Spatiotemporal recruitment of the ubiquitin-specific protease USP8 directs endosome maturation
Figures
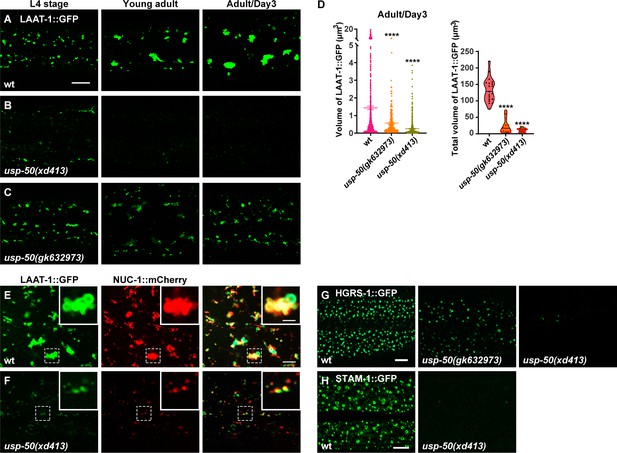
Abnormal lysosome morphology and enlarged multivesicular body (MVB)-like structures in usp-50 mutants.
(A–C) Confocal fluorescence images of hypodermal cell 7 (hyp7) expressing the LAAT-1::GFP marker to highlight lysosome structures in L4 stage, young adult, and 3-day-old adult animals. Scale bar: 5 μm. (D) Quantification of the individual volume and total volume of LAAT-1::GFP vesicles in hyp7 of 3-day-old adults. 19 animals for wild-type, 16 animals for usp-50(xd413), and 13 animals for usp-50(gk632973) were quantified. ****p<0.0001. One-way ANOVA with Tukey’s test. (E, F) The lysosomal membrane marker LAAT-1::GFP co-localizes with the lysosomal hydrolase NUC-1::mCherry in both wild-type (E) and usp-50(xd413) (F) animals. Scale bar represents 5 μm for (E, F) and 2 μm for enlarged inserts. (G) Confocal fluorescence images of hypodermis expressing HGRS-1::GFP in L4 stage worms. Compared to wild-type, HGRS-1::GFP signal is reduced in usp-50(gk632973) and usp-50(xd413) animals. Scale bar: 5 μm. (H) Confocal fluorescence images of hypodermis expressing STAM-1::GFP in L4 stage worms. Compared to wild-type, STAM-1::GFP signal is reduced in usp-50(xd413) animals. Scale bar: 5 μm.
-
Figure 1—source data 1
Excel file containing the quantified data of statistic analysis for Figure 1D.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig1-data1-v1.zip
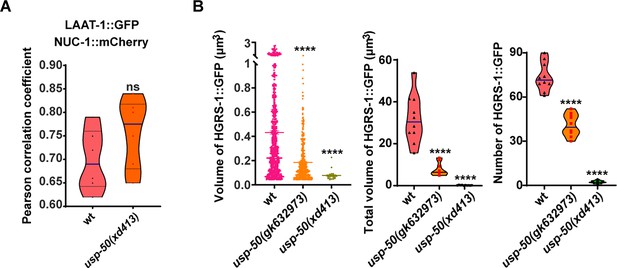
ESCRT-0 components are stabilized by usp-50.
(A) Pearson’s correlation coefficient for co-localization of the lysosome membrane protein LAAT-1 (LAAT-1::GFP) and the lysosomal nuclease NUC-1 (NUC-1::mCherry) in wild-type and usp-50(xd413) animals. ns, not significant. Unpaired Student’s t-test was performed. (B) Quantification of the individual volume, total volume, and number of HGRS-1::GFP puncta in hypodermal cell 7 (hyp7) of L4 stage from 10 animals for wild-type, usp-50(gk632973), and usp-50(xd413), respectively. (Data are presented as mean ± SEM. ****p<0.0001; one-way ANOVA with Tukey’s test.)
-
Figure 1—figure supplement 1—source data 1
Excel file containing the quantified data of statistic analysis for Figure 1—figure supplement 1A and B.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig1-figsupp1-data1-v1.zip
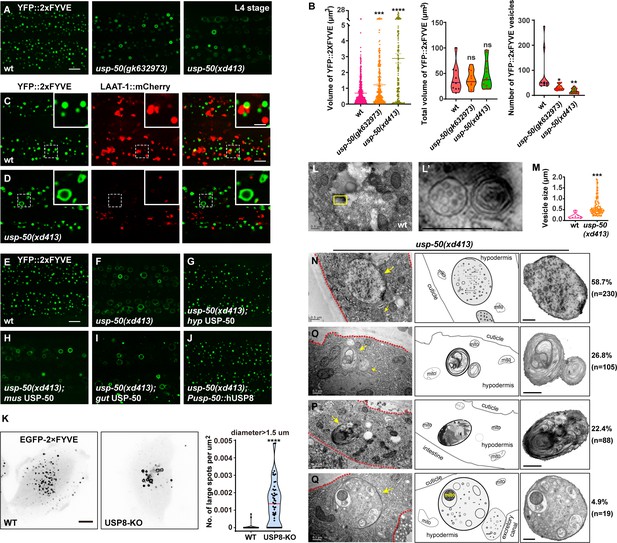
Enlarged early endosomes (EEs) in usp-50/usp8 mutant cells.
