Regulatory genome annotation of 33 insect species
Figures
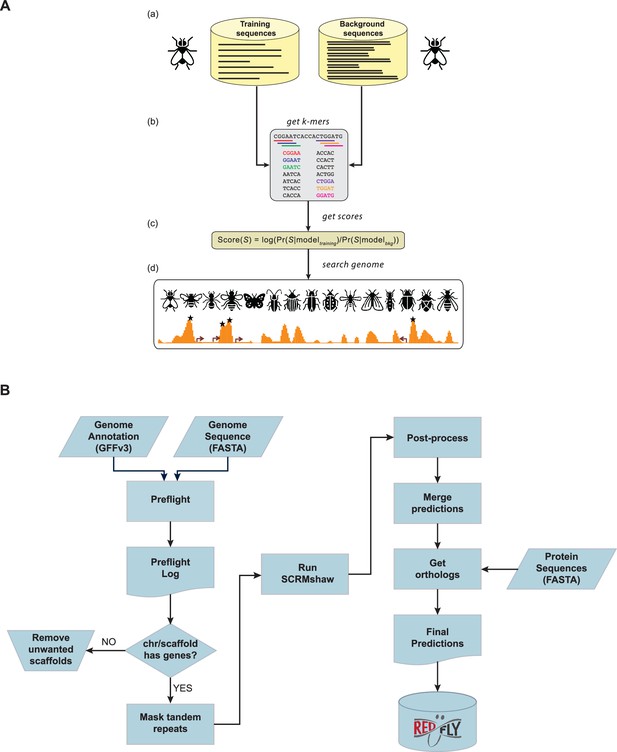
The SCRMshaw method and analysis pipeline.
(A) Supervised motif-blind cis-regulatory module (CRM) discovery (SCRMshaw). (a) SCRMshaw uses a training set of known D. melanogaster enhancers (‘training sequences’), drawn from REDfly, that are defined by common functional characterization, and a 10‐fold larger background set of similarly sized common functional characterization, non‐enhancer sequences (‘background sequences’). (b) The short DNA subsequence (kmer) count distributions of these sequences are then used to train a statistical model. Note that although the pictured example shows 5-mers, kmers of different sizes are used for some of the underlying statistical models (see Methods). The trained model (c) is used to score overlapping windows in the ‘target genome’. (d) High-scoring regions are predicted to be functional regulatory sequences (asterisks). Figure adapted from Asma and Halfon, 2021. (B) The workflow used for the regulatory genome annotation described in this paper. The left side shows pre-processing steps, the right side, post-processing. Input to SCRMshaw consists of the genome sequence and gene annotation. A protein sequence annotation is supplied later for the orthology mapping step. Final results are made available as part of the REDfly regulatory annotation knowledgebase.
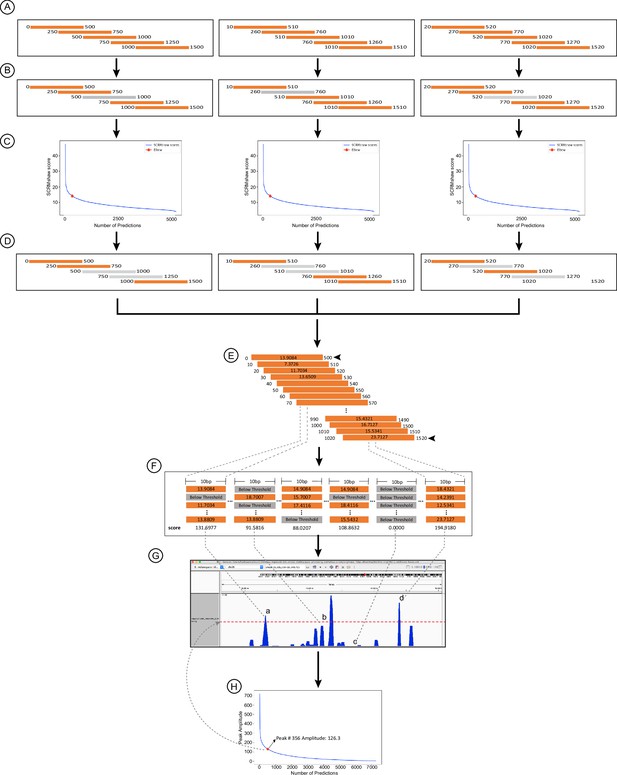
Revised post-processing method used for SCRMshaw.
See Methods for details.
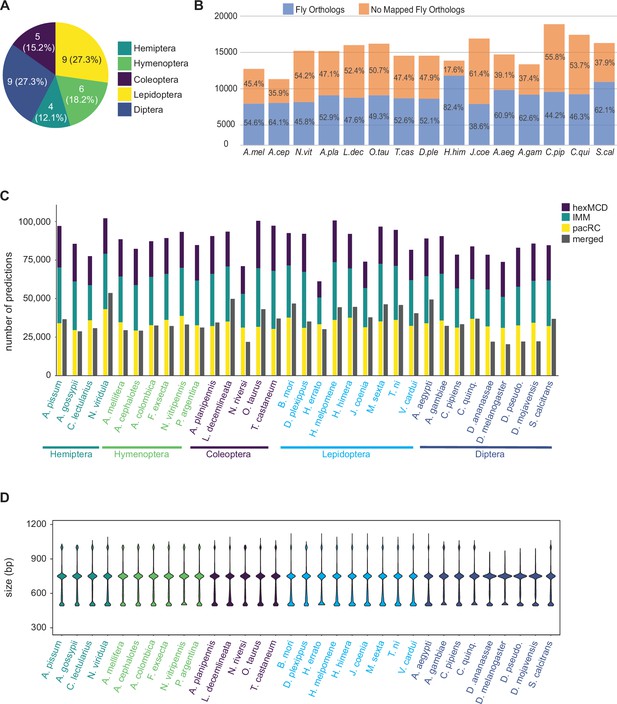
Annotation of 33 insect genomes.
(A) Genomes from five insect orders were annotated in this study (more are ongoing). (B) Percentage of genes with Drosophila orthologs as mapped via our orthology pipeline (see Methods), for the 15 mapped species. ‘No Mapped Fly Orthologs’ indicates that our orthology mapping pipeline did not identify clear D. melanogaster orthologs. For any given gene, this could reflect either a true lack of a respective ortholog, or failure of our procedure to accurately identify an existing ortholog. For complete species names, see Table 1. (C) Total SCRMshaw predictions for each species. For each species, the left-hand column shows cumulative results for each SCRMshaw sub-method summed over each of the 48 training sets. The right-hand column shows the number of unique predictions after merging overlapping predictions from both sub-methods and training sets. Species are displayed alphabetically by taxonomic order (see also Figure 2—figure supplement 1). (D) Size distribution of SCRMshaw predictions, prior to merging overlapping predictions but after removing outlier predictions >2 kb in length. Species are ordered identically to panel C.
-
Figure 2—source data 1
SCRMshaw training sets used in this study.
- https://cdn.elifesciences.org/articles/96738/elife-96738-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Number of predicted enhancers for each species, by method.
- https://cdn.elifesciences.org/articles/96738/elife-96738-fig2-data2-v1.xlsx
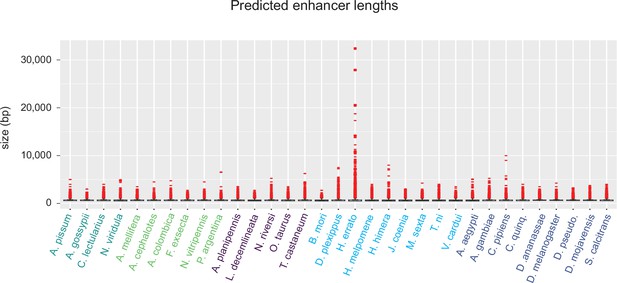
Lengths of predicted enhancers, including long outliers.
Size distribution of SCRMshaw predictions, prior to merging overlapping predictions but without removing outlier predictions. Species are ordered as in Figure 2.
