-5 expression.
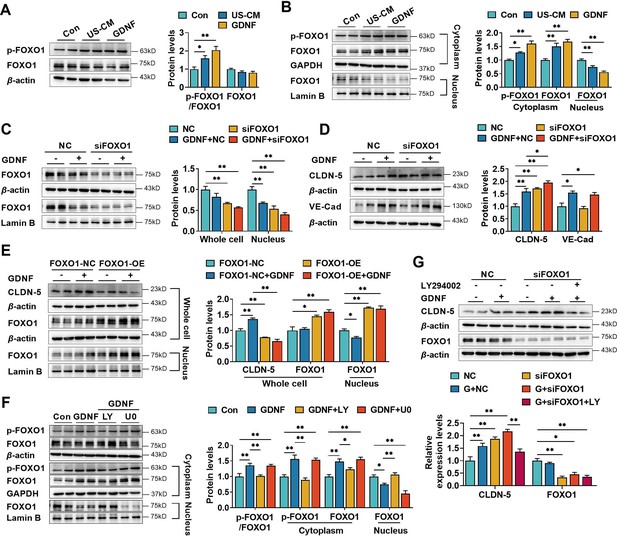
Glial cell line-derived neurotrophic factor (GDNF) induced the claudin-5 expression in hCMEC/D3 cells by activating the PI3K/AKT/FOXO1 pathway.
Effects of US-CM and GDNF on the phosphorylated FOXO1 (p-FOXO1)/FOXO1 ratio, total FOXO1 expression (A), cytoplasmic p-FOXO1, cytoplasmic FOXO1, and nuclear FOXO1 expression (B). The expression levels of total and nuclear FOXO1 (C), claudin-5, and VE-cadherin (D) in hCMEC/D3 cells transfected with FOXO1 siRNA (siFOXO1). NC: negative control. (E) Effects of FOXO1 overexpression (FOXO1-OE) and GDNF on the expression levels of claudin-5, total FOXO1, and nuclear FOXO1. FOXO1-NC: negative control plasmids. (F) Effects of LY and U0 on GDNF-induced alterations of total p-FOXO1/FOXO1 ratio, cytoplasmic p-FOXO1, cytoplasmic FOXO1, and nuclear FOXO1 expression. (G) Effects of LY on the claudin-5 expression upregulated by siFOXO1. The above data are shown as the mean± SEM. Four biological replicates per group. One technical replicate for each biological replicate. *p < 0.05; **p < 0.01 by one-way ANOVA test followed by Fisher’s LSD test, Welch’s ANOVA test, or Kruskal–Wallis test.
-
Figure 4—source data 1
The western blot raw images in Figure 4.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig4-data1-v1.zip
-
Figure 4—source data 2
The labeled western blot images in Figure 4.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig4-data2-v1.zip
-
Figure 4—source data 3
Excel file containing summary data and data analysis of Figure 4.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig4-data3-v1.xlsx
Several reports have demonstrated that nuclear localization of FOXO1 is modulated by multiple pathways, including the PI3K/AKT (Tang et al., 1999) and MAPK/ERK (Asada et al., 2007) pathways. The effects of LY294002 and U0126 on GDNF-induced FOXO1 phosphorylation were measured in hCMEC/D3 cells. LY294002, but not U0126, significantly reversed the GDNF-induced alterations in total p-FOXO1, cytoplasmic p-FOXO1, cytoplasmic FOXO1, and nuclear FOXO1 (Figure 4F). Furthermore, LY294002 reversed the upregulation of claudin-5 expression induced by FOXO1 siRNA and GDNF (Figure 4G).
It was reported that VE-cadherin also upregulates claudin-5 via inhibiting FOXO1 activities (Taddei et al., 2008). Effect of VE-cadherin on claudin-5 was studied in hCMEC/D3 cells silencing VE-cadherin. It was not consistent with Taddei et al. that silencing VE-cadherin only slightly decreased the mRNA level of claudin-5 without significant difference. Furthermore, basal and GDNF-induced claudin-5 protein levels were unaltered by silencing VE-cadherin (Figure 4—figure supplement 1). Thus, the roles of VE-cadherin in regulation of claudin-5 in BBB should be further investigated.
GDNF upregulated VE-cadherin expression in hCMEC/D3 cells by activating the PI3K/AKT/ETS1 and MAPK/ERK/ETS1 signaling pathways
E26 oncogene homolog 1 (ETS1) is a transcription factor that binds to the ETS-binding site located in the proximal region of the VE-cadherin promoter, hence regulating the expression of VE-cadherin (Lelièvre et al., 2000; Luo et al., 2022). Activation of the PI3K/AKT (He et al., 2023; Hui et al., 2018) and MAPK/ERK (Watanabe et al., 2004) pathways was reported to upregulate the expression of ETS1. The previous results showed that GDNF-induced VE-cadherin and claudin-5 expression in hCMEC/D3 cells by activating the PI3K/AKT and MAPK/ERK pathways. Therefore, we hypothesized that GDNF modulated ETS1 levels to promote VE-cadherin and claudin-5 expression through PI3K/AKT and MAPK/ERK signaling pathways. Both US-CM and GDNF significantly increased total (Figure 5A) and nuclear ETS1 expression (Figure 5B). LY294002 and U0126 markedly attenuated GDNF-induced total (Figure 5C) and nuclear ETS1 expression (Figure 5D). To further confirm the involvement of the PI3K/AKT/ETS1 and MAPK/ERK/ETS1 pathways in GDNF-induced VE-cadherin expression, ETS1 in hCMEC/D3 cells was knocked down using ETS1 siRNA. ETS1 silencing saliently declined the expression levels of total (Figure 5E) and nuclear (Figure 5F) ETS1 in hCMEC/D3 cells, demonstrating silencing efficacy. In ETS1 silencing hCMEC/D3 cells, GDNF no longer induced the expression of total (Figure 5E) and nuclear (Figure 5F) ETS1. Moreover, ETS1 silencing substantially downregulated VE-cadherin expression and attenuated GDNF-induced VE-cadherin expression, while having minimal impact on both basal and GDNF-induced claudin-5 expression (Figure 5G).
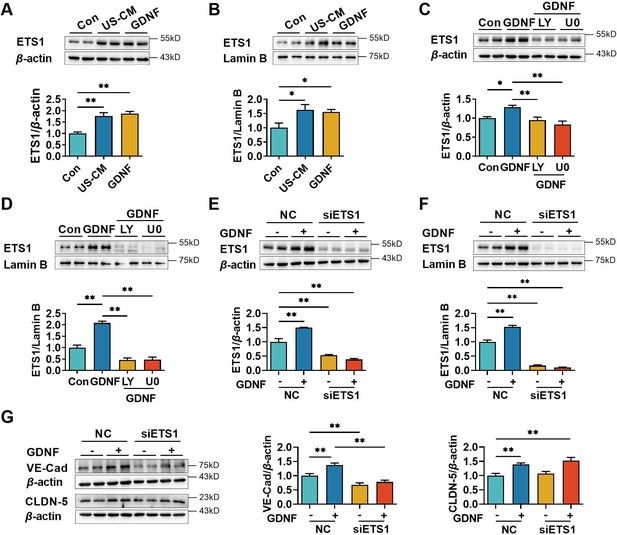
Glial cell line-derived neurotrophic factor (GDNF)-induced VE-cadherin expression in hCMEC/D3 cells by activating the PI3K/AKT/ETS1 and MAPK/ERK/ETS1 pathways.
Effects of US-CM and GDNF on total (A) and nuclear (B) ETS1 expression. Effects of LY and U0 on 200 pg/ml GDNF-induced total (C) and nuclear (D) ETS1 expression. Expression levels of total (E) and the nuclear ETS1 (F) in hCMEC/D3 cells after knocking down ETS1 with siRNA (siETS1). (G) Effects of GDNF and siETS1 on the expression of VE-cadherin and claudin-5. The above data are shown as the mean ± SEM. Four biological replicates per group. One technical replicate for each biological replicate. *p < 0.05; **p < 0.01 by one-way ANOVA test followed by Fisher’s LSD test.
