Lineage-specific intersection of endothelin and GDNF signaling in enteric nervous system development
Figures
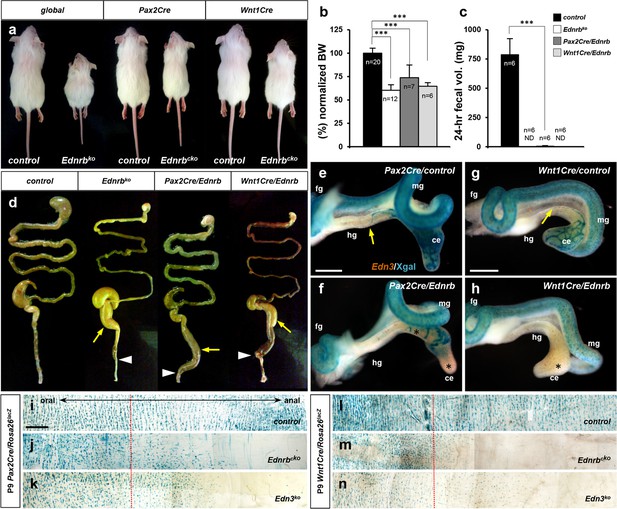
Pax2Cre/Ednrb mutants mice exhibit impaired colonic peristalsis.
(a) External appearances of 2.5-week-old global Ednrb (left), Pax2Cre/Ednrb (middle), and Wnt1Cre/Ednrb (right) mutant mice compared to their littermate controls. (b, c) Compiled representation of normalized body weights (b) and 24 hr fecal elimination (c) from P18 mice of indicated genotypes. Error bars; mean ± s.d. ND, not detected. p-Value: ***p<0.001. (d) External appearances of P18 guts from the indicated genotypes. Arrows indicate distention of colon. Arrowheads point to constriction in the distal colon. (e–h) Wholemount preparations of E12.5 guts isolated from Pax2Cre/Ednrb/Rosa26lacZ (f), Wnt1Cre/Ednrb/Rosa26lacZ (h) mutant embryos and their littermate controls (e and g, respectively) stained with Xgal and labeled by in situ hybridization for Edn3 expression (brown). Yellow arrows denote Pax2Cre-labeled (e) or Wnt1Cre-labeled (g) Xgal+ cells reaching the proximal-distal colon junction in control embryos, whereas Ednrb-deficient Pax2Cre-labeled cells (f) or Wnt1Cre-labeled cells (h) are deficient in caudal migration (asterisks) at the time when the Edn3 gene is normally expressed. (i–n) Wholemount preparations of P9 distal colons isolated from Pax2Cre/Ednrb (j), Edn3 null (k), and control (i) mice, and Wnt1Cre/Ednrb (m), global Edn3 null (n), and control (l) mice stained with Xgal to visualize lineage-labeled cells. Dotted lines indicate the junction between supply by the superior mesenteric and inferior mesenteric arteries, the latter being the vascular source for the distal colon. Abbreviations: ce, cecum; fg, foregut; hg, hindgut; mg, midgut. Scale bars, 500 μm (e–h), 1000 μm (i–n).
-
Figure 1—source data 1
Raw data for panel b (body weight) and panel c (fecal volume).
- https://cdn.elifesciences.org/articles/96424/elife-96424-fig1-data1-v1.docx
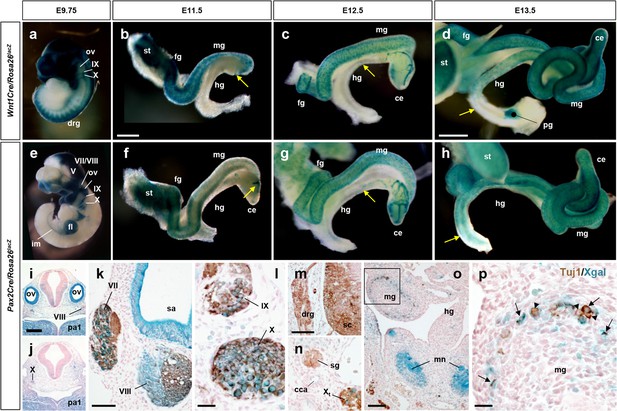
Pax2Cre-derived cells contribute to the enteric nervous system (ENS).
(a–h) Wnt1Cre and Pax2Cre lineage tracing in developing gut. Wholemount Xgal staining of Wnt1Cre/Rosa26lacZ (a) and Pax2Cre/Rosa26lacZ (e) embryos at E9.75. Wholemount Xgal stained guts isolated from Wnt1Cre/Rosa26lacZ (b–d) and Pax2Cre/Rosa26lacZ (f–h) embryos at E11.5 (b, f), E12.5 (c, g), and E13.5 (d, h). Arrows denote migrating wavefronts of Wnt1Cre-labeled (b–d) and Pax2Cre-labeled (f–h) cells on the gut walls. (i, j) Transverse sections of the hindbrain region of a Pax2Cre/Rosa26lacZ embryo at E9.5. (k–p) Transverse sections of E12.5 Pax2Cre/Rosa26lacZ embryos immunolabeled for Tuj1 (brown). Pax2Cre labels the inner ear (sa, which is otic placode-derived and non-neurogenic), and also subsets of CN V, VII, VIII, IX, and X neurons (epibranchial placode-derived) (k, l), and the metanephros (o), but does not label dorsal root (m) or sympathetic ganglion (n) neurons which originate exclusively from trunk neural crest cells. Magnified view of bracketed area in o is shown in (p); arrows in (p) denote Pax2Cre-labeled Tuj1+ ENS neurons. Arrowheads point to Pax2Cre-unlabeled neurons. Abbreviations: cca, common carotid artery; ce, cecum; drg, dorsal root ganglion; fg, foregut; hg, hindgut; mn, metanephros; mg, midgut; ov, otic vesicle; pa, pharyngeal arch; pg, pelvic ganglion; sa, saccule; sc, spinal cord; sg, sympathetic ganglion; st, stomach. V, fifth (trigeminal) ganglion; VII, seventh (facial/geniculate) ganglion; VIII, eighth (vestibulocchlear) ganglion; IX, ninth (glossopharngeal/petrosal) ganglion; X, tenth (vagus/nodose) ganglion. Scale bars, 500 μm (b, c, f–k), 1000 μm (d, h), 200 μm (i–j), 100 μm (k, m–o), 25 μm (l, p).