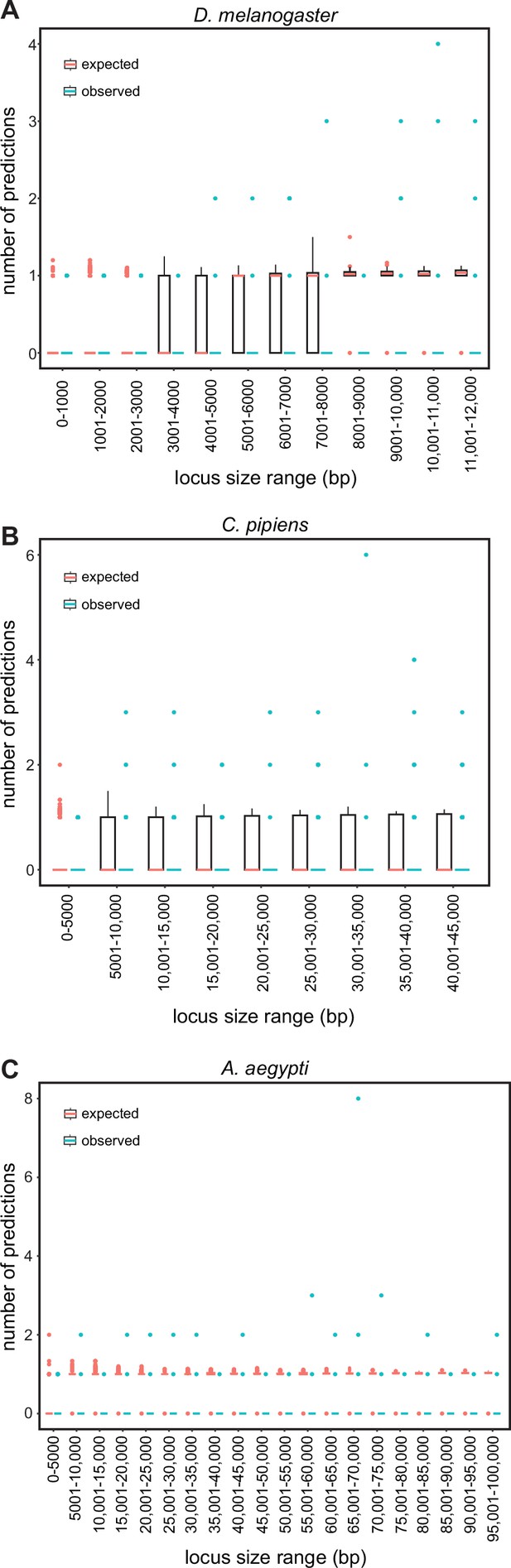
SCRMshaw makes multiple predictions per locus.
The number of SCRMshaw predictions per locus (y-axis) are shown as boxplots for loci falling within the given size ranges (x-axis). Black boxes cover the 25–75th percentiles, bars indicate median values and dots indicate values exceeding 1.5 times the interquartile range (boxes are not visible for all bins due to very low degrees of variation). Values in pink represent expected values drawn from randomization, while values in blue represent observed values from SCRMshaw. All values are from results with the training set ‘mapping1.visceral_mesoderm’; results from other training sets were similar (see Figure 3—source data 1). Shown are results from the genomes of (A) D. melanogaster, (B) C. pipiens, and (C) A. aegypti representing small, medium, and large genomes, respectively.
-
Figure 3—source data 1
Real and simulated predictions per locus.
- https://cdn.elifesciences.org/articles/96738/elife-96738-fig3-data1-v1.xlsx
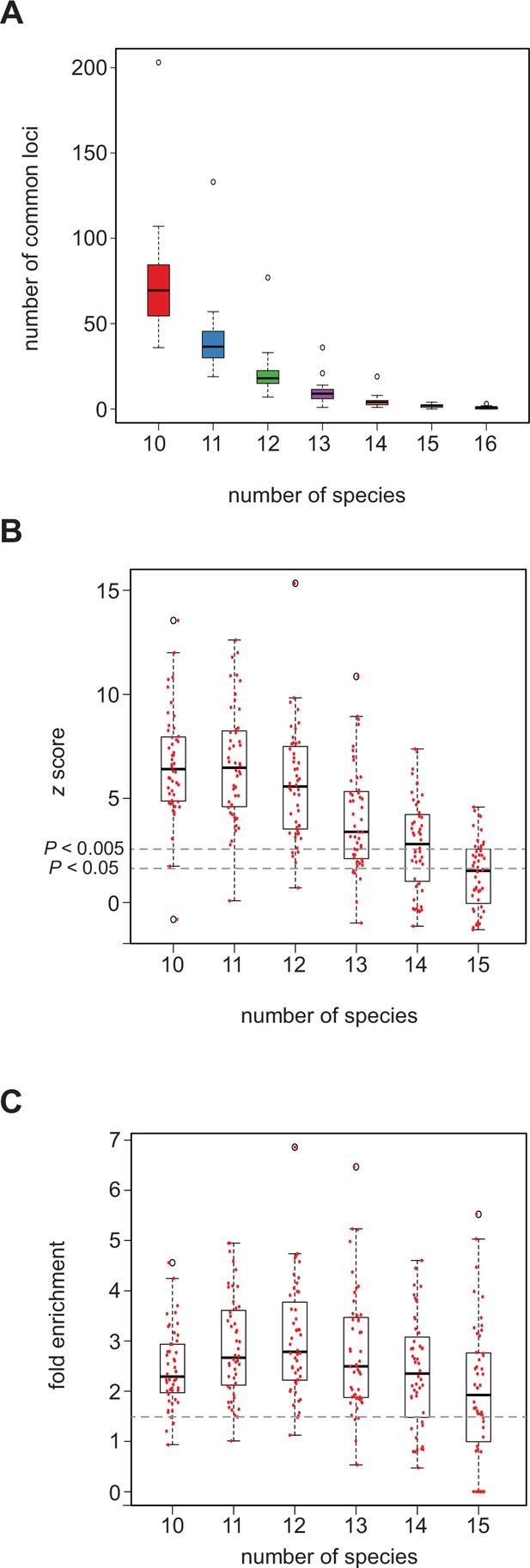
SCRMshaw predicts cis-regulatory modules (CRMs) in orthologous loci across species.
(A) The number of loci in common that contain at least one SCRMshaw prediction, for 10 or more species. (B) z-scores demonstrating that the number of loci in common with one or more SCRMshaw predictions is significantly higher than expectation, based on 360 randomizations. The small number of common predictions for 14–16 species make these statistics unreliable. Dotted lines indicate z-score values representing significance at the (unadjusted) p<0.005 and p<0.05 levels. (C) Fold enrichment values illustrating the excess of loci in common with one or more SCRMshaw predictions compared to expectation. Dotted line shows 1.5× enrichment.
-
Figure 4—source data 1
Real and simulated counts of predictions in orthologous loci.
- https://cdn.elifesciences.org/articles/96738/elife-96738-fig4-data1-v1.xlsx
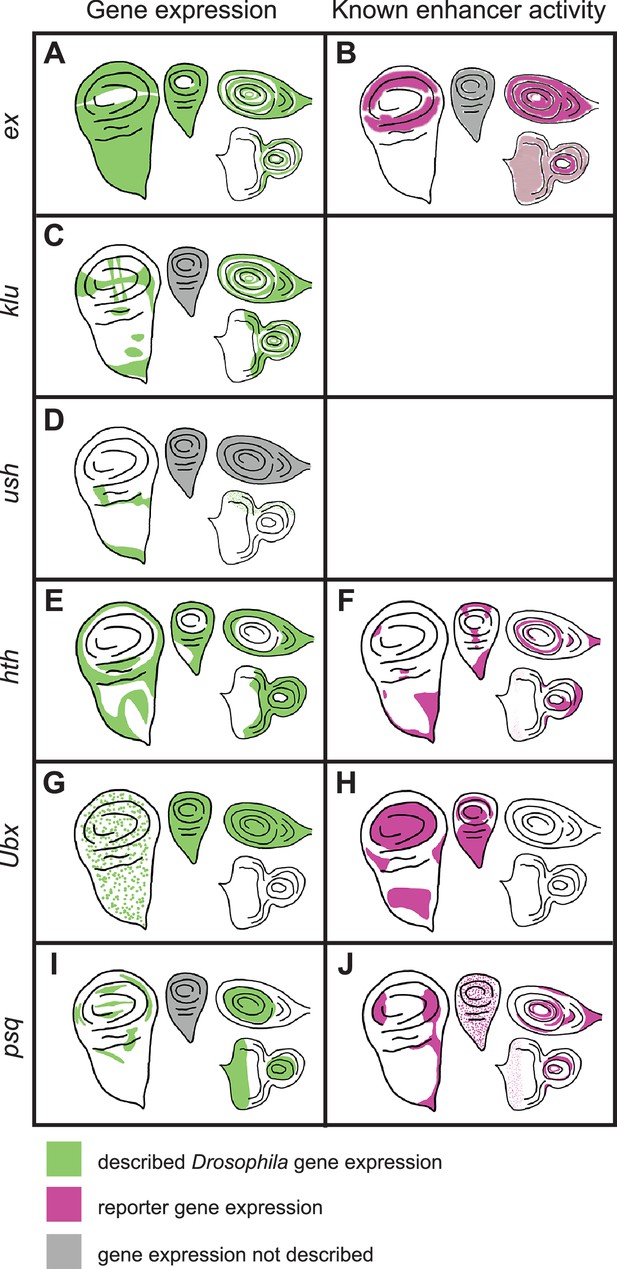
Previously described gene expression and enhancer activity for select D. melanogaster sequences predicted by SCRMshaw.