(A) Confocal fluorescence images of hypodermis expressing YFP::2xFYVE to detect EEs in L4 stage animals. Compared to wild-type, EEs are enlarged in usp-50(gk632973) and usp-50(xd413) mutants. Scale bar: 5 μm. (B) Quantification of the individual vesicle size, total volume, and number of YFP::2xFYVE-marked vesicles in hyp7 in L4 worms (10 animals for wild-type, 11 animals for usp-50(gk632973), 13 animals for usp-50(xd413)). Data are presented as mean ± SEM. *p<0.05. **p<0.01. ***p<0.001. ****p<0.0001. ns, not significant; one-way ANOVA with Tukey’s test. (C, D) The YFP::2xFYVE marker is not co-localized with LAAT-1::mCherry in wild-type (C) or usp-50(xd413) animals (D). Scale bar represents 5 μm for (C, D) and 2 μm for enlarged inserts. (E–J) The YFP::2xFYVE pattern in wild-type (E), usp-50(xd413) mutants (F), usp-50(xd413) with hypodermis-specific USP-50 expression (G), usp-50(xd413) with muscle-specific USP-50 expression (H), usp-50(xd413) with gut-specific USP-50 expression (I), and usp-50(xd413) with expression of human USP8 driven by the usp-50 promoter (J). Scale bar: 5 μm. (K) Distribution of transiently expressed EGFP-2xFYVE in wild-type and USP8-KO SUM159 cells. Both wild-type and USP8-KO SUM159 cells were transiently transfected with EGFP-2xFYVE and then imaged by spinning-disk confocal microscopy. The right panel shows quantification of the number of large endosomes marked by EGFP-2xFYVE from 38 cells for wild-type and 51 cells for USP8-KO (****p<0.0001; unpaired Student’s t-test). Scale bar: 10 μm. (L, L’) High-pressure freezing EM reveals the multivesicular body (MVB) structures in wild-type animals. The yellow boxed area is enlarged in (L’). (M) Quantification of the abnormal enlarged vesicle in usp-50(xd413) animals. For comparative analysis, the quantification of MVB structures in wild-type animals was also included. Data are presented as mean ± SEM. ***p<0.001. Unpaired Student’s t-test was performed. (N–Q) The enlarged abnormal vesicles in usp-50(xd413), including vesicles containing abundant intraluminal vesicles (ILVs) (58.7%) (N), vesicles filled with threadlike membrane structures (26.8%) (O), vesicles loaded with electron-dense material (22.4%) (P), and vesicles containing various cellular organelles (4.9%) (Q). Red dashed lines indicate hypodermal cells. Yellow arrows indicate representative vesicles. mito, mitochondrion. Scale bar represents 0.5 μm for (L–Q).
-
Figure 2—source data 1
Excel file containing the quantified data of statistic analysis for Figure 2B, K, and M.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig2-data1-v1.zip
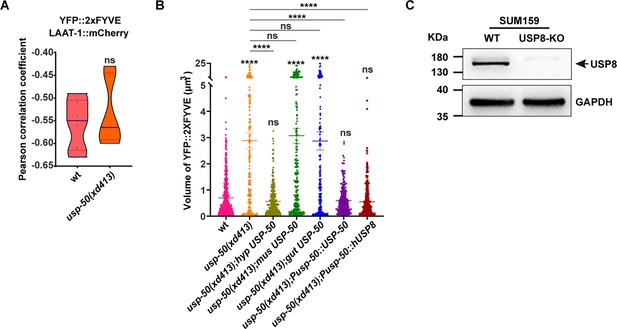
usp-50 functions cell-autonomously.
(A) Pearson’s correlation coefficient for co-localization of early endosomes (EEs) (labeled with YFP::2xFYVE) and lysosomes (labeled with LAAT-1::mCherry) in wild-type and usp-50(xd413) animals. ns, not significant. Unpaired Student’s t-test was performed. (B) Quantification of the volume of individual EEs in various genotypes. Data are presented as mean ± SEM. ****p<0.0001. ns, not significant; one-way ANOVA with Tukey’s test. (C) The CRISPR/Cas9 genome-editing tool was used to knock out the expression of USP8 (USP8-KO) in SUM159 cells. Knockout of USP8 was confirmed by western blot analysis using antibodies against USP8 and GAPDH.
-
Figure 2—figure supplement 1—source data 1
Original file for the western blot analysis in Figure 2—figure supplement 1C (anti-USP8 and anti-GAPDH).
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig2-figsupp1-data1-v1.zip
-
Figure 2—figure supplement 1—source data 2
PDF containing original scans of the relevant western blot analysis (anti-USP8 and anti-GAPDH) with highlighted bands and sample labels for Figure 2—figure supplement 1C.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig2-figsupp1-data2-v1.pdf
-
Figure 2—figure supplement 1—source data 3
Excel file containing the quantified data of statistic analysis for Figure 2—figure supplement 1A and B.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig2-figsupp1-data3-v1.zip

Mutation of usp-50 does not affect other organelles.
(A–C) In usp-50 mutant animals, no obvious alterations are detected in Golgi apparatus (MANS::GFP) (A), recycling endosomes (GFP::RME-1) (B), or retromers (VPS-29::GFP) (C). Confocal fluorescence images of the hypodermis were captured at L4 stage. Scale bars: 5 μm.