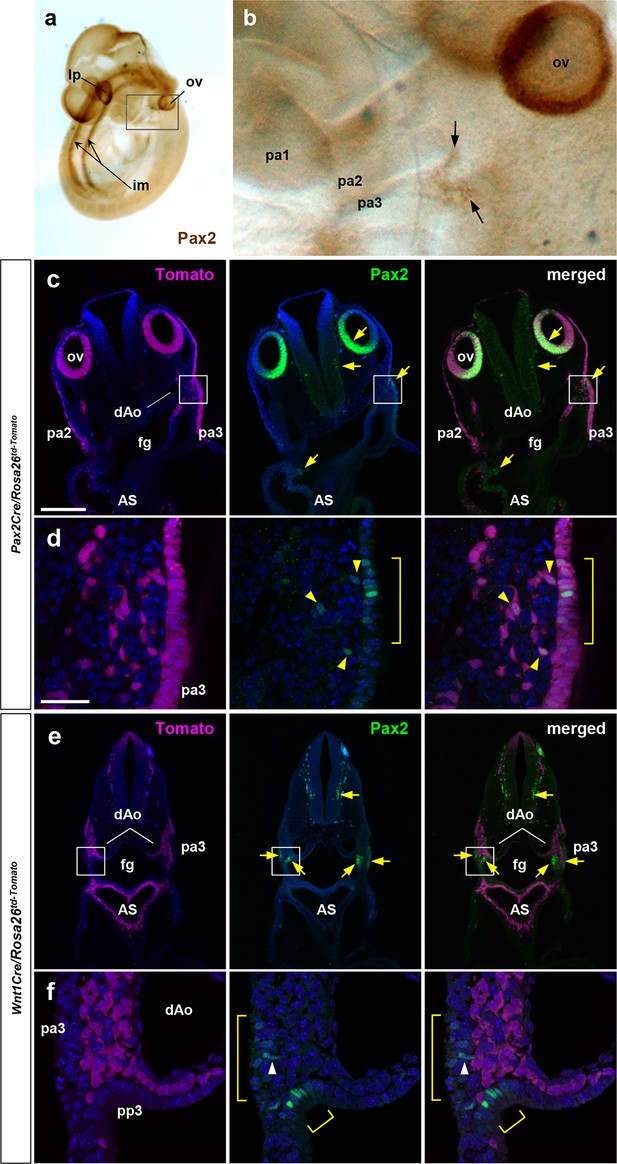
Pax2Cre and Wnt1Cre label distinct early migratory cell populations.
(a, b) An E9.5 wild-type embryo immunostained for Pax2 protein; (b) is a magnified view of the bracketed area including the pharyngeal arch regions in a. Arrows denote Pax2-expressing cells of the third pharyngeal cleft and pouch. (c–f) Vibratome sections of E9.75 Pax2Cre/Rosa26td-Tomato (c, d) and Wnt1Cre/Rosa26td-Tomato (e, f) embryos immunostained for Pax2. Yellow arrows denote Pax2 expression (c, e). Magnified views of bracketed area of c and e are shown in d and f. Yellow brackets denote the Pax2 expressing cells in the third pharyngeal ectoderm and endoderm (d, f). Yellow arrowheads point to Pax2-expressing Pax2Cre+ cells that are delaminated and migrating from the ectodermal layer (d). White arrowhead points to a Pax2-expressing Wnt1Cre-unlabeled cell that has just delaminated from the ectoderm (f). Abbreviations: AS, aortic sac; dAo, dorsal aorta; im, intermediate mesoderm; lp, lens placode; pa, pharyngeal arch; pp, pharyngeal pouch. Scale bars, 200 μm (c, e), 25 μm (d, f).
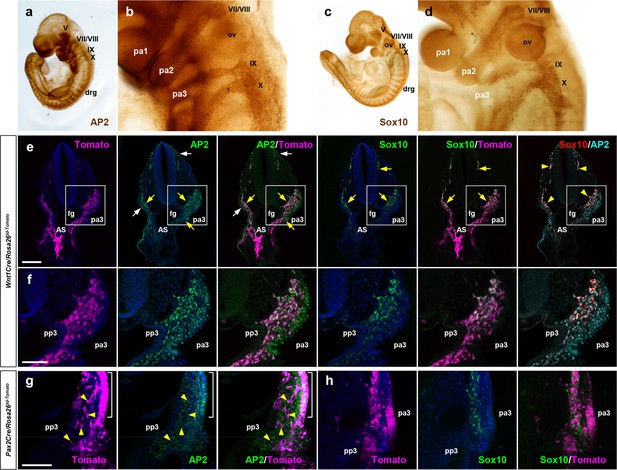
Distinct lineage origin of Pax2Cre-labeled and Wnt1Cre-labeled enteric nervous system (ENS) progenitor cells.
(a–d) E9.5 wild-type embryos stained for AP2 (a, b) and Sox10 (c, d). Magnified views of the bracketed areas including the pharyngeal arch regions in a and c are shown in b and d, respectively. (e, f) Vibratome sections of E9.75 Wnt1Cre/Rosa26td-Tomato embryo immunostained for AP2 and Sox10. White arrows denote AP2+ ectodermal cells. Yellow arrows indicate the Wnt1Cre-labeled neural crest cell population expressing AP2 or Sox10. Magnified views of the bracketed areas in e are shown in f. (g, h) Magnified views of vibratome sections of E9.75 Pax2Cre/Rosa26td-Tomato embryo immunostained for AP2 (g) or Sox10 (h). Yellow arrowheads point to migratory Pax2Cre-derived cells with AP2-negative nuclei. Scale bars, 200 μm (e), 100 μm (f–h).