The left-hand column shows native D. melanogaster gene expression in imaginal discs (green), while the right-hand column shows described enhancer activity (magenta). Gray shading indicates that expression has not been described. Moving clockwise from the left side of each panel are the wing, haltere, leg, and eye-antennal discs. The enhancers whose activities are described in the table are: (B) ex_BCDE (Wang and Baker, 2018), (F) hth_GMR46D04 (Jory et al., 2012), (H) Ubx_GMR39A02 (Jory et al., 2012), (J) psq_GMR41E12 (Jory et al., 2012).
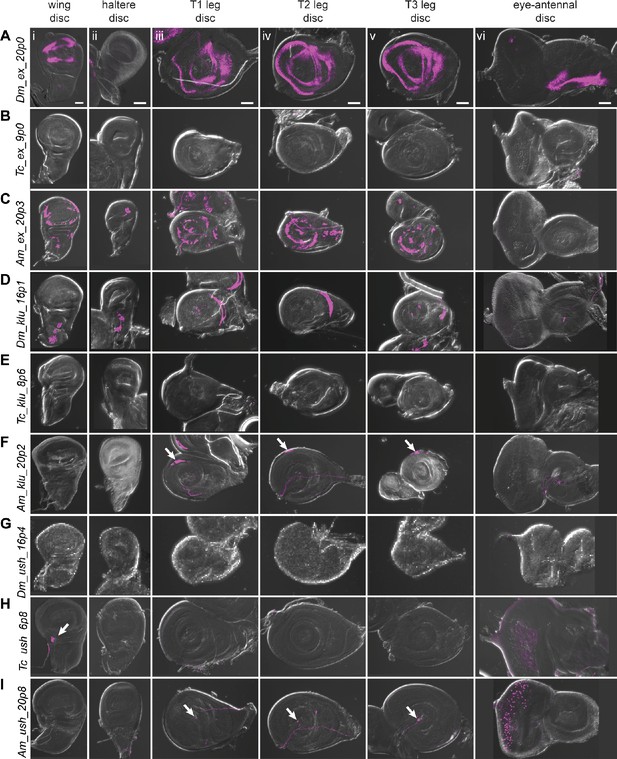
Reporter gene expression for tested ex, klu, and ush predicted enhancer sequences.
Each row shows expression for the indicated construct in (i) wing discs, (ii) haltere discs, (iii) T1 (prothoracic) leg discs, (iv) T2 (mesothoracic) leg discs, (v) T3 (metathoracic) leg discs, and (vi) eye-antennal discs (with eye portion to the left). Positive results were obtained by the enhancers associated with the ex locus of D. melanogaster (wing, legs, antenna, A) and A. mellifera (wing, haltere, legs, C); the klu locus of D. melanogaster (wing, haltere, legs, D) and A. mellifera (legs, arrows in F); the ush locus of T. castaneum (wing, arrow, H (i); eye, H (vi)) and A. mellifera (legs, arrows I (iii, iv, v); eye, I (vi)). Enhancer activities were visualized by UAS-tdTomato that was included in the reporter construct. Scale bar is 50 µm for each column.
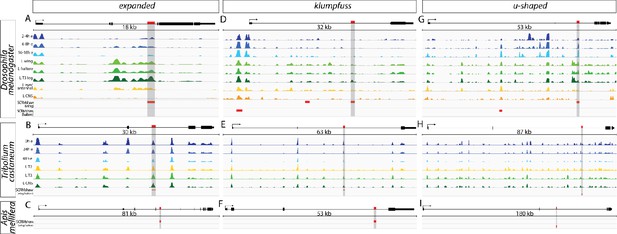
Open chromatin data for predictions in the ex, klu, and ush loci.
SCRMshaw predictions (red bars) are shown for D. melanogaster (A, D, G), T. castaneum (B, E, H), and A. mellifera (C, F, I) at the ex (A–C), klu (D–F), and ush (G–I) loci. For D. melanogaster and T. castaneum, open chromatin profiles are also indicated, for a variety of tissues and timepoints (see text). Vertical gray bars highlight regions chosen for in vivo validation.
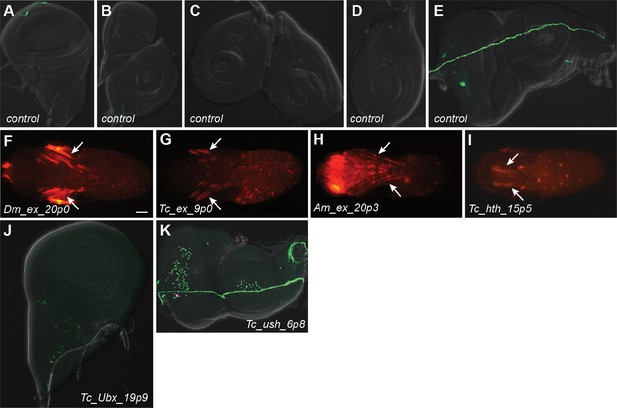
Additional expression observed in selected transgenic reporter lines.
(A–E) Control lines using a regulatory-inactive mock enhancer sequence demonstrate that there is no default expression in the imaginal discs. (A) Wing disc, (B) haltere disc, (C, D) leg discs, (E) eye-antennal disc. (F–I) Pupal expression. Arrows indicate expression in the legs (F–H) or notum (I). For T. castaneum enhancer Tc_ex_9p0, expression was observed in pupal legs but not larval leg imaginal discs (G, compare with Figure 6B). (J) Expression can be observed in migrating adepithelial cells of the wing disc in Tc_Ubx_19p9. (K) Expression can be seen in the peripodial membrane surrounding the eye-antennal disc in several lines, including Tc_ush_6p8 (see also Figures 6 and 7).
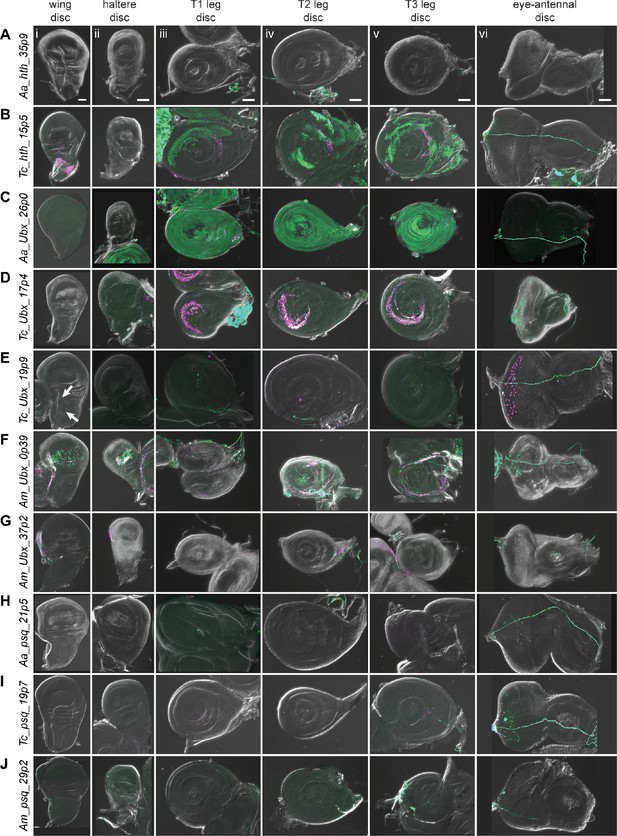
Reporter gene expression for tested hth, Ubx, and psq predicted enhancer sequences.
Each row shows expression for the indicated construct in (i) wing discs, (ii) haltere discs, (iii) T1 (prothoracic) leg discs, (iv) T2 (mesothoracic) leg discs, (v) T3 (metathoracic) leg discs, and (vi) eye-antennal discs (with eye portion to the left). Positive results were obtained by the enhancers associated with the hth locus of T. castaneum (the notum portion of the wing disc, legs, B); the Ubx locus of A. aegypti (legs, C), T. castaneum (legs, D) (myoblast cells in the wing disc, arrows, E (i); legs, eye, E (iii–vi)), and A. mellifera (wing, haltere, legs, F) (wing, haltere, G); the psq locus of T. castaneum (legs, eye, I). Enhancer activities were detected by the G-TRACE system; magenta represents direct enhancer activity detected by dsRed expression, while green indicates lineage-based GFP expression. Scale bar is 50 µm for each column.