-
Figure 5—source data 1
The western blot raw images in Figure 5.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig5-data1-v1.zip
-
Figure 5—source data 2
The labeled western blot images in Figure 5.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig5-data2-v1.zip
-
Figure 5—source data 3
Excel file containing summary data and data analysis of Figure 5.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig5-data3-v1.xlsx
Brain GDNF deficiency increased BBB permeability partly due to the impairment of claudin-5 and VE-cadherin expression
To further demonstrate the positive effects of GDNF on BBB maintenance, GDNF in mice brains was knocked down via intracerebroventricular (i.c.v) injection of AAV-PHP.eB packaged with Gdnf short hairpin RNA (shRNA) (Figure 6A). Knockdown efficiency was confirmed through western blotting (Figure 6B). Consistent with in vitro results, GDNF knockdown greatly downregulated claudin-5 and VE-cadherin expression in the mice brains (Figure 6B). The integrity of BBB was assessed by examining the brain distributions of fluorescein and FITC-Dex. The results showed that specifically knocking down brain GDNF little affected plasma levels of fluorescein (Figure 6C) and FITC-Dex (Figure 6F), but significantly elevated the concentrations of fluorescein (Figure 6D) and FITC-Dex (Figure 6G) in the brains, leading to notable increases in the brain-to-plasma concentration ratios of two probes (Figure 6E, H). These alterations were consistent with the decline in claudin-5 and VE-cadherin expression. In addition, significant reductions in the levels of p-AKT (Figure 6I), p-ERK (Figure 6J), p-FOXO1 (Figure 6K), and ETS1 expression (Figure 6L) were observed in the brains of Gdnf knockdown mice.
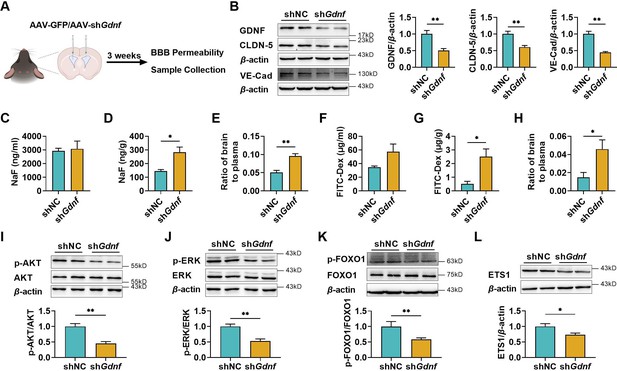
The deficiency of brain glial cell line-derived neurotrophic factor (GDNF) in mice increased the permeability of blood–brain barrier (BBB) and reduced claudin-5 and VE-cadherin expression in mice brains.
(A) Experimental configuration of AAV-GFP (shNC) or AAV-shGdnf (shGdnf) intracerebroventricular injection. (B) Effects of brain-specific Gdnf silencing on the expression levels of GDNF, claudin-5, and VE-cadherin in the brains. Effects of brain-specific Gdnf silencing on NaF levels in plasma (C), brain (D), and the ratio of brain to plasma (E). Effects of brain-specific Gdnf silencing on FITC-Dex levels in plasma (F), brain (G), and the ratio of brain to plasma (H). The expression ratios of p-AKT/AKT (I), p-ERK/ERK (J), and p-FOXO1/FOXO1 (K) in the brains of Gdnf silencing mice. (L) The expression level of ETS1 in the brains of Gdnf silencing mice. The above data are shown as the mean ± SEM. Six biological replicates per group. One technical replicate for each biological replicate. *p < 0.05; **p < 0.01 by unpaired t-test, unpaired t-test with Welch’s correction, or Mann–Whitney test.
-
Figure 6—source data 1
The western blot raw images in Figure 6.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig6-data1-v1.zip
-
Figure 6—source data 2
The labeled western blot images in Figure 6.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig6-data2-v1.zip
-
Figure 6—source data 3
Excel file containing summary data and data analysis of Figure 6.
- https://cdn.elifesciences.org/articles/96161/elife-96161-fig6-data3-v1.xlsx
The triple co-culture BBB model better predicted the permeabilities of drugs across BBB
In this study, 18 drugs were utilized to further investigate the superiority of the triple co-culture BBB model over the hCMEC/D3 mono-culture BBB model. The Papp of 18 drugs from the apical to the basolateral side based on the hCMEC/D3 mono-culture (Papp, Mono) and triple co-culture (Papp, Triple) BBB models are measured and listed in Table 1. The results showed that Papp, Triple values of all tested drugs were lower than the Papp, Mono values. Significant differences were observed in 14 out of 18 drugs. The predicted permeability coefficient-surface area product values (PS) of the tested drugs were, respectively, calculated based on their Papp, Mono values (PSPre, Mono) and Papp, Triple values (PSPre, Triple). The predicted PS values were further compared to their corresponding observations (PSobs). The results showed that the predictive accuracy of PSPre, Triple was superior to PSPre, Mono. Except for verapamil, amitriptyline, fluoxetine, and clozapine, the predicted PSPre, Triple values of the other 14 drugs in the triple co-culture BBB models were within the 0.5- to 2-folds of PSobs (Figure 7B). However, in the hCMEC/D3 mono-culture BBB model, only seven predicted PSPre, Mono values were within the 0.5- to 2-folds range of their observations (Figure 7A).
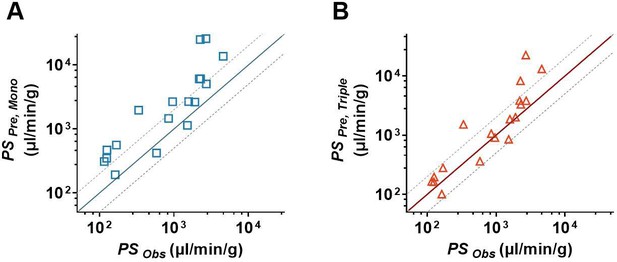
In vitro/in vivo correlation assay of blood–brain barrier (BBB) permeability.
(A) The comparison of the estimated permeability coefficient-surface area product (PSPre, Mono) recalculated from Papp, Mono with the observed in vivo PS values (PSObs). (B) The comparison of the estimated permeability coefficient-surface area product (PSPre, Triple) recalculated from Papp, Triple with the observed in vivo PS values (PSObs). The solid line represents a perfect prediction, and the dashed lines represent the 0.5- to 2-folds of their observations. The PSObs values were determined by in situ brain perfusion in rodents, which were collected from the literature.
The unbound fraction in brain (fu, brain), the observed PSObs, and the predicted PS (PSPre), Papp across the hCMEC/D3 mono-culture model (Papp, Mono) and triple co-culture model (Papp, Triple) of the tested drugs.
| Compounds | fu, brain | PSObsμl/min/g | Papp, Mono cm/s × 10−6 | PSPre, Monoμl/min/g | Papp, Triple cm/s × 10−6 | PSPre, Tripleμl/min/g |
|---|---|---|---|---|---|---|
| Amantadine | 0.1985* | 116.10† | 6.84 ± 0.95 | 310.22 | 3.64 ± 0.26 | 165.23 |
| Amitriptyline | 0.01‡ | 4608.00*** | 15.24 ± 0.64 | 13,716.00 | 14.61 ± 0.27 | 13,149.00 |
| Bupropion | 0.12§ | 1519.20† | 15.19 ± 0.20 | 1139.58 | 11.34 ± 0.44 | 850.58 |
| Carbamazepine | 0.116¶ | 959.40† | 34.37 ± 1.26 | 2666.89 | 11.71 ± 0.15 | 908.69 |
| Clozapine | 0.014** | 2260.80† | 38.97 ± 0.54 | 25,052.57 | 12.87 ± 2.06 | 8272.72 |
| Donepezil | 0.07†† | 1581.30† | 20.91 ± 0.75 | 2688.43 | 14.47 ± 0.84 | 1860.43 |
| Doxepin | 0.025† | 2192.40† | 16.88 ± 1.08 | 6076.80 | 10.66 ± 0.92 | 3837.60 |
| Fluoxetine | 0.004† | 2698.20† | 11.48 ± 0.85 | 25,830.00 | 9.97 ± 1.03 | 22,430.25 |
| Gabapentin | 0.782† | 162.90† | 16.75 ± 1.62 | 192.77 | 8.78 ± 0.23 | 101.05 |
| Lamotrigine | 0.273† | 126.00† | 14.26 ± 0.37 | 470.11 | 5.97 ± 0.11 | 196.88 |
| Metoclopramide | 0.365† | 125.10† | 14.14 ± 1.44 | 348.66 | 6.63 ± 0.42 | 163.41 |
| Midazolam | 0.045‡ ‡ | 2727.00† | 25.30 ± 1.00 | 5060.00 | 19.09 ± 0.24 | 3818.00 |
| Mirtazapine | 0.08† | 1912.50† | 23.44 ± 0.44 | 2637.00 | 17.86 ± 0.21 | 2009.25 |
| Olanzapine | 0.034† | 2279.70† | 22.91 ± 3.80 | 6064.41 | 12.49 ± 0.53 | 3306.18 |
| Prazosin | 0.09§ § | 169.20¶ ¶ | 5.61 ± 0.38 | 560.93 | 2.81 ± 0.52 | 280.99 |
| Risperidone | 0.099† | 849.60† | 16.10 ± 2.87 | 1463.64 | 11.70 ± 0.25 | 1063.64 |
| Venlafaxine | 0.205* | 584.10† | 9.58 ± 0.28 | 420.60 | 8.25 ± 0.36 | 362.02 |
| Verapamil | 0.033 ‡ | 335.70 ‡ | 7.21 ± 0.41 | 1965.24 | 5.56 ± 0.06 | 1517.67 |
-
One technical replicate of four biological replicates per group.