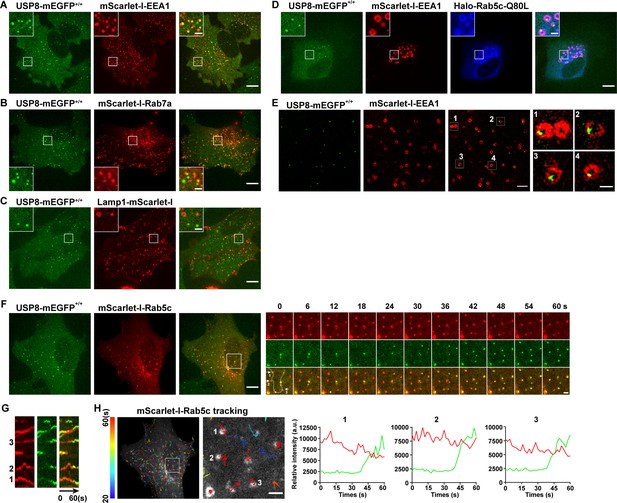
USP8 is recruited to Rab5-positive vesicles.
(A–C) SUM159 cells genome-edited for USP8-mEGFP+/+ were transiently transfected with the indicated mScarlet-I-tagged proteins and then imaged by spinning-disk confocal microscopy. The single-frame image in the figure shows the distribution of USP8 with EEA1, Rab7a, or Lamp1. USP8-mEGFP is co-localized with the early endosome (EE) marker mScarlet-I-EEA1 in genome-edited USP8-mEGFP+/+ SUM159 cells (A). USP8-mEGFP is partly co-localized with the late endosome marker mScarlet-I-Rab7a (B) or the lysosome marker Lamp1-mScarlet-I (C). Scale bar represents 10 μm for (A–C) and 2 μm for enlarged inserts. (D) USP8-mEGFP localization on enlarged EEs in the genome-edited SUM159 cells. SUM159 cells genome-edited for USP8-mEGFP+/+ were transiently transfected with mScarlet-I-EEA1 and Halo-Rab5c-Q80L, labeled with the JF646-HaloTag ligand, and then imaged by spinning-disk confocal microscopy. Scale bar represents 10 μm for (D) and 2 μm for enlarged inserts. (E) Structured illumination microscopy (SIM) images showing the sub-organelle localization of USP8-mEGFP on EEs marked with mScarlet-I-EEA1. USP8-mEGFP+/+ cells were transiently transfected with mScarlet-I-EEA1 and then imaged near the middle plane by SIM. Scale bar represents 2 μm for the left panels and 0.5 μm for the right enlarged panel. (F) SUM159 cells genome-edited for USP8-mEGFP+/+ were transiently transfected with mScarlet-I-Rab5c and then imaged at two planes (starting from the bottom plane, spaced by 0.5 μm) every 2 s (for 1 min) by spinning-disk confocal microscopy. Shown is the first frame of the maximum intensity projection of the two planes. The boxed region is enlarged and the images at the indicated times are shown on the right. Scale bar represents 10 μm for the left images and 2 μm for the right enlarged images. (G) Kymographs along the line (width 3 pixels) on the first merged image of the montage in (F) showing dynamic recruitment of USP8-mEGFP to the pre-existing mScarlet-I-Rab5-positive vesicles. (H) The time-lapse images in (F) were analyzed by single-particle tracking. The trajectories (longer than 20 s, color-coded based on track lifetime) of Rab5c are plotted on the first frame of mScarlet-I-Rab5c. The boxed region is enlarged and shown on the right. The fluorescence intensity traces for mScarlet-I-Rab5c (red) and USP8-mEGFP (green) of the three tracked events are shown. Scale bar: 2 μm.
-
Figure 3—source data 1
Excel file containing the quantified data of statistic analysis for Figure 3H.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig3-data1-v1.xlsx
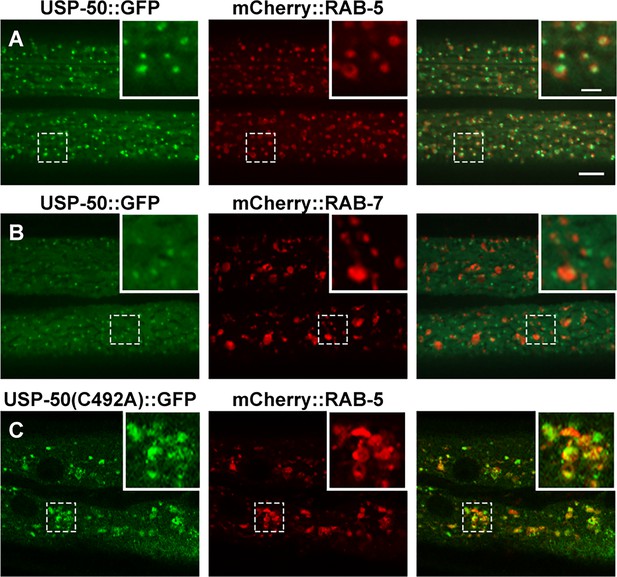
The early endosome (EE) localization of USP-50 and USP8.
(A) USP-50::GFP is co-localized with mCherry::RAB-5 in hypodermal cell 7 (hyp7) of wild-type animals at L4 stage. (B) USP-50::GFP is not co-localized with mCherry::RAB-7 in hypodermal cell 7 (hyp7) of wild-type animals at L4 stage. (C) USP-50::GFP(C492A) is co-localized with mCherry::RAB-5 in hypodermal cell 7 (hyp7) and causes enlarged EEs. Scale bar represents 5 μm for (A–C) and 10 μm for enlarged inserts.