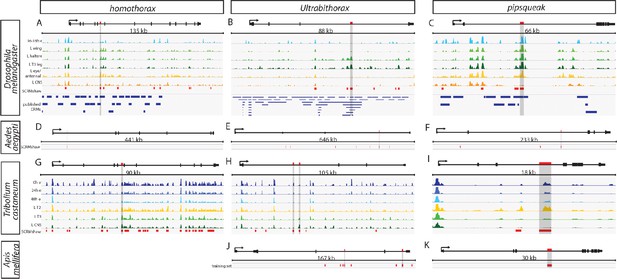
Open chromatin data for predictions in the hth, Ubx, and psq loci.
SCRMshaw predictions (red bars) are shown for D. melanogaster (A–C), A. aegypti (D–F), T. castaneum (G–I), and A. mellifera (J, K) at the hth (A, D, G), Ubx (B, E, H, J), and psq (C, F, I, K) loci. For D. melanogaster and T. castaneum, open chromatin profiles are also indicated, for a variety of tissues and timepoints (see text). Blue bars indicate the positions of known enhancers. Vertical gray bars highlight regions chosen for in vivo validation.
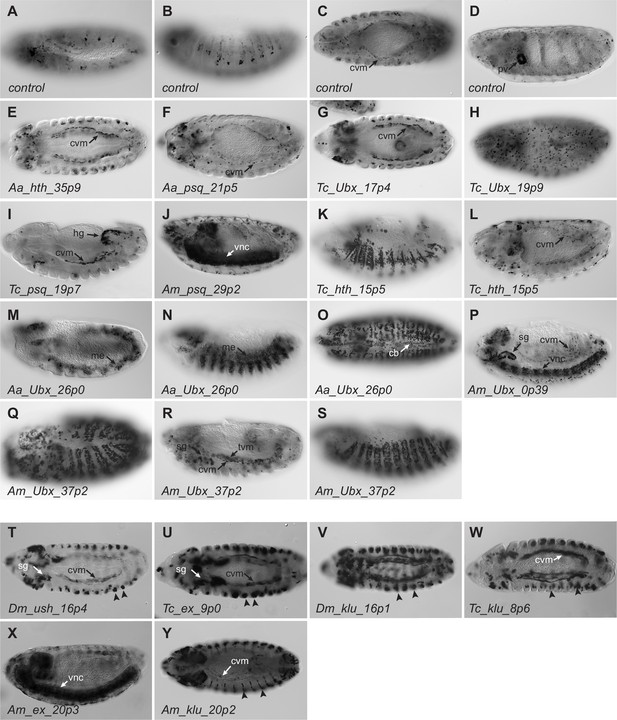
Reporter gene expression observed in embryos.
Panels (A–S) are putative enhancers cloned into piggyPhiGUGd and crossed to G-TRACE. Panels (T–Y) have the putative enhancer inserted into piggyPhiGUGd-TomatoI. All embryos are stained using anti-dsRed antibodies and the ABC-HRP kit and shown with anterior to the left. (A–D) A non-enhancer control sequence was inserted into piggyPhiGUGd to serve as a control for vector-related expression. Limited segmentally repeated expression is observed starting around stage 11 (A) and can be observed in the epidermis and/or peripheral nervous system at stage 14 (B). (C) Expression is observed in the caudal (longitudinal) visceral mesoderm (arrow, ‘cvm’). (D) In older embryos, strong expression is observed in the proventriculus (‘pv’) as well as in migrating hemocytes (individual cells observed throughout the embryo). This ‘default’ expression pattern was also observed in enhancer lines Aa_hth_35p9 (E), Aa_psq_21p5 (F), Tc_Ubx_17p4 (G), and Tc_Ubx_19p9 (H). Note that not all stages are shown for each of these genotypes, as expression was highly similar for each line at all stages. While this expression was seen in the remaining reporter lines too, additional activity, which may therefore represent specific enhancer activity, was also observed. (I) Tc_psq_19p7 had prominent hindgut (‘hg’) expression. (J) Am_psq_29p2 had strong activity in the ventral nerve cord (‘vnc’). (K) Tc_hth_15p5 displayed epidermal expression following germ band retraction; note that the caudal visceral mesoderm expression can also be observed (L). (M–O) Aa_Ubx_26p0 had reporter gene expression throughout the mesoderm (‘me’) starting around stage 10 (M) and persisting throughout embryogenesis. ‘cb’, cardioblasts. (P) Am_Ubx_0p39, in addition to the default visceral muscle expression, was active in the ventral nerve cord (‘vnc’) and salivary glands (‘sg’). (Q–S) Am_Ubx_37 p2 showed expression in the epidermis from stage 11 on, and also in the salivary glands. (T–Y) Although we do not have a no-enhancer control line for piggyPhiGUGd-dTomatoI, all lines generated using this vector had reporter gene activity in the caudal visceral mesoderm (‘cvm’), the salivary glands (‘sg’), and in segmentally repeated epidermal cells (arrowheads). We therefore view this expression as possible vector-specific expression (not all lines shown). However, two lines had additional, unique activity: Am_ex_20p3 (X) was active in the ventral nerve cord, and Am_klu_20p2 (Y) was active in narrow stripes in the epidermis (arrowheads). These activities may therefore be enhancer-specific.
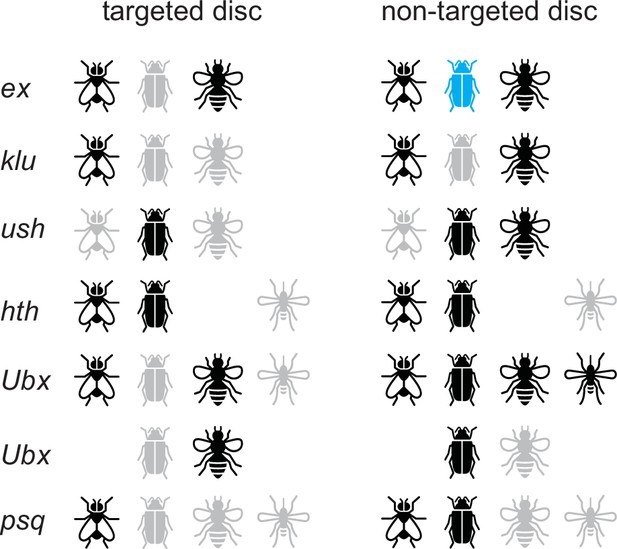
Summary of in vivo validation results.
Results are shown for D. melanogaster, T. castaneum, A. mellifera, and A. aegypti. Black, positive for expression; gray, negative for expression; blue, expression observed in pupae but not in larvae.
Image credits: The insect silhouettes were obtained from The Noun Project (https://thenounproject.com/), artist Georgiana Ionescu, under a Creative Commons CC-BY 3.0 license.
Tables
Species used in this study.