-
*
-
†
-
‡
-
§
-
¶
-
**
-
††
-
‡ ‡
-
§ §
-
¶ ¶
-
***
-
Table 1—source data 1
The apparent permeability coefficients of 18 tested drugs from mono- or triple- culture blood–brain barrier (BBB) model.
- https://cdn.elifesciences.org/articles/96161/elife-96161-table1-data1-v1.xlsx
Discussion
The main findings of the study were to successfully develop an in vitro triple co-culture BBB model consisting of hCMEC/D3, U251, and SH-SY5Y cells and to confirm the involvement of neurons in BBB maintenance as well as the possible mechanisms. Co-culture with U251 and/or SH-SY5Y cells markedly promoted the TEER of hCMEC/D3 cells and reduced the leakage of fluorescein and FITC-Dex due to the upregulation of claudin-5 and VE-cadherin expression.
The roles of claudin-5 and VE-cadherin in the maintenance of BBB function have been demonstrated (Dejana et al., 2008; Hashimoto et al., 2023; Li et al., 2018; Ohtsuki et al., 2007). It was reported that Cld-5-deficient mice exhibited BBB impairment, allowing the transport of small molecules (<800 D) across BBB (Nitta et al., 2003). In contrast, claudin-5 overexpression significantly restricted the permeability of inulin across the conditionally immortalized rat brain capillary endothelial cell monolayer (Ohtsuki et al., 2007). Moreover, the claudin-5 expression in hCMEC/D3 is much lower than in human brain microvessels (Eigenmann et al., 2013; Ohtsuki et al., 2013; Weksler et al., 2013), which may cause the low TEER values. VE-cadherin, a major member of the cadherin family in endothelial cells, is also required for BBB integrity (Dejana et al., 2008; Li et al., 2018). The absence of VE-cadherin resulted in faulty cell-to-cell junctions and disrupted distribution of ZO-1 (Sauteur et al., 2017; Tunggal et al., 2005). Strongly negative correlations between Papp values of fluorescein or FITC-Dex and expression of claudin-5 or VE-cadherin further demonstrated the importance of claudin-5 and VE-cadherin in BBB integrity.
Neurons and astrocytes, as important components of the NVU, may be involved in the formation and maintenance of BBB function. Several reports showed that co-culture with astrocytes CC-2565 or SC-1810 enhanced the TEER of hCMEC/D3 cells (Hatherell et al., 2011), whereas co-culture of RBE4.B cells with primary rat neurons and astrocytes showed a lower permeability of [3H] sucrose than mono-culture of RBE4.B cells (Schiera et al., 2005). Similarly, co-cultured with hCMEC/D3 cells and 1321N1 (astrocytes) or 1321N1+SH-SY5Y cells possess higher TEER values and lower permeability to Lucifer yellow than hCMEC/D3 alone. Compared with double co-culture of hCMEC/D3 and 321N1 cells, the triple co-culture of hCMEC/D3, 1321N1, and SH-SY5Y cells showed higher TEER values (Barberio et al., 2022). Our study also demonstrated that co-culture with U251 and/or SH-SY5Y cells significantly lowered BBB permeability and upregulated VE-cadherin or claudin-5 expression in hCMEC/D3 cells.
Next, we focused on the molecular mechanisms by which neurons and possibly astrocytes upregulated the VE-cadherin and claudin-5 expression in BMECs. Co-culture with SH-SY5Y cells significantly upregulated claudin-5 and VE-cadherin expression in hCMEC/D3 cells. In the double co-culture with SH-SY5Y cells or triple co-culture BBB models, hCMEC/D3 cells were not in direct contact with SH-SY5Y cells, indicating that the interaction between SH-SY5Y and hCMEC/D3 cells depended on the release of some active compounds. It was consistent with the above deduction that the S-CM also markedly induced claudin-5 and VE-cadherin expression. Different from S-CM, U-CM mainly upregulated the VE-cadherin expression and just had a slight impact on claudin-5. In general, neurons but also astrocytes secrete some neurotrophic factors (Lonka-Nevalaita et al., 2010; Sweeney et al., 2019) that could contribute to the maintenance of structural stability of BBB. High levels of bFGF, GDNF, IGF-1, and TGF-β were detected in U-CM, S-CM, and US-CM. Further research showed that only GDNF-induced VE-cadherin and claudin-5 expression in hCMEC/D3 cells in a concentration-dependent manner, and the anti-GDNF antibody attenuated claudin-5 and VE-cadherin expression induced by US-CM or GDNF. These results indicated that neurons upregulated claudin-5 and VE-cadherin expression through GDNF secretion. Levels of GDNF in S-CM were higher than those in U-CM, which seemed to partly explain why S-CM has a stronger promoting effect on claudin-5 than U-CM. The roles of GDNF in BBB maintenance and the regulation of claudin-5 and VE-cadherin expression were further confirmed using brain-specific Gdnf knockdown C57BL/6J mice. Consistent with our in vitro results, brain-specific Gdnf silencing greatly increased BBB penetration of fluorescein and FITC-Dex, accompanied by the downregulation of claudin-5 and VE-cadherin expression.
GDNF is mainly expressed in astrocytes and neurons (Lonka-Nevalaita et al., 2010; Pochon et al., 1997). In adult animals, GDNF is mainly secreted by striatal neurons rather than astrocytes and microglial cells (Hidalgo-Figueroa et al., 2012). The present study also shows that GDNF mRNA levels in SH-SY5Y cells were significantly higher than that in U251 cells. GDNF was also detected in CM from SH-SY5Y cells. All these results demonstrate that neurons may secrete GDNF.
Generally, GDNF activates several signal transduction pathways, such as the PI3K/AKT and MAPK signaling pathways (Fielder et al., 2018) by forming a heterohexameric complex with two GFRα molecules and RET receptors. It was also reported that GDNF improved BBB barrier function due to the activation of MAPK/ERK1 (Dong and Ubogu, 2018) and PI3K/AKT (Liu et al., 2022) signaling. Consistent with previous reports, we found that signaling inhibitors SPP-86, PP2, LY294002, and U0126 markedly attenuated US-CM- and GDNF-induced claudin-5 and VE-cadherin expression in hCMEC/D3 cells, inferring that GDNF promoted claudin-5 and VE-cadherin expression via activating both the PI3K/AKT and MAPK/ERK pathways.
Signal transduction pathways control gene expression by modifying the function of nuclear transcription factors. The nuclear accumulation of FOXO1 negatively regulates claudin-5 expression (Beard et al., 2020; Taddei et al., 2008). FOXO1 is an important target of the PI3K/AKT signaling axis (Zhang et al., 2011), and AKT-induced phosphorylation of FOXO1 results in cytoplasmic FOXO1 accumulation and decreases nuclear FOXO1 accumulation (Zhang et al., 2011). From these results, we inferred that GDNF-induced claudin-5 expression in hCMEC/D3 cells may be involved in the activation of PI3K/AKT/FOXO1 pathway. Similarly, both US-CM and GDNF increased cytoplasmic p-FOXO1 and decreased nuclear FOXO1 in hCMEC/D3 cells, which was reversed by LY29002 rather than U0126. Roles of FOXO1 in GDNF-induced claudin-5 were verified through silencing and overexpressing FOXO1 in hCMEC/D3 cells. FOXO1 silencing enhanced claudin-5 but not VE-cadherin expression, accompanied by a decline in total and nuclear FOXO1. In contrast, FOXO1 overexpression significantly decreased claudin-5 expression. In hCMEC/D3 cells overexpressing FOXO1, GDNF lost its promotion effect on claudin-5 expression. It was noticed that U0126 attenuated the GDNF-induced upregulation of claudin-5 but had minimal impact on GDNF-mediated FOXO1 phosphorylation and the decline of nuclear FOXO1. In Sertoli cells, it was found that testosterone-stimulated claudin-5 expression by activating the RAS/RAF/ERK/CREB pathway (Bulldan et al., 2016). The transcriptional regulation of claudin-5 by CREB was confirmed in bEnd.3 (mouse brain endothelial cell). CREB overexpression significantly increased both gene and protein expression of claudin-5. In contrast, depletion of CREB decreased claudin-5 expression in gene and protein levels (Li et al., 2022a). However, another report showed that in human lung microvascular endothelial cells, U0126 attenuated phosphorylation of ERK and lipopolysaccharide-stimulated claudin-5 damage, indicating activation of MAPK/ERK pathway impaired rather than promoted claudin-5 expression (Liu et al., 2019). Thus, the real mechanisms that GDNF-induced activation of the RET/MAPK/ERK pathway promotes claudin-5 expression need further investigation.