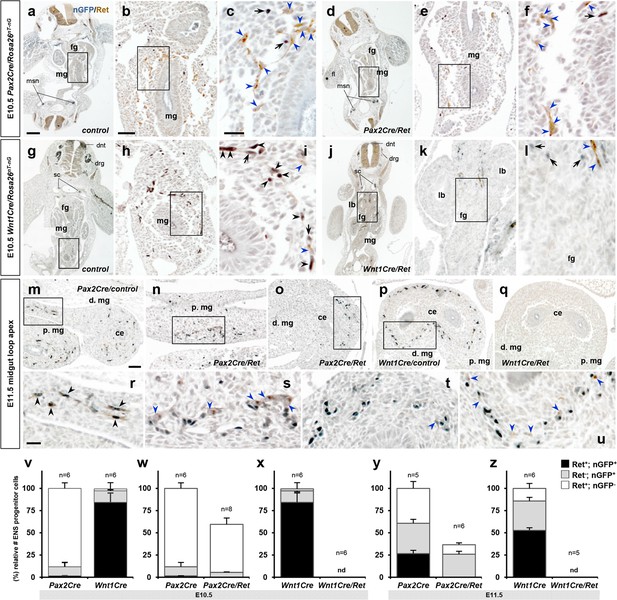
Developmental transition of Ret expression in migratory enteric nervous system (ENS) progenitor cells.
(a–u) Histological sections prepared from E10.5 Pax2Cre/Ret/Rosa26nT-nG mutant (d–f), Wnt1Cre/Ret/Rosa26nT-nG mutant (j–l), and their littermate controls (a–c and g–I, respectively), and E11.5 Pax2Cre/Ret/Rosa26nT-nG mutant (n–o), Wnt1Cre/Ret/Rosa26nT-nG mutant (q), and their littermate control embryos (m and p, respectively) stained for nuclear-GFP (blue) and Ret (brown). Magnified views of the bracketed areas (midgut) in a, d, g, and j are shown in b, e, h, and k, respectively; of the bracketed areas in b, e, h, and k are shown in c, f, I, and l, respectively; of the bracketed areas in m–p are shown in r–u, respectively. Black arrows denotes lineage marker nGFP+ nuclei with no Ret stain, black arrowheads denoted cells that are positive for both nuclear-GFP and Ret, and blue arrowheads denote non-Cre (nGFP-) Ret+ cells along the gut walls. (v–z) Compiled representations of relative fractions of Ret+, nGFP+ (black); Ret-, nGFP+ (gray); and Ret+, nGFP- (white) ENS progenitor cells comparing (v) E10.5 midgut areas of Pax2Cre/Rosa26nT-nG (n=6; 249.7±15.7 cells/embryo) and Wnt1Cre/Rosa26nT-nG (n=6; 178.8±11.4 cells/embryo) embryos, (w) E10.5 midgut areas of Pax2Cre/Ret/Rosa26nT-nG mutants (n=8, 146.9±3.6 cells/embryo) and littermate control embryos, (x) E10.5 midgut areas of Wnt1Cre/Ret/Rosa26nT-nG mutants (n=6, no nGFP+ or Ret+ cell was found) and littermate control embryos, (y) E11.5 midgut loop apex areas of Pax2Cre/Ret/Rosa26nT-nG mutants (n=6, 301.9±35.9 cells/embryo) and littermate control embryos (n=5, 826.2±32.2 cells/embryo), and (z) E11.5 midgut loop apex areas of Wnt1Cre/Ret/Rosa26nT-nG mutants (n=4, no nGFP+ or Ret+ cell was found) and littermate control embryos (n=6, 943.8±63.8 cells/embryo). Data in v are the control mouse groups in w and x, shown together for easier comparison. Ret-, nGFP- (double negative) cell populations were not included in this analysis. Error bars; mean ± s.e.m. nd = not detected. Abbreviations: dnt, dorsal neural tube; drg, dorsal root ganglia; msn, mesonephros; sc, sympathetic chain ganglia. Scale bars, 200 μm (a, d, g, j), 50 μm (b, e, h, k, m–q), 20 μm (c, f, i, l, r–u).
-
Figure 2—source data 1
Raw data for panels v to x and panels y and z (numbers and fractions of linegae-labeled Ret+, lineage-labled Ret-, and lineage-unlabeled Ret+ ENS progenitor cells).
- https://cdn.elifesciences.org/articles/96424/elife-96424-fig2-data1-v1.docx
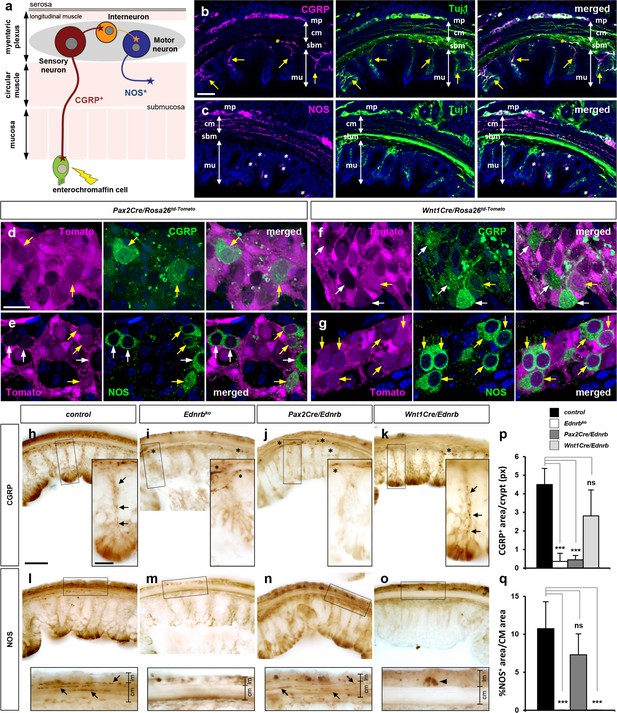
Mechanosensory fate of Pax2Cre lineage and selective loss of CGRP+ myenteric mechanosensory axons in Pax2Cre/Ednrb mutant distal colon.