| Scientific name | Common name | Order | Assembly version/ annotation version | Link/URL for assembly information |
|---|---|---|---|---|
| Acyrthosiphon pisum | Pea aphid | Hemiptera | racon and v3_wdel | Courtesy Jennifer Brisson (University of Rochester) |
| Aedes aegypti | Yellow fever mosquito | Diptera | L5.2 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/7159/ |
| Agrilus planipennis | Emerald ash borer | Coleoptera | Apla_2.0 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/224129/ |
| Anopheles gambiae | African malaria mosquito | Diptera | P4.9 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/7165/ |
| Aphis gossypii | Cotton aphid/melon aphid | Hemiptera | ASM401081v2 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/80765/ |
| Apis mellifera | Honey bee | Hymenoptera | HAv3.1 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/7460/ |
| Atta cephalotes | Leafcutter ant | Hymenoptera | A.ceph_1.0 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/12957/ |
| Atta colombica | Leafcutter ant | Hymenoptera | Acol1.0 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/520822/ |
| Bombyx mori | Silkworm | Lepidoptera | ASM15162v1 | http://ensembl.lepbase.org/Bombyx_mori_asm15162v1/Info/Index |
| Cimex lectularius | Bed bug | Hemiptera | Clec_2.1 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/79782/ |
| Culex pipiens pallens | Northern house mosquito | Diptera | TS_Cpip_V1 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/42434/ |
| Culex quinquefasciatus | Southern house mosquito | Diptera | VIPSU_Cqui_1.0_pri_paternal | https://doi.org/10.1093/gbe/evab005 |
| Danaus plexippus | Monarch butterfly | Lepidoptera | v3 | http://ensembl.lepbase.org/Danaus_plexippus_v3/Info/Index |
| Drosophila ananassae | Fruit fly | Diptera | caf1 | http://ftp.flybase.net/genomes/Drosophila_ananassae/ |
| Drosophila melanogaster | Fruit fly | Diptera | r6 1.8 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/7227/ |
| Drosophila mojavensis | Fruit fly | Diptera | caf1 | https://www.ncbi.nlm.nih.gov/assembly/GCF_000005175.2/ |
| Drosophila pseudoobscura | Fruit fly | Diptera | r3 | http://ftp.flybase.net/genomes/Drosophila_pseudoobscura/dpse_r3.03_FB2015_01/ |
| Formica exsecta | Wood ant | Hymenoptera | ASM365146v1 | https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_003651465.1/ |
| Heliconius erato | Red postman butterfly | Lepidoptera | v1 | http://ensembl.lepbase.org/Heliconius_erato_lativitta_v1/Info/Index |
| Heliconius melpomene | Postman butterfly | Lepidoptera | Hmel2 | http://ensembl.lepbase.org/Heliconius_melpomene_melpomene_hmel2/Info/Index |
| Heliconius himera | False postman butterfly | Lepidoptera | Hed.V1 | Courtesy Robert Reed (Cornell University) |
| Junonia coenia | Common buckeye butterfly | Lepidoptera | JC v1.0 | Courtesy Robert Reed (Cornell University) |
| Leptinotarsa decemlineata | Colorado potato beetle | Coleoptera | Ldec_2.0 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/7539/ |
| Manduca sexta | Tobacco hornworm | Lepidoptera | v1.0 | http://ensembl.lepbase.org/Manduca_sexta_msex1/Info/Index |
| Nezara viridula | Southern green stink bug | Hemiptera | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/85310/ | |
| Nasonia vitripennis | Jewel wasp | Hymenoptera | Psr_1.1 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/7425/ |
| Onthophagus taurus | Dung beetle | Coleoptera | Otau_2.0 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/166361/ |
| Pseudoatta argentina | Leafcutter ant | Hymenoptera | ASM1760752v1 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/621737/ |
| Stomoxys calcitrans | Stable fly | Diptera | Stomoxys_calcitrans-1.0.1 | https://doi.org/10.1186/s12915-021-00975-9 |
| Tribolium castaneum | Red flour beetle | Coleoptera | r5.2 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/7070/ |
| Trichoplusia ni | Cabbage looper | Lepidoptera | tn1 | https://www.ncbi.nlm.nih.gov/datasets/taxonomy/7111/ |
| Vanessa cardui | Painted lady butterfly | Lepidoptera | Vcar_v1 | Courtesy Robert Reed (Cornell University) |
| Nebria riversi | Ground beetle | Coleoptera | v1 | https://doi.org/10.1111/1755-0998.13409 |
Overlap of SCRMshaw predictions with FAIRE-seq and ATAC-seq peaks.
| Species | Training set | Profiled tissue | Overlap(real) | Overlap (random) | s.d.* | z-score | FE† |
|---|---|---|---|---|---|---|---|
| D. melanogaster | haltere_disc | Wing, leg, and haltere third instar discs and pharate appendages; eye-antennal disc; third instar CNS | 41.82% | 14.02% | 10.65 | 21.53 | 2.98 |
| D. melanogaster | blastoderm.mapping1 | Blastoderm | 40.99% | 13.86% | 12.35 | 22.56 | 2.96 |
| D. melanogaster | mapping2.wing | Wing, leg, and haltere third instar discs and pharate appendages; eye-antennal disc; third instar CNS | 35.14% | 10.69% | 10.19 | 23.01 | 3.29 |
| T. castaneum | mapping2.wing | Embryo, larval thoracic epidermis, larval brain | 85.58% | 26.18% | 10.98 | 28.88 | 3.27 |
| T. castaneum | mapping2.ectoderm | Embryo, larval thoracic epidermis, larval brain | 81.68% | 25.48% | 12.48 | 26.29 | 3.21 |
| T. castaneum | mapping1.ventral_ectoderm | Embryo, larval thoracic epidermis, larval brain | 81.17% | 24.76% | 12.56 | 33.87 | 3.28 |
| T. castaneum | mapping1.dorsal_ectoderm | Embryo, larval thoracic epidermis, larval brain | 71.15% | 24.69% | 12.64 | 24.07 | 2.88 |
| T. castaneum | mapping1.ectoderm | Embryo, larval thoracic epidermis, larval brain | 69.53% | 24.66% | 11.92 | 22.35 | 2.82 |
| A. gambiae | embryonic_midgut | Adult midgut, salivary_gland | 40.20% | 11.66% | 7.90 | 21.56 | 3.45 |
| A. gambiae | mapping1.salivary_gland | Adult midgut, salivary_gland | 36.76% | 11.43% | 8.52 | 20.55 | 3.22 |
| D. plexippus | mapping2.wing | Larval forewing, hindwing, head | 60.43% | 30.35% | 6.30 | 8.93 | 1.99 |
| H. himera | mapping2.wing | Larval forewing, hindwing, head | 68.34% | 41.10% | 8.97 | 10.27 | 1.66 |
| J. coenia | mapping2.wing | Larval forewing, hindwing, head | 3.06% | 34.91% | 8.52 | –12.22 | 0.09 |
| V. cardui | mapping2.wing | Larval forewing, hindwing, head | 15.46% | 36.01% | 8.01 | –7.13 | 0.43 |
-
*
Standard deviation.
-
†
FE, fold enrichment.
Gene loci chosen for in vivo validation.
| D. melanogaster | T. castaneum | A. mellifera | A. aegypti | |||
|---|---|---|---|---|---|---|
| Name | Symbol | FlyBaseID | RefSeq | iBeetleBase | RefSeq | VectorBase |
| expanded | ex | FBgn0004583 | gene-LOC657053 | TC012545 | gene-LOC551519 | AAEL001437 |
| u-shaped | ush | FBgn0003963 | gene-LOC659918 | TC013689 | gene-LOC100577801 | AAEL020615 |
| klumpfuss | klu | FBgn0013469 | gene-LOC103312803 | TC002783 | gene-LOC100577692 | AAEL013544 |
| homothorax | hth | FBgn0001235 | gene-Hth | TC008629 | gene-LOC552079 | AAEL011643 |
| pipsqueak | psq | FBgn0263102 | gene-LOC660343 | TC003349 | gene-psq | AAEL021255 |
| Ultrabithorax | Ubx | FBgn0003944 | gene-Ubx | TC000903 | gene-ubx | AAEL014032 |
SCRMshaw predictions chosen for in vivo validation.