ETS1 is a member of the ETS family that plays an important role in cell adhesion, migration, and blood vessel information. ETS1 binds to an ETS-binding site located in the proximal region of the VE-cadherin promoter, controlling VE-cadherin expression (Lelièvre, 2001). Several studies have demonstrated the role of ETS1 in the regulation of VE-cadherin expression. For example, IFN-γ and TNF-α impaired BBB integrity by decreasing ETS1-induced VE-cadherin expression (Luo et al., 2022). ETS1 silencing reduced VE-cadherin expression in umbilical vein endothelial cells (Colás-Algora et al., 2020). In contrast, ETS1 overexpression induced VE-cadherin expression in mouse brain capillary endothelial cells and fibroblasts (Lelièvre et al., 2000). As expected, ETS1 silencing resulted in a decrease in the expression of VE-cadherin in hCMEC/D3 cells, but claudin-5 expression remained unaffected. Additionally, ETS1 silencing removed the inductive effect of GDNF on VE-cadherin expression while unaffecting the upregulation of claudin-5 induced by GDNF. The activation of the PI3K/AKT (He et al., 2023; Hui et al., 2018) and MAPK/ERK (Watanabe et al., 2004) pathways promotes ETS1 expression. In our findings, US-CM and GDNF significantly increased total and nuclear ETS1 levels, which were eliminated by signaling inhibitors LY294002 and U0126. Both our results and previous research provide evidence that GDNF upregulates ETS1 expression via the activation of PI3K/AKT and MAPK/ERK signaling to promote VE-cadherin expression.
Claudin-5 expression is also regulated by VE-cadherin (Taddei et al., 2008). Differing from the previous reports, silencing VE-cadherin with siRNA only slightly affected basal and GDNF-induced claudin-5 expression. The discrepancies may come from different characteristics of the tested cells. Several reports have supported the above deduction. In retinal endothelial cells, hyperglycemia remarkably reduced claudin-5 expression (but not VE-cadherin) (Saker et al., 2014). However, in hCMEC/D3 cells, hypoglycemia significantly decreased claudin-5 expression but hyperglycemia increased VE-cadherin expression (Sajja et al., 2014).
The present study showed that characteristics of in vitro triple co-culture BBB model were superior to those of hCMEC/D3 mono-culture BBB model. The hCMEC/D3 mono-culture and triple co-culture BBB models were used to try to predict the PS values of 18 drugs by comparing them with their observations. The prediction success rate (14/18) of triple co-culture BBB model was greater than that of hCMEC/D3 mono-culture BBB model (7/18). However, poor predictions were observed for verapamil, amitriptyline, fluoxetine, and clozapine, which may partly be due to inaccuracies in their fu, brain values. These four drugs are high protein-binding drugs. Due to the methodological discordance and limitations of historic devices for these drugs, the fu ≤ 0.01 maybe with low confidence and accuracy (Bowman and Benet, 2018; Di et al., 2017). For example, fu, brain values of amitriptyline from different reports display large differences [0.002 (Sánchez-Dengra et al., 2021), 0.0071 (Weber et al., 2013)]. The same large differences of fu, brain values were also found in fluoxetine [0.0023 (Maurer et al., 2005), 0.004 (Summerfield et al., 2007), 0.00094 (Liu et al., 2005)], and clozapine [0.0056 (Bhyrapuneni et al., 2018), 0.014 (Cremers et al., 2012), 0.011 (Summerfield et al., 2007)].
In summary, our triple co-culture BBB model outperformed the mono-culture or double co-culture BBB models, mainly attributing to the fact that neurons and possibly astrocytes upregulated claudin-5 and VE-cadherin expression by secreting GDNF through activating PI3K/AKT and MAPK/ERK pathways. Additionally, the developed in vitro triple co-culture BBB model accurately predicted the in vivo BBB permeability of CNS drugs. This suggests the potential of our triple co-culture BBB model for utilization in CNS candidate screening during the drug development process (Figure 8).
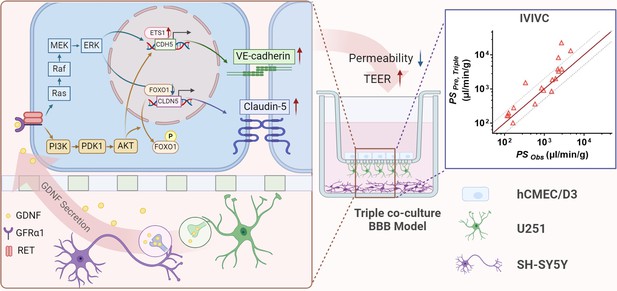
The mechanism of neurons and astrocytes induced the integrity of brain endothelial cells.
Neurons but also astrocytes trigger the activation of PI3K/AKT and MAPK/ERK pathways in brain endothelial cells by glial cell line-derived neurotrophic factor (GDNF) secretion, which in turn regulates transcription factors of claudin-5 (FOXO1) and VE-cadherin (ETS1) to promote claudin-5 and VE-cadherin expression and leads to the enhancement of blood–brain barrier (BBB) integrity. Meanwhile, with the increase in barrier integrity, the in vitro BBB model also obtained a stronger in vivo correlation.
However, the study also has some limitations. In addition to neurons and astrocytes, other cells such as microglia, pericytes, and vascular smooth muscle cells, especially pericytes, may also affect BBB function. How pericytes affect BBB function and interaction among neurons, astrocytes, and pericytes needs further investigation.
Materials and methods
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | β-Actin (mouse monoclonal) | Proteintech | 66009 RRID:AB_2883475 | 1:10,000 |
| Antibody | GAPDH (mouse monoclonal) | Absin | Abs830030ss RRID:AB_2811228 | 1:50,000 |
| Antibody | β-Tubulin (mouse monoclonal) | Fdbio Science | FD0064 RRID:AB_3076327 | 1:10,000 |
| Antibody | Lamin B (mouse monoclonal) | Proteintech | 66095 RRID:AB_2721256 | 1:10,000 |
| Antibody | Claudin-5 (rabbit polyclonal) | Wanleibio | WL03731 RRID:AB_3076320 | 1:1000 |
| Antibody | Occludin (rabbit polyclonal) | Wanleibio | WL01996 RRID:AB_3076325 | 1:500 |
| Antibody | ZO-1 (mouse polyclonal) | Proteintech | 21773-1-AP RRID:AB_10733242 | 1:5000 |
| Antibody | VE-cadherin (rabbit polyclonal) | Wanleibio | WL02033 RRID:AB_3076321 | 1:1000 |
| Antibody | β-Catenin (rabbit polyclonal) | Wanleibio | WL0962a RRID:AB_3076323 | 1:5000 |
| Antibody | BCPR (rabbit polyclonal) | CST | 4477S RRID:AB_10544928 | 1:1000 |
| Antibody | P-gp (rabbit monoclonal) | CST | 13978S RRID:AB_2798357 | 1:1500 |
| Antibody | p-AKT (mouse monoclonal) | Huaan Biotechnology | ET1607 RRID:AB_2940863 | 1:2000 |
| Antibody | AKT (mouse monoclonal) | Huaan Biotechnology | ET1609 RRID:AB_3069857 | 1:2000 |
| Antibody | p-ERK (rabbit polyclonal) | Proteintech | 28733-1-AP RRID:AB_2881202 | 1:1000 |
| Antibody | ERK (rabbit polyclonal) | Proteintech | 11257-1-AP RRID:AB_2139822 | 1:1000 |
| Antibody | p-p38 (rabbit monoclonal) | CST | 4511S RRID:AB_10890701 | 1:250 |
| Antibody | p38 (rabbit monoclonal) | CST | 8690S RRID:AB_10999090 | 1:250 |
| Antibody | p-JNK (rabbit polyclonal) | Wanleibio | WL01813 RRID:AB_2910628 | 1:1000 |
| Antibody | JNK (rabbit polyclonal) | Wanleibio | WL01295 RRID:AB_3064853 | 1:1000 |
| Antibody | FOXO1 (rabbit polyclonal) | Proteintech | 18592 RRID:AB_2934932 | 1:1000 |
| Antibody | p-FOXO1 (rabbit polyclonal) | Wanleibio | WL03634 RRID: AB_3076326 | 1:1000 |
| Antibody | ETS1 (mouse monoclonal) | Santa Cruz | sc-55581 RRID:AB_831289 | 1:500 |
| Antibody | ETS1 (mouse monoclonal) | Proteintech | 66598 RRID:AB_2881958 | 1:3000 |
| Cell line (Homo sapiens) | hCMEC/D3 cells | JENNIO Biological Technology, Guangzhou, China | Cat#JNO-H0520 RRID:CVCL_U985 | Authenticated (STR profiling) |