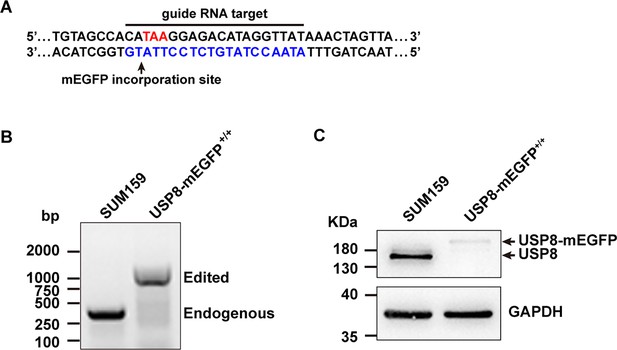
The generation of endogenous tagged USP8.
(A) CRISPR/Cas9 genome-editing strategy used to incorporate mEGFP at the C-terminus of USP8 in SUM159 cells. The target sequence at the genomic USP8 locus recognized by the single-guide RNA is highlighted by the blue nucleotides. The stop codon TAA (red) and the site of mEGFP incorporation upon homologous recombination are highlighted. (B) Genomic PCR analysis showing biallelic integration of mEGFP into the USP8 genomic locus to generate the clonal gene-edited cell line USP8-mEGFP+/+. (C) Western blot analysis of cell lysates probed with antibodies for USP8 and GAPDH. The endogenous and mEGFP-tagged USP8 are indicated by arrows.
-
Figure 3—figure supplement 2—source data 1
Original file for the genomic PCR analysis in Figure 3—figure supplement 2B.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig3-figsupp2-data1-v1.zip
-
Figure 3—figure supplement 2—source data 2
PDF containing original file for the genomic PCR analysis with highlighted bands and sample labels for Figure 3—figure supplement 2B.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig3-figsupp2-data2-v1.pdf
-
Figure 3—figure supplement 2—source data 3
Original file for the western blot analysis in Figure 3—figure supplement 2C (anti-USP8 and anti-GAPDH).
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig3-figsupp2-data3-v1.zip
-
Figure 3—figure supplement 2—source data 4
PDF containing original scans of the relevant western blot analysis (anti-USP8 and anti-GAPDH) with highlighted bands and sample labels for Figure 3—figure supplement 2C.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig3-figsupp2-data4-v1.pdf
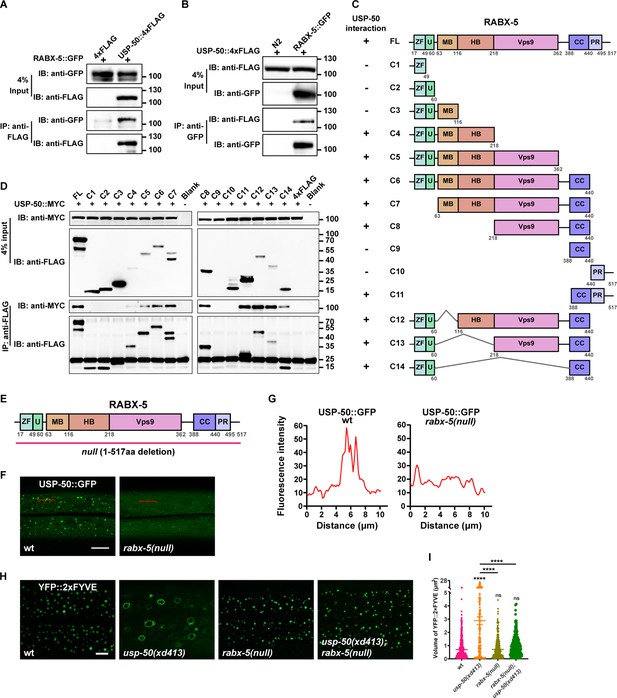
USP-50 interacts with RABX-5.
(A) The affinity-purified RABX-5::GFP from worm lysates is immunoprecipitated by USP-50::4xFLAG purified from HEK293T cells using anti-FLAG beads. Only the area of the blot containing the USP-50::4xFLAG band is displayed. (B) The affinity-purified USP-50::4xFLAG from HEK293T cells is immunoprecipitated by RABX-5::GFP purified from worm lysates using anti-GFP beads. N2 is wild-type. (C) Schematic drawing of RABX-5 showing the domains that interact with USP-50. (D) Immunoprecipitation tests between USP-50::MYC and FLAG-tagged truncated RABX-5. USP-50::MYC and FLAG-tagged truncated RABX-5 were expressed via co-transfection into HEK293T cells, immunoprecipitated with FLAG beads, and immunoblotted with antibodies against MYC and FLAG. (E) Schematic drawing of the RABX-5 protein structure. The molecular lesion of the null mutant is indicated. (F) Confocal fluorescence images of hyp7 expressing USP-50::GFP in wild-type L4 animals (wt), rabx-5(null) L4 mutants. Scale bar represents 5 μm. (G) Line scan analyses show the fluorescence intensity values along the red solid lines in (F). (H) The early endosome (EE) (labeled by YFP::2xFYVE) enlargement phenotype of usp-50(xd413) could be suppressed by rabx-5(null) mutation. (I) Quantification of the volume of individual EEs in various genotypes. 10 or over animals were examined in each genotype. Data are presented as mean ± SEM. ****p<0.0001. ns, not significant. One-way ANOVA with Tukey’s test.