| Coordinates | Max. score | Training set(s) | Method | |
|---|---|---|---|---|
| Set 1 | ||||
| T. castaneum | ||||
| Tc_ex_9p0 | NC_007424.3:11221530..11222170 | 9.04 | mapping2.wing | imm |
| Tc_ush_6p8 | NC_007420.3:5968840..5969370 | 6.84 | haltere_disc | imm |
| Tc_klu_8p6 | NC_007418.3:10416700..10417300 | 8.59 | mapping2.wing | imm |
| D. melanogaster | ||||
| Dm_ex_20p0 | 2L:442110..442810 | 20.04 | mapping2.wing | imm |
| Dm_klu_16p1 | 3L:10991040..10991700 | 16.06 | mapping2.wing | imm |
| Dm_ush_16p4 | 2L:531250..532060 | 16.40 | mapping2.wing | imm |
| A. mellifera | ||||
| Am_ex_20p3 | NC_007075.3:7545750..7546440 | 20.36 | mapping2.wing | imm |
| Am_klu_20p2 | NC_007070.3:720220..720870 | 20.25 | mapping2.wing | imm |
| Am_ush_20p8 | NC_007080.3:10609550..10610200 | 20.80 | haltere_disc | imm |
| Set 2 | ||||
| T. castaneum | ||||
| Tc_hth_15p5 | NC_007422.5:13408990..13409850 | 15.52 | mapping2.wing | hexmcd,imm,pac |
| Tc_psq_19p7 | NC_007418.3:1805000..1806250 | 19.71 | haltere_disc, mapping2.wing | hexmcd,imm,pac |
| Tc_Ubx_19p9 | NC_007417.3:8137250..8138000 | 19.32 | haltere_disc | hexmcd |
| Tc_Ubx_17p4 | NC_007417.3:8153250..8154030 | 19.95 | mapping2.wing | hexmcd,imm |
| A. aegypti | ||||
| Aa_hth_35p9 | 1:149733960..149734710 | 35.96 | mapping2.wing | imm |
| Aa_psq_21p5 | 2:228190750..228191640 | 21.50 | disc.mapping2 | hexmcd,imm |
| Aa_Ubx_26p0 | 1:309747490..309748390 | 26.08 | mapping2.wing | imm |
| A. mellifera | ||||
| Am_psq_29p2 | NC_007078.3:10712000..10712750 | 29.26 | mapping2.wing | hexmcd |
| Am_Ubx_37p2 | NC_007085.3:2921250..2922250 | 37.18 | haltere_disc, mapping2.wing | hexmcd,imm |
| Am_Ubx_0p39 | NC_007085.3:2967750..2968250 | 0.39 | mapping2.wing | pac |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | expanded (ex) | FlyBase | FBgn0004583 | |
| Gene (D. melanogaster) | u-shaped (ush) | FlyBase | FBgn0003963 | |
| Gene (D. melanogaster) | klumpfuss (klu) | FlyBase | FBgn0013469 | |
| Gene (D. melanogaster) | homothorax (hth) | FlyBase | FBgn0001235 | |
| Gene (D. melanogaster) | pipsqueak (psq) | FlyBase | FBgn0263102 | |
| Gene (D. melanogaster) | Ultrabithorax (Ubx) | FlyBase | FBgn0003944 | |
| Gene (Tribolium castaneum) | gene-LOC657053 | iBeetleBase | TC012545 | |
| Gene (T. castaneum) | gene-LOC659918 | iBeetleBase | TC013689 | |
| Gene (T. castaneum) | gene-LOC103312803 | iBeetleBase | TC002783 | |
| Gene (T. castaneum) | gene-Hth | iBeetleBase | TC008629 | |
| Gene (T. castaneum) | gene-LOC660343 | iBeetleBase | TC003349 | |
| Gene (T. castaneum) | gene-Ubx | iBeetleBase | TC000903 | |
| Gene (Apis mellifera) | gene-LOC551519 | RefSeq | ||
| Gene (A. mellifera) | gene-LOC100577801 | RefSeq | ||
| Gene (A. mellifera) | gene-LOC100577692 | RefSeq | ||
| Gene (A. mellifera) | gene-LOC552079 | RefSeq | ||
| Gene (A. mellifera) | gene-psq | RefSeq | ||
| Gene (A. mellifera) | gene-ubx | RefSeq | ||
| Gene (Aedes aegypti) | AAEL001437 | VectorBase | ||
| Gene (A. aegypti) | AAEL020615 | VectorBase | ||
| Gene (A. aegypti) | AAEL013544 | VectorBase | ||
| Gene (A. aegypti) | AAEL011643 | VectorBase | ||
| Gene (A. aegypti) | AAEL021255 | VectorBase | ||
| Gene (A. aegypti) | AAEL014032 | VectorBase | ||
| Antibody | anti-dsRed (Rabbit polyclonal) | Clontech | Cat#632496 | (1:500) |
| Recombinant DNA reagent | piggyPhiGUGd (plasmid) | Deem et al., 2024, PMID:38698030 | ||
| Recombinant DNA reagent | piggyPhiGUGd-TomatoI (plasmid) | Deem et al., 2024, PMID:38698030 | ||
| Sequence-based reagent | Dm_ex_20p0 | This paper | TTCCCAGAACAAACTTGTGGGGGGTGATTAGGTTTGGCAACAAAATATTTTGCTAGTATTCCCTAATCATTTTTTTGAGTGAACCAAACTCGAAGAGCTCTACTCCCCTGGCCATCCACTTGTTGCCACTTCCATTCCAGCTTTGCGTCGACGACGTCGTCATTGATAGGCACTTATTCGGCCGCTGATGATTATTATGATATTGTAGCTGCTGCTGCTGTGTTGTGGATTCGATGCTGAGGTGCCTCTATTCCATGGCCTCCTTCAACCTGCCTGCCTGCTTTTTTCATAATTATTATTTTTCATCTTGCTGCTCTTCATTTTGTATGCAGGAATTCCAATTTTTCGTTCGATGAAGTGTGTGTTGATTTCGCTGTTGTTTTTTCTCTGCCTTCTCGAGCACCGCCGACATGCCCTTGGGCCCTTCTGCTTGGCTCGGGTCGGAGCTATGTAGCGCGGTCCGGTACCGGTCTCGTCTTCGAGCATCAGGCAATGGGCCTCTGACAACCTGACGTGTCGTCATCATCATCGTCTTCATCTGCTGGAGTCTCTGACTCTTGTTGATGTCAATGGGTTGCTTGTTTATTGCCTGACAAACTGACAGAAGTCTGGTCGGGGTCTCGATCCGATTTGAGCCCGATTTGGGACGCAAGAGGAGCGCTCCCTCTTGCATAGCCGAAAGTTCATTTAAAATTTTGAT | |
| Sequence-based reagent | Dm_klu_16p1 | This paper | GACCAGGCTGTTGCAGTTTCGTGTTGAAACCAGTTTGAATATATTTATTTTTATTTCCTGCGTCCCCTTCCCAATTTCTGTGGCCCTTTTAGGCGCCTCAGTTAGTCGGCAACGATAAGGCGGCAATGGTTTAATTTAGCTGCACCAGCGGCAGCAGCAGATGACGACAGGATCGTTGGGCCGGTCTACGTGCAACAGAAGTTGCTGCGCCGGCAGAAGCAACGGCAGCGGCAGCAACAGCAACAGCAAAACAAATGTGTCTGTATATCGCAGCTAAATTGACTTTGATCACGCGATCCCGAATCCCCCCCCCCATTTGGTCCGAGTTATATGGCCGATTCCAGGTTGCAGGCTCCAGGTTCTCGGGCGCGGCCTTTTGTGGCACAAACGGAAGTATGCTAAGCAACTTGTTGCTGCCGCAAAGGCAAAGCAGCAAAAGCAGCTGAAGGTGTATATTGCAAAAATAATTACATTTGATTGTAAAAGGCCAGCGTCTCTAGGCTGGGGACTCGAGATCGGACCTCGGCCTGCCAGAGAAAAATGTGCAACATGATTGCAGTTTACAGCCCCAGCAGCAGCAGCAGCAGCTAAAGCAGCAACAACAACAGCAGCAGCAGCAGCAAAACCAACAGATAAAATGCATTTACAATTGAATTTGAA | |