(a) Schematic illustration of neuronal subtypes in the colonic myenteric ganglia and their end organs. Mechanosensory neurons (red) extend their peripheral axons to the mechanosensory (enterochromaffin) cells while their other axons synapse to interneurons (orange). Motor neurons (blue) innervate the colonic circular muscle to generate colonic peristalsis. (b, c) Confocal images of transverse sections of the distal colon co-immunostained for CGRP and Tuj1 (b) or NOS and Tuj1 (c) show selective label of sensory and motor axons by CGRP (b) and NOS (c), respectively. Yellow arrows in b denote CGRP+ axons that project through each crypt to reach enteric epithelium. Yellow asterisks in b indicate CGRP+ axons from pelvic ganglia along the submucosal layer. White asterisks in c point to NOS-reactive non-neuronal epithelial cells of the mucosa. (d–g) Confocal images of the myenteric ganglia of P9 distal colons isolated from Pax2Cre/Rosa26td-Tomato (d, e) and Wnt1Cre/Rosa26td-Tomato (f, g) mice immunostained for CGRP (d, f) and NOS (e, g). Low-magnification views from which these images were taken are shown in Figure 4. Compared to b and c, this wholemount preparation emphasizes cell bodies in the myenteric plexus rather than axon projections. Yellow arrows point to lineage marker-positive CGRP+ or NOS+ neurons, whereas white arrows indicate lineage marker-negative CGRP+ or NOS+ neurons of the myenteric ganglia. (h–o) Serial transverse vibratome sections of the transition areas (see Figure 6c and h) of the distal colons isolated from P9 control (h, l), global Ednrb (i, m), Pax2Cre/Ednrb (j, n), and Wnt1Cre/Ednrb (k, o) mutants immunostained for CGRP (h–k) or NOS (l–o). Magnified views of bracketed areas are shown in the insets. Arrows in h and k denote CGRP+ axonal projections toward mucosal epithelium in a single crypt. Arrows in l and n denote NOS+ axonal projections perforating throughout the circular muscle layer. Arrowhead in o indicates a NOS+ presumed myenteric interneuron (because it lacks motor axons in the circular muscle layer) of Wnt1Cre/Ednrb mutant colon. Scale bars, 50 μm (b, c, h–o), 20 μm (d–g, h–o insets). (p) Complied representation of the average pixel values of CGRP+ axon area per single crypt. (q) Complied representation of the average fraction of NOS+ axon area in the circular muscle area. Analysis includes 24 crypts from 8 distal colon sections per genotype (four controls, two Ednrb, three Pax2Cre/Ednrb, and three Wnt1Cre/Ednrb mutants). Error bars; mean±s.d. p-Values: ***p<0.001, ns = not significant. Abbreviations: cm, circular muscle; lm, longitudinal muscle; mp, myenteric plexus; mu, mucosa; sbm, submucosa.
-
Figure 3—source data 1
Raw data for panel p (CGRP+ pixel area/crypt) and panel q (NOS+ pixel area/circular muscle ROI area).
- https://cdn.elifesciences.org/articles/96424/elife-96424-fig3-data1-v1.docx
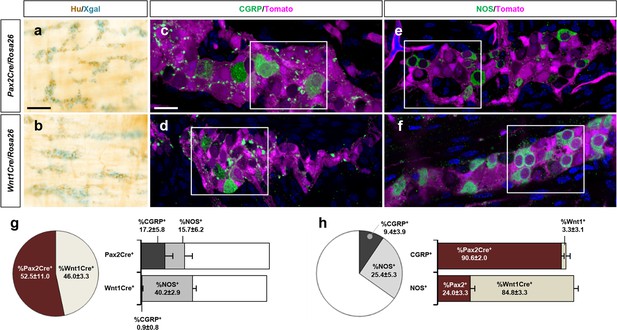
Pax2Cre-derived cells preferentially give rise to CGRP+ myenteric neurons of the distal colon.
(a, b) Wholemount preparations of P9 distal colons isolated from Pax2Cre/Rosa26lacZ (a) and Wnt1Cre/Rosa26lacZ (b) mice were Xgal stained and immunolabeled for Hu (a, b). (c–f) Wholemount preparations of P9 distal colons isolated from Pax2Cre/Rosa26td-Tomato (c, e) and Wnt1Cre/Rosa26td-Tomato (d, f) were immunofluorescence labeled for CGRP (c, d) or NOS (e, f) and counterstained by DAPI. Magnified views of bracketed areas in c–f are shown in Figure 3d–g. Scale bars, 100 μm (a, b), 20 μm (c–f). (g, h) Compiled representations of the proportion of Pax2Cre+ and Wnt1Cre+ neurons (g) or the proportion of CGRP+ and NOS+ neurons (h) among a total of 5585 myenteric neurons counted from the P9 distal colons isolated from nine Pax2Cre/Rosa26td-Tomato and eight Wnt1Cre/Rosa26td-Tomato mice (mean±s.d). The proportion of the CGRP+ and NOS+ neurons within the Pax2Cre+ or the Wnt1Cre+ population are presented in the bar graph of (g). The percentage of Pax2Cre+ and Wnt1Cre+ neurons in the CGRP+ or NOS+ populations are shown in the bar graph of (h). The pie charts and bar graphs do not add to 100% because these are compiled from separate and independent measurements that were compiled together graphically for ease of comparison. Error bars; mean±s.d.
-
Figure 3—figure supplement 1—source data 1
Raw data for panels g and h (numbers and fractions of of lineage labeled, CGRP+ and/or NOS+ neurons in the ENS of the distal colon).
- https://cdn.elifesciences.org/articles/96424/elife-96424-fig3-figsupp1-data1-v1.docx
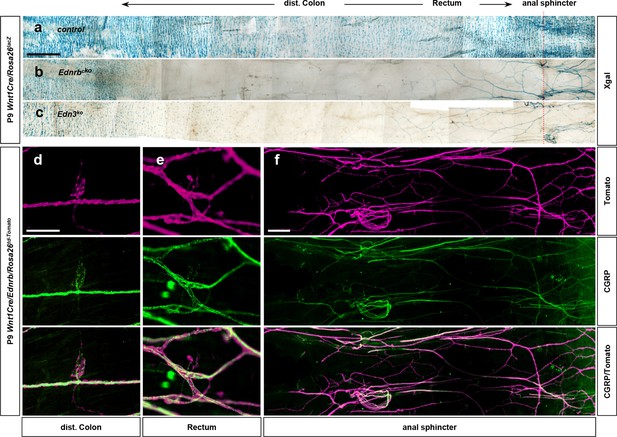
Pelvic ganglion neurons and their projections express CGRP.