-
Figure 4—source data 1
Original file for the western blot analysis in Figure 4A (anti-GFP and anti-FLAG).
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig4-data1-v1.zip
-
Figure 4—source data 2
PDF containing original scans of the relevant western blot analysis (anti-GFP and anti-FLAG) with highlighted bands and sample labels for Figure 4A.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig4-data2-v1.pdf
-
Figure 4—source data 3
Original file for the western blot analysis in Figure 4B (anti-GFP and anti-FLAG).
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig4-data3-v1.zip
-
Figure 4—source data 4
PDF containing original scans of the relevant western blot analysis (anti-GFP and anti-FLAG) with highlighted bands and sample labels for Figure 4B.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig4-data4-v1.pdf
-
Figure 4—source data 5
Original file for the western blot analysis in Figure 4D (anti-MYC and anti-FLAG).
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig4-data5-v1.zip
-
Figure 4—source data 6
PDF containing original scans of the relevant western blot analysis (anti-MYC and anti-FLAG) with highlighted bands and sample labels for Figure 4D.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig4-data6-v1.pdf
-
Figure 4—source data 7
Excel file containing the quantified data of statistic analysis for Figure 4G and I.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig4-data7-v1.zip
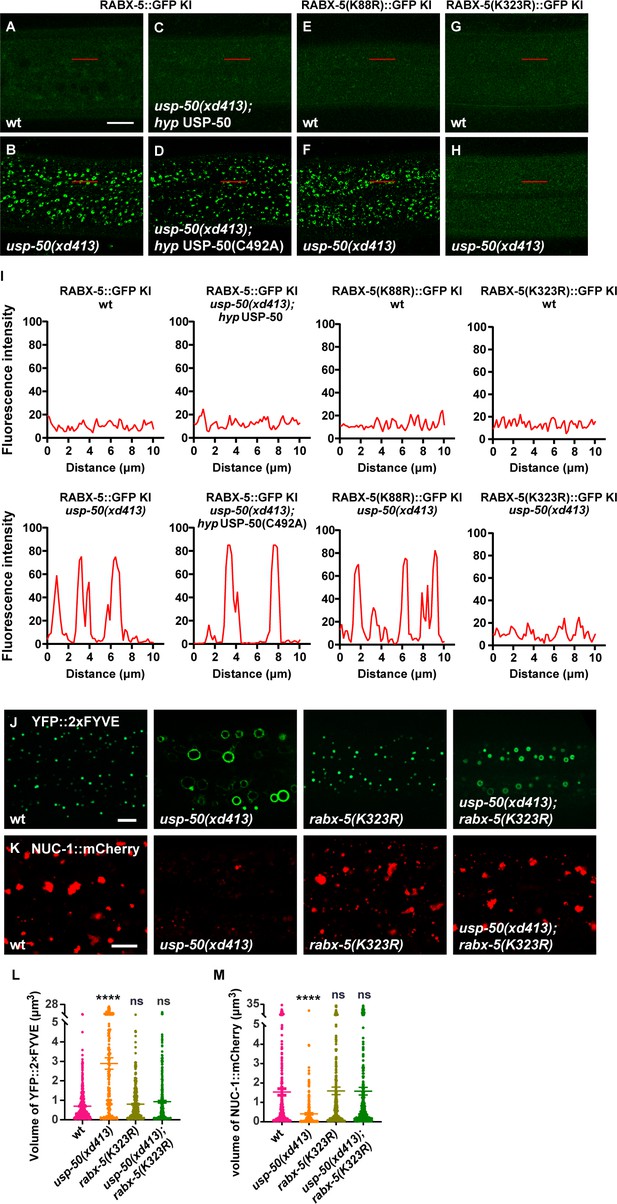
USP-50 dissociates RABX-5 from endosomes.
(A–D) The punctate distribution of RABX-5::GFP KI (knock-in) in wild-type (A) and usp-50(xd413) (B). The increased RABX-5::GFP KI signal in usp-50(xd413) is rescued by expressing wild-type (C) but not C492A mutant usp-50 (D). (E, F) The usp-50(xd413) mutation increases the GFP intensity of RABX-5(K88R)::GFP KI. (G, H) The usp-50(xd413) mutation does not alter the GFP intensity of RABX-5(K323R)::GFP KI. (I) Line scan analyses show the fluorescence intensity values along the red solid lines in (A–H). (J, K) Both the enlarged early endosome (EE) (YFP::2xFYVE) and the diminished late endosome (LE) (NUC-1::mCherry) phenotypes of usp-50(xd413) can be suppressed by rabx-5(k323R). (L, M) Quantification of the volume of individual EEs (labeled by YFP::2xFYVE) and LEs (labeled by NUC-1::mCherry) in various genotypes. Data are presented as mean ± SEM. ****p<0.0001. ns, not significant; one-way ANOVA with Tukey’s test. Scale bar represents 10 μm for (A–H), 5 μm for (J, K).
-
Figure 5—source data 1
Excel file containing the quantified data of statistic analysis for Figure 5I, L, and M.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig5-data1-v1.zip

USP8 inhibits the early endosome (EE) localization of Rabex5.