| Sequence-based reagent | Dm_ush_16p4 | This paper | GAATGTTGCTGCGGTGGCATGTTGTTGCTCGTCGAAGTTCAGCCGATGTTGCTGCTGCTGCTGCAGCTGCTGTTGCTGCCATTCCCACTATCAACCGATGGTAATCGAAGGAAGCGACATTTATGCAAATGCCAGGTTGTTTAATAAACGCAAATTATGAGCCCGGCAGCAACATGTTGCAGCAACAGTCGATGGCAGATTAGCGACATTCATACTTGCACTTGGGTCAATTTAAATTTGTGCAACAGTGGCAGCACGGCACGGCAGCAACTCCTTGCCGCAGCAGCAGCCGCTCCAGCAGCACATGAGATTGTGGGAGCAACAGGCAGCATTATTGTGTTCGGCCAAGATCGCAATTGATCAGTGTGTTGGTGCTGGTGTTGGTGTTGCAGTTGCAGTTGCAGTTGCAGTTGCAGTCGCTGTTGCTGTTGCTGTTGCTACCCGACGACAACAGTTGCTGCTGTGCTGGTGCTAGTGCTTGTGCTTGTGCTTGTGCTTGTGTTGCTGCTGATCAAGCGATCAAGCACCGCAGCCAAAAACAATCGGCGCTGAGCGTGCTCACACGAAATTTTCAAGTACTGCGACAATTTCCATGCCCCCAGCCGCTGCCTTGTTATCAGCGCGCCATGCAACAGCAACAGCGACTGCAGCACAGGCAACAGCAGCAACACATCTCAACAGTTGCATCAATTGCTCAACATTGAACTCTGGGCATGGGCCCAGACGATCACCCCTCCTGGGGACCCCCTTCGGTCGCCCCTGCCCCAGTCCCTTGTATCATTTGCACGTTTTTTAATTAAGACATCAAAA | |
| Sequence-based reagent | Tc_ex_9p0 | This paper | ATAGTTCTAAAGTTCTAAACTATTTGCAAATGTAAACAACGACCGACATTTTCAACATGTTCGGGTGTACGTCGCTTTGAATATGGAAAATGTGATTTTGTAGAGAAATTTGGTGGCGTAGCCGTTGCCAGCTCCAGTTTCTTAAGGCAGGCATCTGGTAGCGCGCATCACAGCAGGCCGGGCCAGGTCAATAAAAAATCGAGTCAGCCGTGGGCTAAAAAACGCGACTAAAAAAACAATGCAGAGCCGCGGTTAAAGACAGGTAGCCGAGCTAAGCGGTAGGGGAGAGGGGGATCAGGATCATTTGTATACTCGGGGAATGTGCCCGACCCGGTACACTCGATGCCAAAACGAACACGCCGACTTATACAGATGCCTATCTGACAATACGGACACTTTTAAAAAACACTTTTTCGGTTTTTAAAAGTTAAAGCCAACAAGCGCTGTGTTTTACACAATTCTTCCAATTTCCATTCCCAAGTTGCAAAAGTGAAACGTCCCAAAATATTTTGTCGCGCAAATCAAAAGTATATTTTATTCATGAGGAGCTCGTGTAATTTTTATGTAAATTTTAATTATTGATAACAAGGGACCATTGTTTGACGACACTTTCTTCGGCATGCGGAGCTCCTTGTTTTGT | |
| Sequence-based reagent | Tc_klu_8p6 | This paper | TTATGAGTTTGGTTATCGTCGGAATCGGGCATTCTTCCTTTGTATCCGATTTTTAGGTAACAAGCTAGAAATTCCAGAGCTACACCTACGATCCATCAAGTCGGAGCCGTTCTAATTGGCCGCTCCTATCTATCGTCTGAAGGAGGCAGCAAGCAGCACACGAACACCTGGCTGCCAATGCACCTGATGCGGTCGTCCGTCGCTGCTATCAACTCACAATGTTTACTTGTGCTTACGCCAAAATTATGACGAATATTAATGCGGCCTCGTACGCAGGCAGGCATGCGGCGTAATTACTACCGGAGGGACCTTATCTCGAATTATTATGCAGCAAGCAGCAGTGAAAAATAGCGCAACGCCTGCTGCACTCATCCAGATCTGACAAAGATGAAGACGTCGCTGACATCTTTATGATTGTGCTTTTATTGCACCTTTTCGCGGAATTCCGACCATTCGAAGCGACCTTTGCGCTACGGAGAAGAGGAAATTTACACCGGGAGTTGACTTATGATGGGAGAACCACTCTCAACGAACGCAACTACTTTCCAGGAATATGAGAAGAGTGCTTAACTGACAAGTCCAAATTCGAACTTGAGGTTA | |
| Sequence-based reagent | Tc_ush_6p8 | This paper | TAAATAATCCAGACATGCATTGCATGTAAGTATCAGAGATACACGGTAAGAGTGGAGCTTTTCGAGAATCCGGAAACCGATCAGATAAGTTCTGAAAATGACTCGTCCACGAATAGATTTAGGATCGGAGCTGTTTTCCATTCGCCGAGATAAGTCGATAAGTTTCAATAAGTCCGAGTTCTGGCAACAGCCAGCACGGTACGGGCTGCCGCCGTCTTGGTTTCCAGTTTTCTCCAATGTCGTGGTATTAATCAGGGCGTTATCTCTAGCACATAAACACACGTATGTGTATGTGGGGCGATGTCGGTGGCGATACCGTTCCATGTGGGGGTGTAGCTGTTGGGGGTATACGGGCCTGTTCGCCGTCCGATAGCGCGAAAGATACGACCTGGAAGTAGGAAACGAGACAGCGAGATAAGAAAGTAATATGGCGGCTGCTGCAAAGAGATAACGACTGATACGCGCCTGCACCTTTCCCGACCTGCAACTCTACGTGCCCATTATTTTTGGAAAATTCAATGAGAAATCCG | |
| Sequence-based reagent | Am_ex_20p3 | This paper | TCTTGAGATCTTTCTGCATATAGCCGTGGTCTTCTTGCCCTCCCTCGCCCTCTGGCCCCGGACACCATCCACGGAGCTCCTTCCCTTCCCTTCACCGAATATACTCGGCTGTGCAGCGCCTGCTACCCTCTCGCTCTACTCTCTTGCCTTTCCACCAGTCATGACAAGCCGGTCCGACTGGTACCCCCACCAACGCGGCCGGACGGACCCTTCTTGGGCCTCGCAAGGGCCCTGCGGGACCCCTCCCTCCTACATTCCAGCGGGGCCCCATCACGGCGAGGCTGAGCTGGCGGGTTTTGAGGCGCGCGAGCCATGCCACGACAGGAAAAAAATGCATCTGAAAAACGAAAACAAGTAGAAAAAGGTGGCTCACACCCCTGCATGCGTGCGTCGGTTTGCGTGAACGTTGCCCGGACCCCGTACCGAGGCCTCCTCCTCCTCCTCCTTTCTCCTCCTTTCTTCCTTCCTTTCTTTTCGCTCCTCTACCTCCTCTGCGCGCCTCTTTACGCTCCTCTTCTTGCTCTACGCTCTCGCGTACGTGCCCGCAAACTGCTGCCTGCCTGTTCAACGCTTCTTCTTCGTTTCCTTCTTCTTCTTCTTCTTCTTCCTCCTCTCTGCTTCGTCCTTGCGTTTCTCTCGATTCGCGTCACATCTCCGCTCCCCCAAATACGTTTCCTTTCTAAGATCGTTT | |
| Sequence-based reagent | Am_klu_20p2 | This paper | TTATTTATCGCCCTCGAAAGCGCTCGTCCTCTGCAGATTTCGATCGAGTCGTTCGACTTCGATATAAGAATTTCAGTGTAAACGCGATACACGTTAAATAACGAATATTTACGGACAAAGTCGGGCGAACGACGCGATCCTGGCCGCTCGTGGCCGATGCGCAGGAAGGTAGGAGAGCGGAGAGGTTTTACGCTGTTCGCGGAGAGGAGGATAGGTTCGTATAGCTCCTTAAATCAACCCTAGTTGGCCTGTCAACCGAGTTGGCGCGCGCGCGCATTCCCTTTCGCGGCGCACAAATTACCACGCGTTTAATTACCGTCCGATATACGAAGCAGGCTCATTAATCACCACGCCGATAACCCGTAATTTTCCAGCAACGATAAAATCTATCGCGCGACACCGGCTCTCGCGACTTTCCTCTCTCTCTCTCTCCCTCTCTCTCTCTCGTTCGAGGAGAAAGGAGAAAAGGAAGCAGGAGGACGGAGGAGGTGTGCAGAGCGATCCTGTCGCCGCTTCCATATAGATTTTTTTTCTCCCTTCCGCGCTCTTTCTCGCGCCAGTTCTCTTCGTGCGGCGGAAAATAGAGCGGCGCAACTCCCCTTCTCGCGACTCACGGAGGGCGAACAGCTGAAGCCGGCCGATCGATACGAA | |