(a–c) Distal-most end of Xgal-stained colons of the control (a), Wnt1Cre/Ednrb (b), and global Edn3 (c) mutants shown in Figure 1l–n, respectively. In the absence of vagal neural crest-derived intrinsic enteric nervous system (ENS) ganglia (b, c), sacral neural crest-derived pelvic ganglia and their axons become noticeable with Xgal staining. (d–f) Epiflurorecence microscope images of wholemount preparations of the distal colons isolated from P9 Wnt1Cre/Ednrb/Rosa26td-Tomato mutant immunofluorescently labeled for CGRP shows sacral neural crest-derived CGPR+ neurons and axons in the distal colon (d), rectum (e), and anal sphincter (f) areas. Scale bars, 1000 μm (a–c), 100 μm (d, e), 500 μm (f).
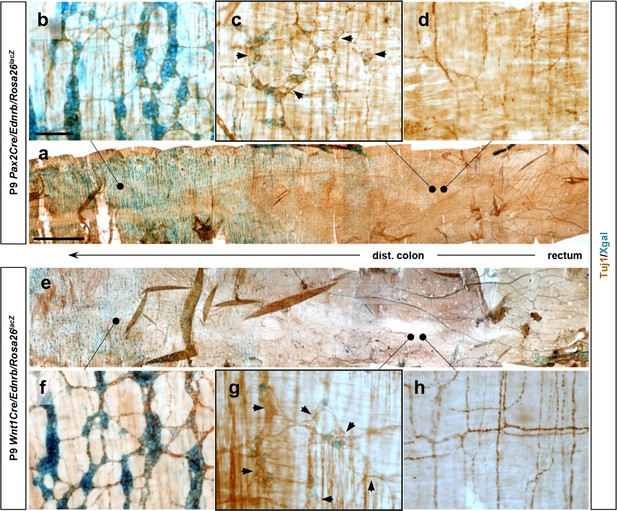
Transition areas in lineage-specific Ednrb mutant distal colon also contain lineage-unlabeled neurons.
(a, e) Wholemount preparations of P9 distal colons isolated from Pax2Cre/Ednrb (a) and Wnt1Cre/Ednrb (e) stained with Xgal and for Tuj1 to visualize lineage-labeled and lineage-unlabeled neurons and their axons. (b, c, f–h) Magnified views showing myenteric plexus morphology in the areas that are normally colonized (b, f), at the location of terminal migration (c, g), and at a location immediately beyond the point of terminal migration (d, h). Residual Tuj1 signal in axons in d and h are extrinsic nerves. Histological analysis for CGRP and NOS in Figure 3h–o was performed with segments equivalent to the location of panels c and g. Arrows in c and g point to lineage-unlabeled cells of myenteric ganglia. Scale bars, 1000 μm (a, e), 100 μm (b–d, f–h).
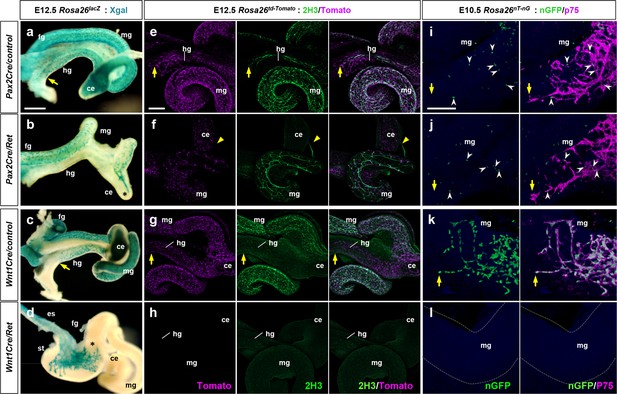
Pax2Cre-specific deletion of Ret results in defective enteric nervous system (ENS) progenitor migration and differentiation.
(a–d) Wholemount preparations of E12.5 guts isolated from Pax2Cre/Ret/Rosa26lacZ (b), Wnt1Cre/Ret/Rosa26lacZ (d) mutant embryos and their littermate controls (a and c, respectively) stained with Xgal (blue). Arrows denote the wavefronts of lineage-labeled cells in the hindguts of the controls. Asterisks denote halted migration of Ret-deficient Pax2Cre-labeled cells at the cecum (b) and of Ret-deficient Wnt1Cre-labeled cells at the caudal end of the stomach (d). (e–h) Wholemount preparations of E12.5 guts isolated from Pax2Cre/Ret/Rosa26td-Tomato (f), Wnt1Cre/Ret/Rosa26td-Tomato (h) mutant embryos and their littermate controls (e and g, respectively) stained for neuronal 2H3 (green). Arrows denote the wavefronts of lineage-labeled cells in the hindguts of the control embryos. Arrowheads denote halted migration of Ret-deficient Pax2Cre-labeled cells at the cecum (f). (i–l) Wholemount preparations of E10.5 guts isolated from Pax2Cre/Ret/Rosa26nT-nG (j), Wnt1Cre/Ret/Rosa26nT-nG (l) mutant embryos and their littermate controls (i and k, respectively) stained for nuclear-GFP (green) and ENS progenitor marker p75 (magenta). Arrows denote the wavefronts of lineage-labeled cells in the midguts. Arrowheads denote p75+ Pax2Cre-labeled cells at the midgut areas (i, j). Abbreviations: es, esophagus; st, stomach. Scale bars, 500 μm (a–d), 250 μm (e–h), 50 μm (i–l).
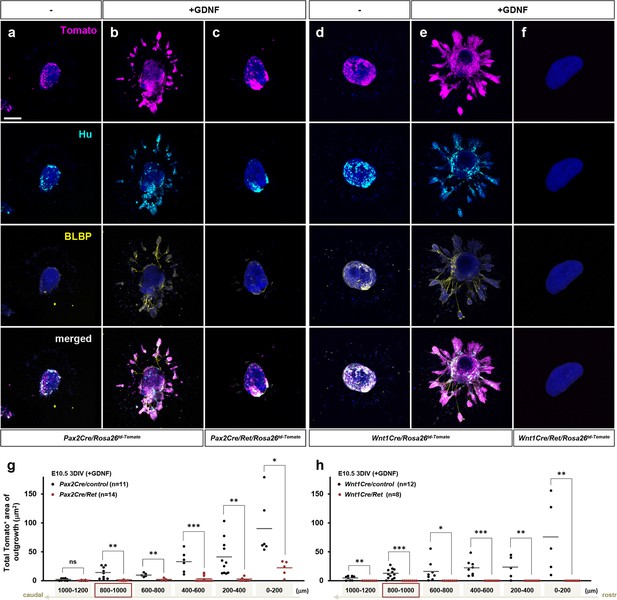
Defective GDNF-induced outgrowth and differentiation of Pax2Cre/Ret mutant enteric nervous system (ENS) progenitors in vitro.