| Cell line (H. sapiens) | U251 cells | Cellcook Biological Technology, Guangzhou, China | Cat#CC1701 RRID:CVCL_0021 | Authenticated (STR profiling) |
| Cell line (H. sapiens) | SH-SY5Y cells | Cellcook Biological Technology, Guangzhou, China | Cat#CC2101 RRID:CVCL_0019 | Authenticated (STR profiling) |
| Software, algorithm | GraphPad Prism | Version 8.0.2 | RRID:SCR_002798 | |
| Software, algorithm | BioTek Cytation 5 Cell Imaging Multi-Mode Reader | BioTek Cytation 5 | RRID:SCR_019732 | |
| Software, algorithm | QuantStudio 3 Real Time PCR System | QuantStudio 3 | RRID:SCR_018712 | |
| Software, algorithm | FACS Celesta Flow Cytometer | BD Biosciences | RRID:SCR_019597 | |
| Software, algorithm | Flowjo software | Version 10.4 | RRID:SCR_008520 | |
| Commercial assay or kit | GDNF-Elisa kit | R&D system RRID:SCR_006140 | Cat#212-GD | |
| Commercial assay or kit | bFGF-Elisa kit | Elabscience RRID:SCR_025982 | Cat#E-EL-H6042 | |
| Commercial assay or kit | IGF-1-Elisa kit | Elabscience RRID:SCR_025982 | Cat#E-EL-H0086 | |
| Commercial assay or kit | TGF-β-Elisa kit | Elabscience RRID:SCR_025982 | Cat#E-EL-0162 | |
| Peptide, recombinant protein | GDNF | R&D system RRID:SCR_006140 | Cat#212-GD | |
| Peptide, recombinant protein | bFGF | MedChemExpress RRID:SCR_025062 | Cat#HY-P7331 | |
| Peptide, recombinant protein | IGF-1 | MedChemExpress RRID:SCR_025062 | Cat#HY-P70783 | |
| Peptide, recombinant protein | TGF-β | MedChemExpress RRID:SCR_025062 | Cat#HY-P70543 | |
| Chemical compound, drug | SPP-86 | MedChemExpress RRID:SCR_025062 | Cat#HY-110193 | |
| Chemical compound, drug | PP2 | MedChemExpress RRID:SCR_025062 | Cat#HY-13805 | |
| Chemical compound, drug | LY294002 | MedChemExpress RRID:SCR_025062 | Cat#HY-10108 | |
| Chemical compound, drug | U0126 | MedChemExpress RRID:SCR_025062 | Cat#S1102 | |
| Chemical compound, drug | SP600125 | Selleck RRID:SCR_003823 | Cat#HY-12041 | |
| Chemical compound, drug | SB203580 | MedChemExpress RRID:SCR_025062 | Cat#HY-10256 |
Cell culture and viability assay
Request a detailed protocolRat BMECs were isolated from Sprague-Dawley rats (male, 7–10 days old, Sino-British Sippr/BKLaboratory Animal Ltd, Shanghai, China) as the described method (Ji et al., 2013; Li et al., 2013) and cultured in Dulbecco’s Modified Eagle Media (DMEM)/F12 (#12500-039, Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS) (#10100147C, Gibco, Carlsbad, CA, USA) and 62.5 µg/ml penicillin and 100 µg/ml streptomycin (SunShine Biotechnology Co., Ltd, Nanjing, China). Then, hCMEC/D3, U251, and SH-SY5Y cells were cultured in DMEM/F12 containing 10% FBS, 62.5 µg/ml penicillin and 100 µg/ml streptomycin. Cell viability was assessed using a CCK-8 kit (Beyotime Biotechnology, Shanghai, China), and the results were expressed as the fold of control.
Establishment of the triple co-culture model
Request a detailed protocolAlthough hCMEC/D3 cells have poor barrier properties and low TEER compared to human physiological BBB, the use of human BMECs may be restricted by the acquisition of materials and ethical approval. Isolation and purification of primary BMECs are time-consuming and laborious. Moreover, culture conditions can alter transcriptional activity (Qi et al., 2023). All limit the establishment of BBB models based on primary human BMECs for high-throughput screening. Here, hCMEC/D3 cells were selected to establish an in vitro BBB model.
The establishment process of the triple co-culture model is illustrated in Figure 9. SH-SY5Y cells were seeded at a density of 4.5 × 104 cells/cm2 in plates and differentiated with 10 μM retinoic acid (Sigma-Aldrich, St. Louis, MO, USA) for 72 hr. The differentiated SH-SY5Y cells were cultured in the fresh DMEM/F12 medium containing 10% FBS. U251 cells were seeded at 2 × 104 cells/cm2 on the bottom of Transwell inserts (PET, 0.4 µm pore size, SPL Life Sciences, Pocheon, Korea) coated with rat-tail collagen (Corning Inc, Corning, NY, USA). Next, the inserts were suspended in plate wells containing the culture medium after 5 hr of incubation. After 24 hr of incubation, hCMEC/D3 cells were seeded on the apical side of the inserts at 3 × 104 cells/cm2 and cultured for another 48 hr. Then, the inserts seeded with U251 and hCMEC/D3 cells were suspended in plates seeded with differentiated SH-SY5Y cells and co-cultured for another 6 days. The culture medium was replaced every 24 hr. TEER was periodically measured using a Millicell-ERS (MERS00002) instrument (Millipore, Billerica MA, USA) to monitor cell confluence and development of TJs.
EdU incorporation assay
Request a detailed protocolThe cells were incubated with medium containing 10 μM Edu for 2 hr. Then cells were washed by phosphate-buffered saline (PBS) and harvested by 0.25% trypsin–ethylenediaminetetraacetic acid (#25200072, Gibco, Carlsbad, CA, USA). The EdU incorporation assay was measured using the BeyoClick EdU Cell Proliferation Kit (Beyotime Biotechnology, Shanghai, China) according to the manufacturer’s instructions. The samples were determined on the FACSCelesta flow cytometer (Becton, Dickins on and Company, USA), and data were analyzed by Flowjo 10.4 software.
In vitro BBB permeability study
Request a detailed protocolOn day 7, the TEER values of BBB models showed a decreasing trend. Therefore, the subsequent experiments were all completed on day 6. The culture medium was removed from the apical and basolateral sides of the inserts and washed twice with preheated Hank’s balanced salt solution (HBSS). Fresh HBSS was then added to both the apical and basolateral chambers. After 15 min of preincubation, HBSS in the apical and basolateral chambers was replaced with HBSS containing FITC-dextran 3–5 kDa (FITC-Dex) (Sigma-Aldrich, St. Louis, MO, USA), fluorescein sodium (Sigma-Aldrich, St. Louis, MO, USA), or other tested agents and blank HBSS, respectively. Next, 200 μl aliquots were collected from the basolateral chamber after 30 min of incubation at 37°C. The concentrations of the tested agents in the basolateral chamber were measured.
The apparent permeability coefficient (Papp, cm/s) values of the tested agents across the in vitro BBB model were calculated using the equation (Tavelin et al., 2002):
where S is the surface area of the insert membrane (0.33 cm2 for 6.5 mm inserts, 4.46 cm2 for 24 mm inserts), Q is the transported amount of the tested agents transported from the donor chamber to the receiver chamber for 30 min (1800 s), and C0 is the initial concentration of the tested agents in the donor chamber.
The quantification methods of prazosin, verapamil, lamotrigine
Request a detailed protocolPrazosin, verapamil, and lamotrigine (Aladdin, Shanghai, China) were analyzed by high-performance liquid chromatography (Shimadzu, Kyoto, Japan) with YMC-Triart C18 column (5 μm, 150 × 4.6 mm, YMC America Inc, Allentown, PA, USA). Prazosin and verapamil were detected using the RF-20A fluorescence detector. Lamotrigine was detected using the SPD-20A ultraviolet detector. Samples were centrifugated at 12,000 rpm for 10 min, then 150 μl supernatant was taken and used for analysis. The run temperature was set at 40°C, the injection volume was 20 μl and the flow rate was 1 ml/min. Initial concentrations in donor chamber and other chromatographic conditions of drugs are summarized in Table 2.