(A) USP8-KO leads to enlarged EEs and enhanced Rabex5 (Rabex5-mEGFP+/+) and Rab5 (mScarlet-I-Rab5c) signals. Rabex5-mEGFP+/+ SUM159 cells with or without USP8 expression were transiently transfected with mScarlet-I-Rab5c and then imaged by spinning-disk confocal microscopy. Images show the intracellular distribution of Rabex5-mEGFP and mScarlet-I-Rab5c. Scale bar: 10 μm. (B) Quantification of the number of Rabex5-mEGFP spots in Rabex5-mEGFP+/+ cells. 32 wild-type and 32 USP8-KO cells were scored (*p<0.05; unpaired Student’s t-test). (C) Quantification of the number of large Rabex5-mEGFP spots (diameter >1.5 μm) in Rabex5-mEGFP+/+ cells. 30 wild-type and 29 USP8-KO cells were scored (****p<0.0001; unpaired Student’s t-test). (D) Enlarged EEs in USP8-KO SUM159 cells contain both Rabex5 (detected via knock-in of Rabex5-mEGFP) and EEA1 (detected by anti-EEA1). The wild-type and USP8-KO Rabex5-mEGFP+/+ cells were immunostained by anti-EEA1 antibody, and then imaged by spinning-disk confocal microscopy. Scale bar: 10 μm. Enlarged inserts: 2 μm. (E) The CRISPR/Cas9 genome-editing tool was used to knock out of USP8 expression (USP8-KO) in Rabex5-mEGFP+/+ SUM159 cells. Knockout of USP8 was confirmed by western blot analysis using antibodies against USP8 and GAPDH.
-
Figure 5—figure supplement 1—source data 1
Original file for the western blot analysis in Figure 5—figure supplement 1E (anti-USP8 and anti-GAPDH).
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
PDF containing original scans of the relevant western blot analysis (anti-USP8 and anti-GAPDH) with highlighted bands and sample labels for Figure 5—figure supplement 1E.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig5-figsupp1-data2-v1.pdf
-
Figure 5—figure supplement 1—source data 3
Excel file containing the quantified data of statistic analysis for Figure 5—figure supplement 1B and C.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig5-figsupp1-data3-v1.zip

Loss of usp-50/USP8 leads to more activated RAB-5.
(A, B) The GFP::RAB-5 KI signal in wild-type (A) and usp-50(xd413) (B). Scale bar: 5 μm. (C) Quantification of the volume of individual RAB-5::GFP KI vesicles in L4 animals (10 animals for wild-type and 11 animals for usp-50(xd413)). Data are presented as mean ± SEM. ****p<0.0001; unpaired Student’s t-test. (D) Quantification of the relative GFP::RAB-5 KI signal on membrane vs. cytosol in hypodermal cell 7 (hyp7) of L4 animals. 10 animals for wild-type, and 10 animals for usp-50(xd413). ***p<0.001; unpaired Student’s t-test. (E) Coomassie blue staining of purified GST-EEA1-NT protein. (F) More GFP::RAB-5 is pulled down by GST-EEA1-NT protein in usp-50(xd413) mutants. (G) The GFP::RAB-5 protein level is increased in usp-50(xd413) mutants. (H) Quantification of GFP::RAB-5 protein level in wild-type and usp-50(xd413) mutant animals from three independent experiments. ****p<0.0001; unpaired Student’s t-test. (I) Knock-down of USP8 (USP8-KD) leads to enlarged early endosomes (EEs) in genome-edited SUM159 cells expressing EGFP-Rab5c+/+. EGFP-Rab5c+/+ cells were treated with control siRNA (NC) or siRNA targeting USP8, and then imaged by spinning-disk confocal microscopy. Scale bar: 10 μm. Enlarged insertions: 2 μm. (J) The number of Rab5c-positive endosomes in (I) with diameters greater than 1.5 μm were quantified in 31 cells for wild-type and 32 cells for USP8-KD. ****p<0.0001; unpaired Student’s t-test.
-
Figure 5—figure supplement 2—source data 1
Original file for the Coomassie blue staining of purified GST-EEA1-NT protein in Figure 5—figure supplement 2E.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig5-figsupp2-data1-v1.zip
-
Figure 5—figure supplement 2—source data 2
PDF containing the Coomassie blue staining of purified GST-EEA1-NT protein with highlighted bands and sample labels in Figure 5—figure supplement 2E.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig5-figsupp2-data2-v1.pdf
-
Figure 5—figure supplement 2—source data 3
Original file for the western blot analysis in Figure 5—figure supplement 2F (anti-GFP and anti-GST).
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig5-figsupp2-data3-v1.zip
-
Figure 5—figure supplement 2—source data 4
PDF containing original scans of the relevant western blot analysis (anti-GFP and anti-GST) with highlighted bands and sample labels for Figure 5—figure supplement 2F.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig5-figsupp2-data4-v1.pdf
-
Figure 5—figure supplement 2—source data 5
Original file for the western blot analysis in Figure 5—figure supplement 2G (anti-GFP and anti-Tub).
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig5-figsupp2-data5-v1.zip
-
Figure 5—figure supplement 2—source data 6
PDF containing original scans of the relevant western blot analysis (anti-GFP and anti-Tub) with highlighted bands and sample labels for Figure 5—figure supplement 2G.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig5-figsupp2-data6-v1.pdf
-
Figure 5—figure supplement 2—source data 7
Excel file containing the quantified data of statistic analysis for Figure 5—figure supplement 2C, D, H, and J.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig5-figsupp2-data7-v1.zip

The ubiquitin modification sites on RABX-5.