(a–f) Confocal images of 200 mm midgut sections isolated from an E10.5 Pax2Cre/Ret/Rosa26td-Tomato mutant (c) and its littermate control (a, b), and a Wnt1Cre/Ret/Rosa26td-Tomato mutant (f) and its littermate control (d, e) cultured 3 days in vitro (3DIV) in the absence (a, d) or presence of GDNF (b, c, e, f), then immunolabeled for neuronal Hu (cyan) and glial BLBP (yellow). Scale bars, 250 μm. (g) Complied representation of the total area of outgrowth (Tomato+ area around the explanted gut tube) from Pax2Cre/control (black dots; n=11) and Pax2Cre/Ret mutant (red dots; n=14) embryos in the presence of GDNF. (h) Compiled representation of the total outgrowth area from Wnt1Cre/control (black dots; n=12) and Wnt1Cre/Ret mutant (red dots; n=8) embryos in the presence of GDNF. See Figure 5—figure supplement 1a for rostrocaudal orientation of the midgut slice preparations. Each dot represents the outgrowth of each gut slice prepared from each embryos used for data acquisition (lost individual midgut sections are not included as data points). The images shown in a–f represent sections that are 800–1000 μm caudal to the foregut-midgut junction (as indicated by the red boxes in g–h). p-Values: *p<0.05, **p<0.01, ***p<0.001, ns = not significant.
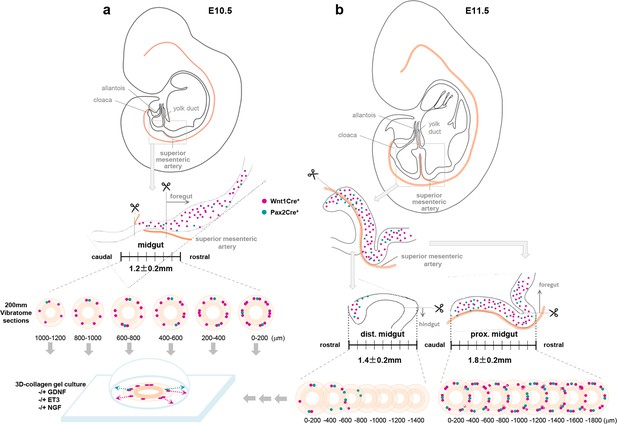
Schematic illustrations of rostrocaudal orientation of gut slice explant culture.
(a) E10.5 midgut slice preparation. The midgut segment was isolated based on the origin of the superior mesenteric artery from the dorsal aorta (cranial end) to where it meets the yolk (vitelline) duct. 200 mm vibratome sections were prepared in a rostral to caudal direction and individually cultured in collagen gel in the absence or presence of GDNF, EDN3, or NGF. (b) E11.5 midgut slice preparation. First, the whole gut tissue (from stomach through cloaca) was excised from E11.5 embryo, and then the midgut loop was isolated. The proximal midgut segment was dissected by separating the midgut loop along the superior mesenteric artery and then by removing the foregut portion at the cranial end of the superior mesenteric artery. The superior mesenteric artery was kept with the proximal midgut segment for the rest of the procedure. The hindgut was then removed from the distal midgut segment based on the position of the inferior mesenteric artery. For both proximal and distal midgut segments, 200 μm vibratome sections were prepared in ventral to dorsal direction and individually cultured in collagen gel in the absence or presence of GDNF, EDN3, or NGF. E10.5 midgut tissues that were larger or smaller than 1200±200 μm, and E11.5 gut tissues with proximal midgut segments larger or smaller than 1800±200 μm, were considered as over- or underdeveloped and excluded from further analysis. See Methods section for details.
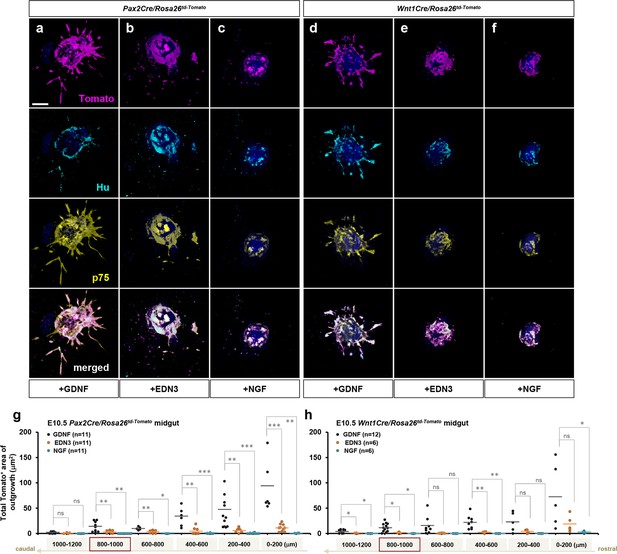
EDN3 exhibited marginal effects on E10.5 enteric progenitor outgrowth in vitro.
(a–f) Confocal z-stack images of 200 mm midgut sections isolated from E10.5 Pax2Cre/Rosa26td-Tomato (a–c) and Wnt1Cre/Rosa26td-Tomato embryos cultured for 3 days in the presence of GDNF (a), EDN3 (b), or NGF (c) and stained for neuronal Hu (cyan) and ENS progenitor marker p75 (yellow). Scale bar, 250 mm. (g, h) Compiled representations of the total area of outgrowth (Tomato+ area around the explanted gut tube) from Pax2Cre/Rosa26td-Tomato and Wnt1Cre/Rosa26td-Tomato midgut slice explants in the presence of GDNF (black), EDN3 (orange), or NGF (blue). The data from GDNF-treated groups here are shown as the controls in Figure 5. See Figure 5—figure supplement 1a for rostrocaudal orientation of these midgut slice preparations. Each dot represents one gut slice isolated from the indicated axial level; lost sections were not included as data points. The images shown in a–f represent sections that are 800–1000 μm caudal to the foregut-midgut junction (red boxes in g and h). p-Values: *p<0.05, **p<0.01, ***p<0.001, ns = not significant.