Initial concentrations in donor chamber and chromatographic conditions of prazosin, verapamil, and lamotrigine.
| Compound | Concentration (μM) | Wavelength (nm) |
|---|---|---|
| Prazosin | 5 | Ex: 250 |
| Em: 390 | ||
| Verapamil | 5 | Ex: 280 |
| Em: 310 | ||
| Lamotrigine | 6 | 220 |
The quantification methods of clozapine, venlafaxine, bupropion, amantadine, carbamazepine, fluoxetine, amitriptyline, gabapentin, midazolam, risperidone, olanzapine, mirtazapine, metoclopramide, doxepin, donepezil
Request a detailed protocolAll compounds were purchased from Aladdin (Shanghai, China). Except for prazosin, verapamil, and lamotrigine, the other compounds were analyzed by using liquid chromatography–mass spectrometry (Shimadzu, Kyoto, Japan) with YMC-Triart C18 column (5 μm, 150 × 2.0 mm, YMC America Inc, Allentown, PA, USA). Each sample was mixed with 10 µl internal standard. Then 1 ml extraction was added to each sample. The samples were vortex vibrated on the oscillator for 10 min, and then centrifuged at 4°C and 12,000 rpm for 10 min. The supernatant solvent was evaporated with nitrogen flow, then redissolved with 100 µl 40% (vol/vol) acetonitrile, and centrifuged at 4°C and 15,000 rpm for 10 min. The supernatants were injected into LC–MS for analysis. The injection volume of each sample was 5 µl. The mass charge ratio, extraction, and initial concentrations in donor chamber of drugs are summarized in Table 3.
The summary of mass charge ratio, extraction, initial concentrations in donor chamber.
| Compound | Concentration (μM) | Mass charge ratio[M+H]+ | Extraction |
|---|---|---|---|
| Amantadine | 3 | 181 | Water-saturated N-butanol |
| Amitriptyline | 1.5 | 278 | Ethyl acetate |
| Bupropion | 3 | 240 | Ethyl acetate |
| Carbamazepine | 3 | 237 | Ethyl acetate |
| Clozapine | 4 | 327 | Ethyl acetate |
| Donepezil | 3 | 380 | Methyl tert-butyl ether |
| Doxepin | 4 | 317 | Methyl tert-butyl ether |
| Fluoxetine | 3 | 310 | Ethyl acetate |
| Gabapentin | 10 | 172 | Ethyl acetate |
| Metoclopramide | 4 | 301 | Ethyl acetate |
| Midazolam | 3 | 327 | Ethyl acetate |
| Mirtazapine | 3 | 266 | Methyl tert-butyl ether |
| Olanzapine | 3 | 313 | Methyl tert-butyl ether |
| Risperidone | 4 | 427 | Methyl tert-butyl ether |
| Venlafaxine | 10 | 278 | Ethyl acetate |
Cell density analysis
Request a detailed protocolOn day 6 of co-culture, hCMEC/D3 cells were fixed with 4% paraformaldehyde for 15 min and washed with PBS for three times. Next, the fixed cells were blocked with 5% goat serum for 2 hr and washed with PBS for four times. The blocked cells were incubated with 4’,6-diamidino-2-phenylindole (DAPI) (Invitrogen, Carlsbad, CA, USA) and washed with PBS for four times. Cell numbers were counted using Cytation5 (BioTek, Winooski, VT, USA).
Reverse transcription and qPCR
Request a detailed protocolTotal RNA of cells was extracted using RNAiso Plus reagent (Takara Bio Inc, Otsu, Shiga, Japan) and reverse transcribed using HiScript III RT SuperMix (Vazyme, Shanghai, China) as the described method (Yang et al., 2023). Paired primers were synthesized by Tsingke Biotech Co., Ltd (Beijing, China), and their sequences are listed in Table 4. The SYBR Master Mix was purchased from Yeasen (Shanghai, China). Then, qPCR was performed on the Applied Biosystems QuantStudio 3 real-time PCR system (Thermo Fisher Scientific, Waltham, MA, USA). The mRNA levels of related genes were normalized to ACTB or GAPDH using the comparative cycle threshold method.
Primer sequences for quantitative real-time PCR (qPCR) for indicted genes.
| Gene (protein) | Forwards primer, 5′→3′ | Reverse primer, 3′→5′ |
|---|---|---|
| ACTB (β-actin) | GGACTTCGAGCAAGAGATGG | AGCACTGTGTTGGCGTACAG |
| GAPDH (GAPDH) | TGTGGGCATCAATGGATTTGG | ACACCATGTATTCCGGGTCAAT |
| CLDN5 (claudin-5) | CTCTGCTGGTTCGCCAACAT | CAGCTCGTACTTCTGCGACA |
| OCLN (occludin) | ACAAGCGGTTTTATCCAGAGTC | GTCATCCACAGGCGAAGTTAAT |
| TJP1 (ZO-1) | ACCAGTAAGTCGTCCTGATCC | TCGGCCAAATCTTCTCACTCC |
| CDH5 (VE-cadherin) | AAGCGTGAGTCGCAAGAATG | TCTCCAGGTTTTCGCCAGTG |
| ABCB1 (P-gp) | TTGCTGCTTACATTCAGGTTTCA | AGCCTATCTCCTGTCGCATTA |
| ABCG2 (BCRP) | ACGAACGGATTAACAGGGTCA | CTCCAGACACACCACGGAT |
| SLC22A1 (OCT1) | ACGGTGGCGATCATGTACC | CCCATTCTTTTGAGCGATGTGG |
| SLC22A2 (OCT2) | CATCGTCACCGAGTTTAACCTG | AGCCGATACTCATAGAGCCAAT |
| SLC22A8 (OAT3) | ATGGCCCAGTCTATCTTCATGG | GACGGTGCTCAGGGTAATGC |
| SLCO1A1 (OATP1A1) | TAATGTGGGTGTACGTCCTAGT | GCTCCTGTTTCTACAAGCCCAA |
| GDNF (GDNF) | GCAGACCCATCGCCTTTGAT | CCACACCTTTTAGCGGAATGC |
| BDNF (BDNF) | CTACGAGACCAAGTGCAATCC | AATCGCCAGCCAATTCTCTTT |
| NGF (NGF) | TGTGGGTTGGGGATAAGACCA | GCTGTCAACGGGATTTGGGT |
| IGF1 (IGF-1) | GCTCTTCAGTTCGTGTGTGGA | GGTCATGGATGGACCTTACTGT |
| VEGFA (VEGF) | CCCACTGAGGAGTCCAACAT | AAATGCTTTCTCCGCTCTGA |
| FGF2 (bFGF) | AGAAGAGCGACCCTCACATCA | CGGTTAGCACACACTCCTTTG |
| TGFB1 (TGF-β) | GGCCAGATCCTGTCCAAGC | GTGGGTTTCCACCATTAGCAC |
Western blotting analysis
Request a detailed protocolWhole-cell and tissue lysates, nucleoprotein, and cytoplasmic protein were prepared using RIPA Lysis Buffer (Beyotime, Shanghai, China) as the described method (Wu et al., 2021). Proteins were separated through sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto nitrocellulose or polyvinylidene difluoride membranes. The membranes were blocked with 5% skim milk and incubated with corresponding primary antibodies at 4°C overnight. After being washed with Tris-buffered saline Tween buffer, the membranes were incubated with secondary antibodies (Cell Signaling Technology, MA, USA) at 1:3000 dilution: Anti-mouse IgG, HRP-linked Antibody (#7076), Anti-rabbit IgG, HRP-linked Antibody (#7074). Protein levels were visualized using a highly sensitive ECL western blotting substrate and a gel imaging system (Tanon Science & Technology, Shanghai, China).
Preparation of CM
Request a detailed protocolCM of U251 cells (U-CM), SH-SY5Y cells (S-CM), or co-culture of U251 and SH-SY5Y cells (US-CM) was prepared. U251 cells were seeded at the top of the insert membrane and suspended on 6-well plates seeded with differentiated SH-SY5Y cells to co-culture U251 cells with SH-SY5Y cells. The medium was collected every 24 hr. The CMs were subsequently used for hCMEC/D3 cell culture after filtrating with 0.2 μm filters for 144 hr. The levels of GDNF, bFGF, IGF-1, and TGF-β in CMs were measured using corresponding ELISA kits according to the manufacturers’ instructions.
Neutralization of GDNF with anti-GDNF antibody
Request a detailed protocolExogenous and endogenous GDNF in the medium was neutralized with anti-GDNF antibody (#AF-212-NA, R&D system, Minneapolis, MN, USA). 0.25, 0.5, and 1.0 μg/ml anti-GDNF antibody was added into the US-CM or medium containing 200 pg/ml GDNF, and then the medium was preincubated at 4°C for 1 hr. The hCMEC/D3 cells were incubated with 200 pg/ml GDNF or US-CM containing anti-GDNF antibody or not for 6 days, and then cell lysate was collected for western blot. The medium was replaced every 24 hr.