(A) Peptide sequences identified from the ubiquitination proteomics analysis. (B) The ubiquitinated sites of RABX-5 detected by ubiquitination proteomics analysis.
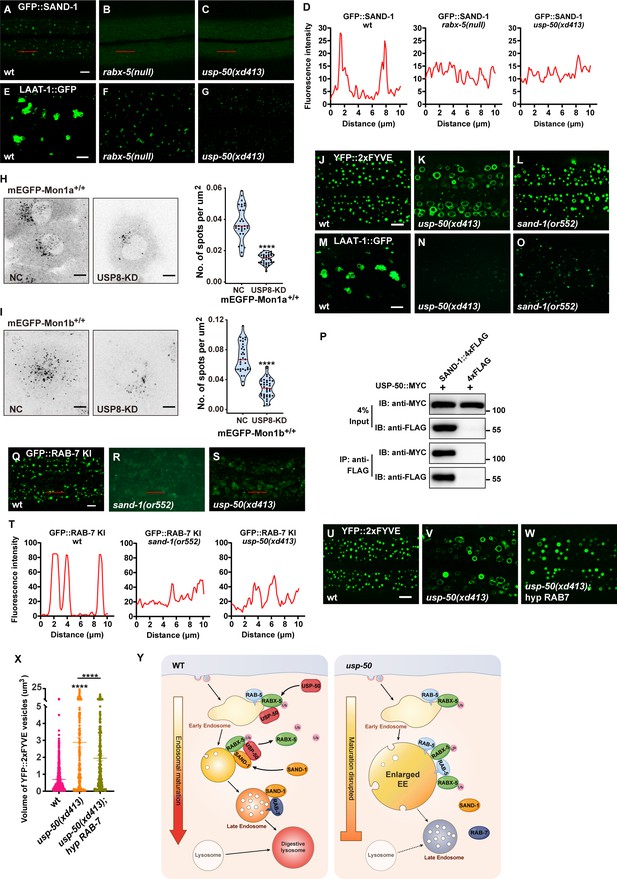
Loss of usp-50/usp8 disrupts SAND-1/Mon1 localization.
(A–C) The reduction of punctate GFP::SAND-1 signals in rabx-5(null) and usp-50(xd413) mutant animals. (D) Line scan analyses for (A–C). (E–G) The reduced lysosomes (labeled by LAAT-1::GFP) in rabx-5(null) and usp-50(xd413) mutant animals. (H, I) SUM159 cells genome-edited for mEGFP-Mon1a+/+ (H) or mEGFP-Mon1b+/+ (I) were treated with control siRNA or siRNA targeting USP8 and then imaged by spinning-disk confocal microscopy (starting from the bottom plane, spaced by 0.35 μm). The single-frame image shows the distribution and area of Mon1a- or Mon1b-labeled endosomes near the middle plane. The right panel shows quantification of the numbers of mEGFP-Mon1a+/+ positive spots from 31 cells for wild-type and 33 cells for USP8-KD, and mEGFP-Mon1b+/+ positive spots from 33 cells for wild-type and 39 cells for USP8-KD (****p<0.0001; unpaired Student’s t-test.) Scale bar: 10 μm. (J, L) Early endosome (EEs) (labeled by YFP::2xFYVE) are enlarged in sand-1(or552) mutants. (M–O) Late endosomes/lysosomes (labeled by LAAT-1::GFP) are smaller in sand-1(or552) mutants. (P) USP-50::MYC and SAND-1::4xFLAG were expressed in HEK293T cells via co-transfection, then immunoprecipitated with anti-FLAG beads and immunoblotted with antibodies against MYC and FLAG. (Q–S) The reduction of punctate GFP::RAB-7 signals in sand-1(or522) and usp-50(xd413) animals. (T) Line scan analyses for (Q–S). (U–W) Overexpressing rab-7 suppresses the enlarged EE phenotype of usp-50 mutants. EEs were detected with YFP::2xFYVE. (X) Quantification of the individual volume of YFP::2xFYVE vesicles in hypodermal cell 7 (hyp7) of L4 stage from 10 animals for wild-type, 13 animals for usp-50(xd413), and 10 animals for the rab-7-expressing line. Data are presented as mean ± SEM. ****p<0.0001; one-way ANOVA with Tukey’s test. (Y) Working models showing the role of USP-50 in endosome maturation. RABX-5-depdendent recruitment of USP-50 promotes the dissociation of RABX-5 from EEs and enhances the recruitment of SAND-1, thereby promoting the endosome maturation process. Scale bar represents 5 μm for (A–C, E–G, J–L, M–O, Q–S, U–W), 10 μm for (H, I).
-
Figure 6—source data 1
Original file for the western blot analysis in Figure 6P (anti-MYC and anti-FLAG).
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig6-data1-v1.zip
-
Figure 6—source data 2
PDF containing original scans of the relevant western blot analysis (anti-MYC and anti-FLAG) with highlighted bands and sample labels for Figure 6P.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig6-data2-v1.pdf
-
Figure 6—source data 3
Excel file containing the quantified data of statistic analysis for Figure 6D, T, H, I, and X.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig6-data3-v1.zip

The distribution of endogenous Mon1a and Mon1b proteins.