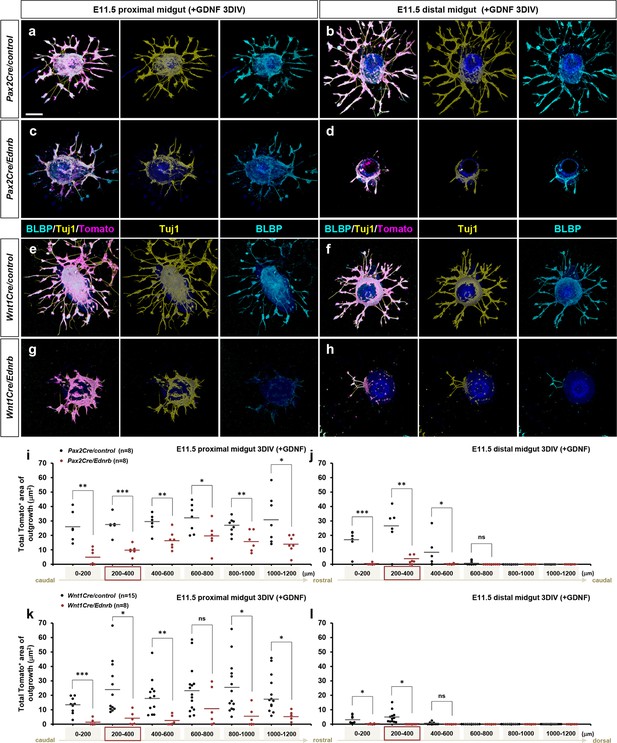
Ednrb-deficient enteric nervous system (ENS) progenitor cells populating the migratory wavefront exhibit attenuated response to GDNF in vitro.
(a–h) Confocal z-stack images of 200 μm proximal (a, c, e, g) and distal midgut (b, d, f, h) sections isolated from an E11.5 Pax2Cre/Ret/Rosa26td-Tomato mutant (c, d) and its littermate control (a, b), and a Wnt1Cre/Ret/Rosa26td-Tomato mutant (g, h) and its littermate control (e, f) cultured 3 days in vitro (3DIV) in the presence of GDNF and then immunolabeled for neuronal Tuj1 (yellow) and glial BLBP (cyan). Scale bars, 250 μm. (i, j) Compiled representation of the total area of outgrowth (Tomato+ area around the explanted gut tube) from proximal and distal midguts of Pax2Cre/control (black dots; n=8) and Pax2Cre/Ret mutant (red dots; n=8) embryos in the presence of GDNF. (k, l) Compiled representation of the total area of outgrowth from Wnt1Cre/control (black dots; n=15) and Wnt1Cre/Ret mutant (red dots; n=8) embryos in the presence of GDNF. See Figure 5—figure supplement 1b for rostrocaudal orientation of the midgut slices. The proximal midgut images shown in a, c, e, g represent sections that are 200–400 μm rostral to the apex of the midgut loop; the distal midgut images shown in b, d, f, h represent sections that are 200–400 μm caudal to the apex of the midgut loop (as indicated by the red boxes in i–l). p-Values: *p<0.05, **p<0.01, ***p<0.001, ns = not significant.
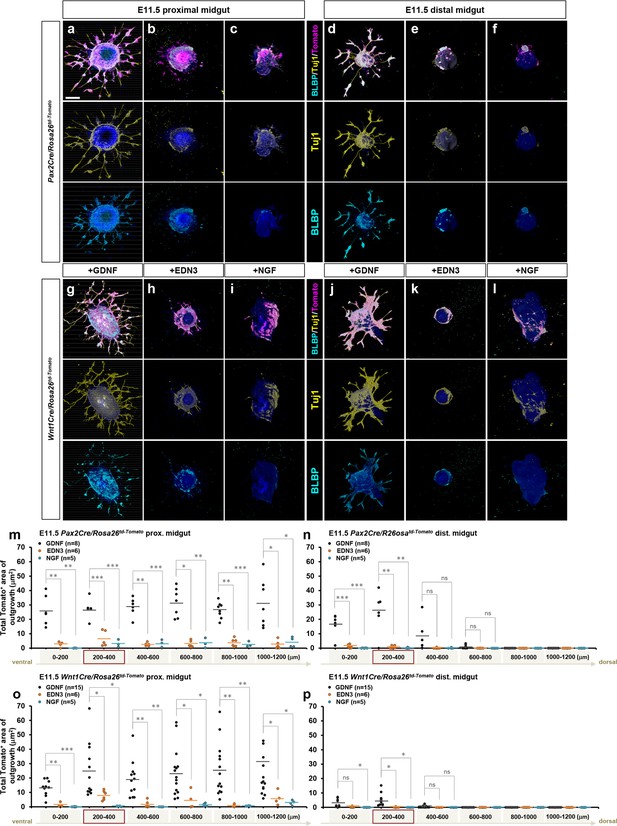
EDN3 exhibits minimal effect on E11.5 enteric progenitor outgrowth in vitro.
(a–h) Confocal z-stack images of 200 μm proximal (a–c, g–i) and distal midgut (d–f, j–l) sections isolated from E11.5 Pax2Cre/Rosa26td-Tomato (a–f), Wnt1Cre/Rosa26td-Tomato (g–l) embryos cultured for 3 days in the presence of GDNF (a, d, g, j), EDN3 (b, e, h, k), or NGF (c, f, i, l), and then stained for neuronal Tuj1 (yellow) and glial BLBP (cyan). Scale bars, 250 μm (a–f). (m–p) Compiled representations of the total area of outgrowth in the presence of GDNF, EDN3, vs. NGF from proximal (m, o) and distal (n, p) midgut slice explants prepared from E11.5 Pax2Cre/Rosa26td-Tomato (m, n) and Wnt1Cre/Rosa26td-Tomato (o, p) embryos. The data from GDNF-treated groups here are shown as the controls in Figure 6. See Figure 5—figure supplement 1b for rostrocaudal orientation of the midgut slice preparation. The proximal midgut images shown in a–c and g–i represent sections that are 200–400 μm rostral to the apex of the midgut loop, the distal midgut images shown in d–f and j–l represent sections that are 200–400 μm caudal to the apex of the midgut loop (red boxes in m–p). p-Values: *p<0.05, **p<0.01, ***p<0.001, ns = not significant.