Transfection of hCMEC/D3
Request a detailed protocolHere, hCMEC/D3 cells were plated in the plates or culture dishes at 6 × 104 cells/cm2 and transfected with 10 nM of negative control or human FOXO1, ETS1, and CDH5 siRNA (Tsingke Biotechnology, Beijing, China) for 12 hr using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) reagent according to the manufacturer’s instructions. Cells were then incubated with a medium containing GDNF for 72 hr. The siRNA sequences of human FOXO1, ETS1, and CDH5 are summarized in Table 5.
The target sequences for small interfering RNA (siRNA) or short hairpin RNA (shRNA).
| Gene | Target sequence |
|---|---|
| ETS1 | CGCTATACCTCGGATTACT |
| FOXO1 | AATCTCCTAGGAGAAGAGCTG |
| Gdnf | GCCAGTGTTTATCTGATAC |
| CDH5 | GCCTCTGTCATGTACCAAA |
FOXO1 overexpression by plasmids
Request a detailed protocolThe plasmids encoding FOXO1 (EX-Z7404-M02) were constructed by GeneCopoeia (Rockville, MD, USA). The hCMEC/D3 cells were plated in plates or dishes at 6 × 104 cells/cm2. They were subsequently transfected with 1 µg of negative control or plasmids encoding FOXO1 6 hr using Lipofectamine 3000 reagent according to the manufacturer’s instructions. Transfected cells were then incubated with the medium containing GDNF for 72 hr.
Animals
C57BL/6J mice (male, 4–5 weeks old, 16–18 g, 12 mice) were obtained from Sino-British Sippr/BKLaboratory Animal Ltd (Shanghai, China). Mice were maintained in groups under standard conditions with free access to food and water. Animal studies were performed in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health) and approved by the Animal Ethics Committee of China Pharmaceutical University (Approval Number: 202307003).
Brain-specific Gdnf knockdown and evaluation of BBB permeability
Request a detailed protocolMice were randomly divided into control (shNC) and Gdnf silencing (shGdnf) groups (6 mice each group). The shRNA sequence of mice Gdnf is listed in Table 5. The 2 × 109 viral genome each of pAAV-U6-shRNA (NC2)-CMV-EGFP or pAAV-U6-shRNA (Gdnf)-CMV-EGFP (OBio Technology, Beijing, China) was injected into the bilateral lateral ventricle area (relative to the bregma: anterior–posterior −0.3 mm; medial–lateral ±1.0 mm; dorsal–ventral –3.0 mm) through i.c.v infusion. Three weeks following i.c.v injection, BBB permeability and expression of corresponding targeted proteins were measured in the mice.
A mixture of FITC-Dex (50 mg/kg) and fluorescein sodium (10 mg/kg) was intravenously administered to experimental mice. Thirty minutes after the injection, the mice were euthanized under isoflurane anesthesia, and brain tissue and plasma samples were obtained quickly. The concentrations of FITC-Dex and fluorescein in the plasma and brain were measured as previously described (Li et al., 2022b; Zhou et al., 2019). No blinding was performed in animal studies.
The prediction of drug permeability across BBB using the developed in vitro BBB model
Request a detailed protocolThe Papp values of 18 drugs – prazosin, verapamil, lamotrigine, clozapine, venlafaxine, bupropion, amantadine, carbamazepine, fluoxetine, amitriptyline, gabapentin, midazolam, risperidone, olanzapine, mirtazapine, metoclopramide, doxepin, and donepezil – across the hCMEC/D3 cells mono-culture and triple co-culture models were measured. The predicted in vivo permeability-surface area product (PSPre, μl/min/g brain) values across BBB were calculated using the following equation:
where VSA is the luminal area of the vascular space of brain, which was set to 150 cm2/g (Fenstermacher et al., 1988), and fu, brain is the unbound fraction of brain. The published in vivo brain permeability values were unified to observed PS (PSObs) by multiplying by VSA equal to 150 cm2/g. If PSPre values were within 0.5- to 2.0-folds of observations, the prediction was considered successful.
Statistical analyses
Request a detailed protocolAll results are presented as mean ± SEM. The average of technical replicates generated a single independent value that contributes to the n value used for comparative statistical analysis. The data were assessed for Gaussian distributions using Shapiro–Wilk test. Brown–Forsythe test was employed to evaluate the homogeneity of variance between groups. For comparisons between two groups, statistical significance was determined by unpaired two-tailed t-test. The acquired data with significant variation were tested using unpaired t-test with Welch’s correction, and non-Gaussian distributed data were tested using Mann–Whitney test. For multiple group comparisons, one-way ANOVA followed by Fisher’s LSD test was used to determine statistical significance. The acquired data with significant variation were tested using Welch’s ANOVA test, and non-Gaussian distributed data were tested using Kruskal–Wallis test. p < 0.05 was considered statistically significant. The simple linear regression analysis was used to examine the presence of a linear relationship between two variables. Data were analyzed using GraphPad Prism software version 8.0.2 (GraphPad Software, La Jolla, CA, USA).
Data availability
All data are available in the manuscript and supporting files; source data files have been provided for Figures 1–6 and Table 1. Source data 1 and Source data 2 files of Figures 1–6 contain original files of western blots. Source data 3 files of Figures 1–6 and Table 1—source data 1 contain the numerical data used to generate the figures.
References
-
Blood–brain barrier structure and function and the challenges for CNS drug deliveryJournal of Inherited Metabolic Disease 36:437–449.https://doi.org/10.1007/s10545-013-9608-0
-
The blood–brain barrier and blood–tumour barrier in brain tumours and metastasesNature Reviews Cancer 20:26–41.https://doi.org/10.1038/s41568-019-0205-x
-
From blood–brain barrier to blood–brain interface: new opportunities for CNS drug deliveryNature Reviews Drug Discovery 15:275–292.https://doi.org/10.1038/nrd.2015.21
-
A human-derived neurovascular unit in vitro model to study the effects of cellular cross-talk and soluble factors on barrier integrityFrontiers in Cellular Neuroscience 16:1065193.https://doi.org/10.3389/fncel.2022.1065193
-
AKT2 maintains brain endothelial claudin-5 expression and selective activation of IR/AKT2/FOXO1-signaling reverses barrier dysfunctionJournal of Cerebral Blood Flow and Metabolism 40:374–391.https://doi.org/10.1177/0271678X18817512
-
Glial-derived neurotrophic factor in human airway smooth muscleJournal of Cellular Physiology 236:8184–8196.https://doi.org/10.1002/jcp.30489
-
Population model analysis of chiral inversion and degradation of bupropion enantiomers, and application to enantiomer specific fraction unbound determination in rat plasma and brainJournal of Pharmaceutical and Biomedical Analysis 195:113872.https://doi.org/10.1016/j.jpba.2020.113872
-
A definite measure of occupancy exposures, seeking with non-radiolabeled in vivo 5-HT2A receptor occupancy and in vitro free fractionsJournal of Receptor and Signal Transduction Research 38:359–366.https://doi.org/10.1080/10799893.2018.1531888
-
An examination of protein binding and protein-facilitated uptake relating to in vitro-in vivo extrapolationEuropean Journal of Pharmaceutical Sciences 123:502–514.https://doi.org/10.1016/j.ejps.2018.08.008
-
The special relationship: glia-neuron interactions in the neuroendocrine hypothalamusNature Reviews. Endocrinology 14:25–44.https://doi.org/10.1038/nrendo.2017.124
-
Compensatory increase of VE-cadherin expression through ETS1 regulates endothelial barrier function in response to TNFαCellular and Molecular Life Sciences 77:2125–2140.https://doi.org/10.1007/s00018-019-03260-9
-
Microdialysis evaluation of clozapine and N-desmethylclozapine pharmacokinetics in rat brainDrug Metabolism and Disposition 40:1909–1916.https://doi.org/10.1124/dmd.112.045682
-
The role of adherens junctions and VE-cadherin in the control of vascular permeabilityJournal of Cell Science 121:2115–2122.https://doi.org/10.1242/jcs.017897
-
Species independence in brain tissue binding using brain homogenatesDrug Metabolism and Disposition 39:1270–1277.https://doi.org/10.1124/dmd.111.038778
-
Comparative study of four immortalized human brain capillary endothelial cell lines, hCMEC/D3, hBMEC, TY10, and BB19, and optimization of culture conditions, for an in vitro blood-brain barrier model for drug permeability studiesFluids and Barriers of the CNS 10:33.https://doi.org/10.1186/2045-8118-10-33
-
Computational model to predict the fraction of unbound drug in the brainJournal of Chemical Information and Modeling 59:3251–3261.https://doi.org/10.1021/acs.jcim.9b00180