(A) Quantification of the individual volume of LAAT-1::GFP vesicles in hyp7 of 3-day-old adults. Data are presented as mean ± SEM. ****p<0.0001; one-way ANOVA with Tukey’s test. (B) Western blotting analysis of SUM159 cells genome-edited to express mEGFP-Mon1a+/+ using antibodies against GFP and GAPDH. (C, D) Genome-edited mEGFP-Mon1a+/+ SUM159 cells were transiently transfected with mScarlet-I-Rab5c (C) and mScarlet-I-Rab7a (D), and then imaged by spinning-disk confocal microscopy. mEGFP-Mon1a localizes to both early endosomes (EEs) (mScarlet-I-Rab5c) and late endosomes (mScarlet-I-Rab7a). (E) Western blotting analysis of genome-edited mEGFP-Mon1b+/+ SUM159 cells using antibodies against GFP and GAPDH. (F, G) mEGFP-Mon1b is not localized on EEs (E) but is localized on late endosomes (G). Genome-edited mEGFP-Mon1b+/+ SUM159 cells were transiently transfected with mScarlet-I-Rab5c (F) and mScarlet-I-Rab7a (G), and then imaged by spinning-disk confocal microscopy. Scale bar: 10 μm. (H) Quantification of the individual volume of YFP::2xFYVE vesicles in hypodermal cell 7 (hyp7) of L4 worms. 10 animals for wild-type, 13 animals for usp-50(xd413), and 18 animals for sand-1(or552). Data are presented as mean ± SEM. ****p<0.0001; one-way ANOVA with Tukey’s test. (I) Quantification of the individual volume of LAAT-1::GFP vesicles in hyp7 of 3-day-old adults (19 animals for wild-type, 16 animals for usp-50(xd413), and 13 animals for sand-1(or552)). Data are presented as mean ± SEM. ****p<0.0001; one-way ANOVA with Tukey’s test.
-
Figure 6—figure supplement 1—source data 1
Original file for the western blot analysis in Figure 6—figure supplement 1B and E (anti-mEGFP and anti-GAPDH).
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig6-figsupp1-data1-v1.zip
-
Figure 6—figure supplement 1—source data 2
PDF containing original scans of the relevant western blot analysis (anti-mEGFP and anti-GAPDH) with highlighted bands and sample labels for Figure 6—figure supplement 1B and E.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig6-figsupp1-data2-v1.pdf
-
Figure 6—figure supplement 1—source data 3
Excel file containing the quantified data of statistic analysis for Figure 6—figure supplement 1A, H, and I.
- https://cdn.elifesciences.org/articles/96353/elife-96353-fig6-figsupp1-data3-v1.zip

(A) Confocal fluorescence images of hypodermis expressing YFP::2xFYVE to detect EEs in L4 stage animals in wild type and stam-1(ok406) mutants. Scale bar: 5 μm. (B) Confocal fluorescence images of hypodermal cell 7 (hyp7) expressing the LAAT-1::GFP marker to highlight lysosome structures in 3-day-old adult animals. Compared to wild type, LAAT-1::GFP signal is reduced in stam-1(ok406) animals. Scale bar: 5 μm. (C) The reduction of punctate endogenous GFP::RAB-7 signals in stam-1(ok406) animals. Scale bar: 10 μm.

ESCRT-0 is adjacent to both early endosomes and late endosomes.
(A) Confocal fluorescence images of wild-type and usp-50(xd413) hypodermis at L4 stage co-expressing HGRS-1::GFP (hgrs-1 promoter) and endogenous wrmScarlet::RAB-5. (B) HGRS-1 and RAB-5 puncta were analyzed to produce Manders overlap coefficient M1 (HGRS-1/RAB-5) and M2 (RAB-5/HGRS-1) (N=10). (C) Confocal fluorescence images of wild-type and usp-50(xd413) hypodermis at L4 stage co-expressing HGRS-1::GFP (hgrs-1 promoter) and endogenous wrmScarlet::RAB-7. (D) HGRS-1 and RAB-7 puncta were analyzed to produce Manders overlap coefficient M1 (HGRS-1/RAB-7) and M2 (RAB-7/HGRS-1) (N=10). Scale bar: 10 μm for (A) and (C).

The endogenous RABX-5 protein level is increased in usp-50 mutants.
(A) The RABX-5::GFP KI protein level is increased in usp-50(xd413). (B) Quantification of endogenous RABX-5::GFP protein level in wild type and usp-50(xd413) mutant animals.

(A-C) Over-expression wild type RABX-5 causes enlarged EEs (labeled by YFP::2xFYVE) while RABX-5(K323R) mutant form does not. (D) Quantification of the volume of individual YFP::2xFYVE vesicles. Data are presented as mean ± SEM. ****P<0.0001. ns, not significant. One-way ANOVA with Tukey’s test.

(A) RABX-5::GFP protein was purified from worm lysates using anti-GFP antibody. FLAG-tagged USP-50 was purified from HEK293T cells using anti-FLAG antibody. Purified RABX-5::GFP was incubated with USP-50::4FLAG for indicated times (0, 15, 30, 60 mins), followed by immunoblotting using antibody against ubiquitin, FLAG or GFP. In the presence of USP-50::4xFLAG, the ubiquitination level of RABX-5::GFP is decreased. (B) Quantification of RABX-5::GFP ubiquitination level from three independent experiments. (C) HEK293T cells were transfected with HA-Ub or indicated mutants and 4xFLAG tagged RABX-5 or RABX-5 K323R mutant for 48h. The cells were subjected to pull down using the FLAG beads, followed by immunoblotting using antibody against HA or Flag.