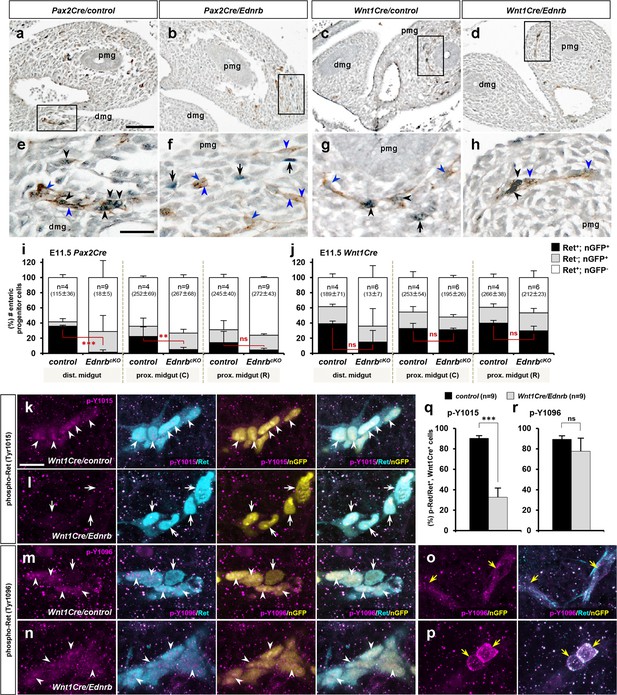
Lineage selective endothelin-dependent regulation of Ret in migratory enteric nervous system (ENS) progenitor cells.
(a–d) Histological sections showing the apex of the midgut loop of E11.5 Pax2Cre/Ednrb/Rosa26nT-nG mutant (b), Wnt1Cre/Ednrb/Rosa26nT-nG mutant (d), and their littermate control embryos (a and c, respectively) stained for nuclear-GFP (blue) and Ret (brown). Magnified views of the bracketed areas (migratory wavefronts) in a–d are shown in e–h, respectively. Abbreviations: dmg, distal midgut; pmg, proximal midgut. (i, j) Compiled representations of the fractions of Ret+, nGFP+ (black); Ret-, nGFP+ (gray), and Ret+, nGFP- (white) ENS progenitor cells in the indicated areas of the embryos of the indicated genotype; (C) and (R) indicate caudal and rostral halves of the proximal midgut. The average total number of countable cells (labeled by either or both markers) in each gut segment is indicated in the parentheses. Error bars; mean ± s.d. (k–p) Confocal z-stack images of wavefront cells in wholemount preparations of E11.5 guts isolated from Wnt1Cre/Ednrb/Rosa26nT-nG mutants (l, n, p) and their littermate control embryos (k, m, o) stained for nuclear-GFP (yellow), Ret (cyan), and phospho-Ret (Tyr1015) (magenta; k, l) or phospho-Ret(Tyr1096) (magenta; m–p). White arrowheads denote the presence of phosphorylated Ret in the Wnt1Cre-labeled cells, white arrows point to the absence of phosphorylated Ret in the Wnt1Cre-labeled cells, and yellow arrows denote cytoplasmic localization of phosphorylated Ret in nonlineage Ret+ cells. (q, r) Compiled representations of phospho-Ret-(Tyr1015)+ fractions (q) and phospho-Ret-(Tyr1096)+ fractions (r) at the wavefront (apex of the midgut loop) in Wnt1Cre/Ednrb mutants (n=9; gray bars) and their littermate control embryos (n=9; black bars). Error bars; mean ± s.e.m. p-Values: *p<0.05, **p<0.01, ***p<0.001, ns = not significant. Scale bars, 50 μm (a–d), 20 μm (e–h, k–p).
-
Figure 7—source data 1
Raw data for panels i and j (numbers and fractions of Ret+ lineage-labeled, Ret- lineage -labeled, and Ret+ lineage-unlabeled cells in the distal midgut, caudal half of the proximal midgut and rostral half of the proximal midgut segments) and for panels q and r (numbers and fractions of pY1015+ or pY1096+ in Wnt1Cre-derived ENS progenitors at the migratory wavefront).
- https://cdn.elifesciences.org/articles/96424/elife-96424-fig7-data1-v1.docx
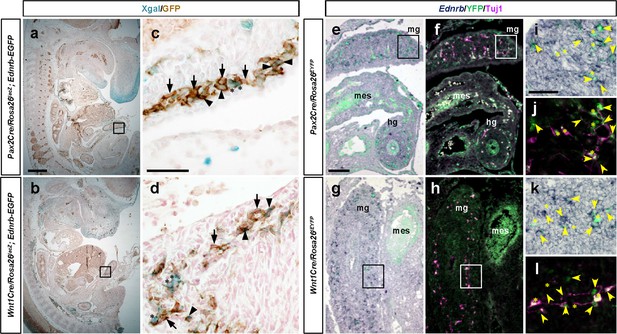
Ednrb is expressed in subsets of Pax2Cre- and Wnt1Cre-derived enteric progenitor cells.
(a–d) Histological sections of Xgal-stained E12.5 Pax2Cre/Rosa26lacZ; Ednrb-EGFP (a) and Wnt1Cre/Rosa26lacZ; Ednrb-EGFP embryos immunolabeled for GFP. Magnified views of bracketed areas in a and b are shown in c and d, respectively. Arrowheads point to Xgal+, GFP+ cells, asterisks denote Xgal+, GFP- cells, and arrows point to Xgal-, GFP+ cells in c and d. (e–h) Cryosections of E12.5 Pax2Cre/Rosa26EYFP (e, f) and Wnt1Cre/Rosa26EYFP (g, h) embryos at the midgut-hindgut area labeled by in situ hybridization for Ednrb gene (purple), and immunolabeled for GFP (green) and Tuj1 (magenta). Brightfield images of Ednrb transcripts overlaid with lineage marker YFP are shown in e and g, and their fluorescence images of YFP and Tuj1 reactive signals are shown in f and h. Magnified views of bracketed areas in e, g are shown in I, k, and in f, h in j, l. mes, mesentery. Arrows denote YFP+, Ednrb+ cells, asterisks indicate YFP+, Ednrb- cells, and arrowheads point to YFP, Ednrb+ ENS cells. Scale bars, 250 μm (a, b), 50 μm (c, d, i–l), 100 μm (e–h).