-
Structural and functional variations in capillary systems within the brainAnnals of the New York Academy of Sciences 529:21–30.https://doi.org/10.1111/j.1749-6632.1988.tb51416.x
-
Blood-brain barrier models: Rationale for selectionAdvanced Drug Delivery Reviews 176:113859.https://doi.org/10.1016/j.addr.2021.113859
-
The CLDN5 gene at the blood-brain barrier in health and diseaseFluids and Barriers of the CNS 20:22.https://doi.org/10.1186/s12987-023-00424-5
-
Glial cell line-derived neurotrophic factor induces barrier function of endothelial cells forming the blood-brain barrierBiochemical and Biophysical Research Communications 261:108–112.https://doi.org/10.1006/bbrc.1999.0992
-
Modulation of P-glycoprotein in rat brain microvessel endothelial cells under oxygen glucose deprivationThe Journal of Pharmacy and Pharmacology 65:1508–1517.https://doi.org/10.1111/jphp.12122
-
IGF-1 released by corneal epithelial cells induces up-regulation of N-cadherin in corneal fibroblastsJournal of Cellular Physiology 221:254–261.https://doi.org/10.1002/jcp.21850
-
Quantitative evaluation of the impact of active efflux by p-glycoprotein and breast cancer resistance protein at the blood-brain barrier on the predictability of the unbound concentrations of drugs in the brain using cerebrospinal fluid concentration as a surrogateThe Journal of Pharmacology and Experimental Therapeutics 339:935–944.https://doi.org/10.1124/jpet.111.180398
-
The Ets family contains transcriptional activators and repressors involved in angiogenesisThe International Journal of Biochemistry & Cell Biology 33:391–407.https://doi.org/10.1016/S1357-2725(01)00025-5
-
Bile duct ligation impairs function and expression of mrp1 at rat blood–retinal barrier via bilirubin-induced p38 mapk pathway activationsInternational Journal of Molecular Sciences 23:7666.https://doi.org/10.3390/ijms23147666
-
Use of a physiologically based pharmacokinetic model to study the time to reach brain equilibrium: an experimental analysis of the role of blood-brain barrier permeability, plasma protein binding, and brain tissue bindingThe Journal of Pharmacology and Experimental Therapeutics 313:1254–1262.https://doi.org/10.1124/jpet.104.079319
-
Unfractionated heparin alleviates sepsis-induced acute lung injury by protecting tight junctionsThe Journal of Surgical Research 238:175–185.https://doi.org/10.1016/j.jss.2019.01.020
-
Relationship between exposure and nonspecific binding of thirty-three central nervous system drugs in miceDrug Metabolism and Disposition 33:175–181.https://doi.org/10.1124/dmd.104.001222
-
Regulation of dendritic branching and spine maturation by semaphorin3A-Fyn signalingThe Journal of Neuroscience 26:2971–2980.https://doi.org/10.1523/JNEUROSCI.5453-05.2006
-
The neurovascular unit - concept reviewActa Physiologica 210:790–798.https://doi.org/10.1111/apha.12250
-
A new blood-brain barrier model using primary rat brain endothelial cells, pericytes and astrocytesNeurochemistry International 54:253–263.https://doi.org/10.1016/j.neuint.2008.12.002
-
Size-selective loosening of the blood-brain barrier in claudin-5-deficient miceThe Journal of Cell Biology 161:653–660.https://doi.org/10.1083/jcb.200302070
-
Exogenous expression of claudin-5 induces barrier properties in cultured rat brain capillary endothelial cellsJournal of Cellular Physiology 210:81–86.https://doi.org/10.1002/jcp.20823
-
In vitro cerebrovascular modeling in the 21st century: current and prospective technologiesPharmaceutical Research 31:3229–3250.https://doi.org/10.1007/s11095-014-1464-6
-
Neuronal GDNF expression in the adult rat nervous system identified by in situ hybridizationThe European Journal of Neuroscience 9:463–471.https://doi.org/10.1111/j.1460-9568.1997.tb01623.x
-
Tissue engineering 3d neurovascular units: a biomaterials and bioprinting perspectiveTrends in Biotechnology 36:457–472.https://doi.org/10.1016/j.tibtech.2018.01.003
-
A review on in vitro model of the blood-brain barrier (BBB) based on hCMEC/D3 cellsJournal of Controlled Release 358:78–97.https://doi.org/10.1016/j.jconrel.2023.04.020
-
In vitro model for predicting the access and distribution of drugs in the brain using hCMEC/D3 cellsEuropean Journal of Pharmaceutics and Biopharmaceutics 163:120–126.https://doi.org/10.1016/j.ejpb.2021.04.002
-
Synergistic effects of neurons and astrocytes on the differentiation of brain capillary endothelial cells in cultureJournal of Cellular and Molecular Medicine 7:165–170.https://doi.org/10.1111/j.1582-4934.2003.tb00215.x
-
Permeability properties of a three-cell type in vitro model of blood-brain barrierJournal of Cellular and Molecular Medicine 9:373–379.https://doi.org/10.1111/j.1582-4934.2005.tb00362.x
-
Central nervous system drug disposition: the relationship between in situ brain permeability and brain free fractionThe Journal of Pharmacology and Experimental Therapeutics 322:205–213.https://doi.org/10.1124/jpet.107.121525
-
Blood-brain barrier: from physiology to disease and backPhysiological Reviews 99:21–78.https://doi.org/10.1152/physrev.00050.2017
-
Negative regulation of the forkhead transcription factor fkhr by aktJournal of Biological Chemistry 274:16741–16746.https://doi.org/10.1074/jbc.274.24.16741
-
BookApplications of epithelial cell culture in studies of drug transportIn: Wise C, editors. Epithelial Cell Culture Protocols. Totowa, NJ: Humana Press. pp. 233–272.https://doi.org/10.1385/1-59259-185-x:233
-
Transcription factor ets-1 mediates ischemia- and vascular endothelial growth factor-dependent retinal neovascularizationThe American Journal of Pathology 164:1827–1835.https://doi.org/10.1016/S0002-9440(10)63741-8
-
The hCMEC/D3 cell line as a model of the human blood brain barrierFluids and Barriers of the CNS 10:16.https://doi.org/10.1186/2045-8118-10-16
-
A novel brain neurovascular unit model with neuronsAstrocytes and Microvascular Endothelial Cells of Rat. International Journal of Biological Sciences 9:174–189.https://doi.org/10.7150/ijbs.5115
-
Akt, FoxO and regulation of apoptosisBiochimica et Biophysica Acta (BBA) - Molecular Cell Research 1813:1978–1986.https://doi.org/10.1016/j.bbamcr.2011.03.010
-
The effect of breast cancer resistance protein and P-glycoprotein on the brain penetration of flavopiridol, imatinib mesylate (Gleevec), prazosin, and 2-methoxy-3-(4-(2-(5-methyl-2-phenyloxazol-4-yl)ethoxy)phenyl)propanoic acid (PF-407288) in miceDrug Metabolism and Disposition 37:946–955.https://doi.org/10.1124/dmd.108.024489
-
Increase in P-glycoprotein levels in the blood-brain barrier of partial portal vein ligation /chronic hyperammonemia rats is medicated by ammonia/reactive oxygen species/ERK1/2 activation: In vitro and in vivo studiesEuropean Journal of Pharmacology 846:119–127.https://doi.org/10.1016/j.ejphar.2019.01.005
Article and author information
Author details
Funding
National Natural Science Foundation of China (82373943)
- Xiaodong Liu
National Natural Science Foundation of China (82173884)
- Li Liu
National Natural Science Foundation of China (82204511)
- Hanyu Yang
Double First Class University Plan (CPU2022QZ21)
- Li Liu
Jiangsu Funding Program for Excellent Postdoctoral Talent (2022ZB305)
- Hanyu Yang
China Postdoctoral Science Foundation (2023M733897)
- Hanyu Yang
Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX24_1019)
- Lu Yang
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
The authors would like to give special thanks to the support of the China Pharmaceutical University Pharmaceutical Animal Laboratory Center.
Ethics
This study was performed in strict accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health). The protocol was approved by the Animal Ethics Committee of China Pharmaceutical University (Approval Number: 202307003). All surgery was performed under isoflurane anesthesia, and every effort was made to minimize suffering.
Version history
- Sent for peer review:
- Preprint posted:
- Reviewed Preprint version 1:
- Reviewed Preprint version 2:
- Version of Record published:
Cite all versions
You can cite all versions using the DOI https://doi.org/10.7554/eLife.96161. This DOI represents all versions, and will always resolve to the latest one.
Copyright
© 2024, Yang et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,921
- views
-
- 150
- downloads
-
- 18
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 16
- citations for umbrella DOI https://doi.org/10.7554/eLife.96161
-
- 1
- citation for Reviewed Preprint v2 https://doi.org/10.7554/eLife.96161.2
-
- 1
- citation for Version of Record https://doi.org/10.7554/eLife.96161.3