| Sequence-based reagent | Am_ush_20p8 | This paper | GGGGCTCCCTCCTCCTTCCCTCCTCCTCCGTCTGTCCCAGTTGGTCAGCCACGGTATCGTTTCGACGTCGATTCATCCCTCTTTTTCTCCCCCCTCTTCTCTCTCTCCTTCTGCCCCTCCCCCTCTCCCCTCCTCGCCATCGGCTTCGAGAGCCACGAGGCGATCGAGAGAGAGAGAGAGAGAGAGAGAAAGGGTACCCCATCGATGGATCGATCTATCGATCCACACGGGGATCCACCACGCTCTTCTGCCCTCTCCTCCTCCTCGCCACAATTTCTCTCCTCTTTACGTACGCTTCTCTCGTCCTTCGTGCCGCTCTTCGTCGCCATCGAGATTACGGCGAGCGAGGGGCCGACAGCCGAGGGGCTTCTTCCAATACTTTGTAAGTTTATTTGTATGATCCGCCAATACTTTGTATCTTTATTTATATGAAATCGGATGGCGGATCGAGATTGCTCTCTCTCTCTCTCTCGTGTCGCTCGTGTCTCGTCTCGCTTCTCCCCCCGTTTCCTCTTAAAATTAATTATACGTCCAAGGTGGGCGTAAGAGAGAGAGAGAGAGAGAAAGAAGTCGCAATGAAACCGGAAGGATAAAGAGAATCCGATGGTGCGCACACGCACGTGTATGTACACGTCCACTTTATAACACTCG | |
| Sequence-based reagent | Aa_hth_35p9 | This paper | TTCCGAACACCTTGATTCAAATCCGAACAGTAGGTACGAATAAATCATACCGTTTTGCTTCGAAATCTGGACACCTAAGACGAAGTGTATTTTCAAACTTGAATATATATATAGTAGAGATGGTCGGGTTTCACATTTTTCAAACCCGAACCCGACCCGTACCCGACTTATTTTATTTCTTCGAACCCGGACCCGACCCGAACCCGAGACCATAATTGAAAAGCAAACCCGGACCCGACCCGAACCCGAAAATTTTTCACAGTGCAAACCCGAACCCGACCCGAAACCCGAAAAATGTTTGTAAAAAAACCCGAATACAACCCGAGTTTGAAAAGATGGTAAATTCATCGTTTCTGATGCATAAAGAAGCTTTTAGATTGTTACTCTGTTCACAATTTTCACCAAACCCGACCTGAACCCGATTCAAACCCGACTTTTTGTAAGCCCGAACCCGACCCGTACCCGATAATTTCGTAGCCTACAAACCCGACCCGAACCCGAACCCGAAAAATTTCAAATATTCAAACCCGAACCCGACCCGAACCCGTCGGGTTCGGGTTCGGGTCGGGTTTCGCGTTTGAAAACCCGAGACCCGACCATCTCTAATATATAGGCAAATTCATACATACTAAAAATCCAATGGTGATTCTTGCTTCGAAATCCGGACAGCATGTGAGAGCCGATTCAAATATTGGACACATTTGCTTCGAATTCCGGACACTTCTATTTATCTAGTTTGTTCAAAGATTC | |
| Sequence-based reagent | Aa_ubx_26p0 | This paper | AAGTCGGGTTTAGTCGGGTTTGTGTCGGGTTTGGTGAGAATTGTGAACAGAGTGACAAACTAAAACCTTCCCTATGTATCAGAAACGATGAATTTATCATCTTTTAGAACTCGGGCTTTATTCGGGTTTTCCTTAGAAAACTTTTTCGGGTTTCGGGTCGGGTTTGGGTTTCAACAGCGAAAATTTTTCGGGTTCGGGTCGGGTCCGGGTTTGATTTTCAATTATTGTCTCGGGTTCGGGTCGGGTCCGGGTTTGAAGAAATAAAATAAGTCGGGTACGGGTCGGGTTCGGGTTTGAAAAATGTGAAACCCGACCATCTCTACTCTTCAGGTAGTCGAGAGTTGTTTTTTTTTATCTTTTATTTTTATTTTAAAGGCACTCTGTGCTCGTGCCCACTACTATGCCGAAATCAGTTCATCTGTATCTTCTTCACCGATTAAGATCTATTTTTAACTAATCTATATTTAAATCTACTTTCACTCTCTTCTACTCGTTTGCTCTCATACCGAGCAGGTAGGAGAGTGCTCTGCTGATAGTCCAATCGATTTCCATAAGCCATAGTTCCATTGCTCTTGCGGTGGTTCGTTTTGCCATGTTCCTGAGTCGTTTGAGGCTAGCTGCCTGCGAAGTGGGTCAGTTTGTCTCAG | |
| Sequence-based reagent | Aa_psq_21p5 | This paper | CCGAATTTGTGAGAGAGATAGGAGCCAATGTTTGAGTGATTCCCGCGAAGAATTGAAACCTATAAACGATTCCCACTAATTTTTGCAACATCTGTGATTTTTGATTTGATTTGAAACTGCAACTGACAGAAGATAATCAAAATACACTTTTTTCGCATTCGTACATCAATTGACAACCATCACTTGACACACCTGGCGATATGGACCAATAGGTCTGTGCCACAATAAGGGAGAGAAAAAAAAAAAAAAAGTGTGAAGCAAAAACACGCACATGTAACTTAAAGCACCACAAGAACCCTTTCAGCACCGGCCGCTTATGCTGATTTTATTAAAAAGCTTTATGCATACATGTACATAAGAGTGAGCATGCCGAAGCTCGAAAGTGTGTGTATGTGCGAATGCGCCAAAACACGATTATGTTTTCGTTTGTATTTCTTTCTTTTGCCGGCAAAAATTCTGTGTTTCGTTTTTTGATAGTAGGTAACTATGCCCACACAGTTACGGATCACACATAGTCATGGATCACTTTGGCGTTCAACATCGGATAACTCGCTCAAAACATATTTGCATGTGATGTAAACATATTTTTGCCAAGTCATAATATTTGTCTTCTGTCATTTGTAATATCAAATAATAAGCATAACATTAAACCGCAAAATAATGGTGTTTTTGAAAAATGTTTAGTTTGTATTGCCAAGCTATCATTAAATAGTCATTTATTGTAAGAAGTGCCATCAGCATTCTCCTATGCTTTTAGGTGATAAAATTCAAATATTATACATAATAGTTCCTCGTTCTCTTGAATTCAGTATGATTTCTTTGTTAGAAAACATTTTTCTTGTTTGTTGATACTGAATATTAGCAATTCCAACTAGTGATATTAGCAATTC | |
| Sequence-based reagent | Tc_hth_15p5 | This paper | TAATCTTTTAATTTAAAGCGTAGCTGAGCAGCTGGCTCTAATTCCACTTTCCTTATTTGGTTTCGTTGGTGTGGATTTTTGAAACGGATTATTTCGAGAAATAATAGTTTATTAGTGGTGGAAATAATGAATGGGTCTGGAGCGAGTTCCAGAGTGCGATTGGTTGGTTAGCGGGTAAATTTTTAAAAAGTGGGTGTCTTCTCCGACGGCAATTTAACGATCGTAACGACGTCGTCGCTAATTAGGCTCGTTGAGGCCGTCGCTAGATCGATAACACAGGCTGCGACATCGTCACAATGCACCGGTCGGGTTACACATCGGAGTCCGTCTCCCGGGGGCCCGTCTCAGATTCTCCGTATTAAAACACCGACATGTAAAAATATGGAAATTGCGCGCGGCAGAATGCGGTCCGATCAACCGGATGGCCATCGCGCATCGCTTTGCATTCGCAGCCGCATTTAAATTGCTAAAAGGGGACACTATCGAGCGGTCCATCTCTCTCGCAGCGTTGCGATATTATAATCTTGTTGCAAGGTAAATGCACATAACCGGTTACCCCAGACAGACGACGTCTTTGACACGAAAAAACCTGCCATCTATGTACAGCGGATCCTAATTTACGGCCTTATTCCATGTCATTAAGAGCATACGGGACGGACACGTTTTTAGGAACTTCGGACCCGACTTATCTCCGCGGACCGATAAGGAAATGTGCCTCTGGACACCTAACTTTGCCGACCAACAAAATCATAACGCTCGCTCTATGCCCATTGGGCAACACGAAAAAACCTGCC | |
| Sequence-based reagent | Tc_Ubx_17p4 | This paper | TGCATGTATGTCGAGTGGGTCCGGATGATGCGAACTCCCGCCGATTTCTTCGCAATCTGCAAATTCGCTCAAGTAGCTTAATAACAATGACAAAAGTGAGGCGGTATATTTCCGGCCGTCCGTTGAAAATTGTAATGATGTTATTAAAATTATGACGTGGCCGTGATGGTCGCCGAATTCTGGCGAAACGGCCGCGTAAAAACGGCACATAATTGGCTGACATTAAGATGTATCTGGAGATGTTTTTCGAATGCCTTCGTCCGGCGCGAATGCCTGAATAAGCGGCAAAGCTCGGAAAGCTCTTATAAATAAAAATGTACGGAGCCAATCAGATCGGCGAGTAAAAAGTACGTCTTTTCTTACACCAGAGGATCGCAGCTGCCGCAGAATCCGGTCGCGGATAAGAAATAAGAAGTGCTGCATAAATGCATTGATCATTCGCCGGGTCTCCGTCTGCTGTTCCTCAGCGAGAAAACGGGTTTAAGTCTGGATACTTTTGGCTCTCTGGAAAGTGCTTTTTGCATTAAGCTGCCGAGAGAGAATAAAGACGTTTGCGGTGTCGGACGGTGACCAATGCTGCTGCTGCTGCTCTGCCTTCCAAGTGCGTGCTTTAAATCTTCCACTTTGCAAGTAAATCGAGACGAACGCTGAATATTTTACACGAACACTGTTTATAGCCCAAATAACAGCCTTCCAAGGGCGGCCACCATCAAAAAATGGAGCGCTCAAACCCGAAATATGGGCGGGCGAAAATTATTCAAACCACAAAGCGAGGAAATCAGAAATTCAAAAATTGACGGCTTTCAACTCAGGACTGAATTTTTATAAATTTTTGTTCGCTACTGCAATTTGGGACAGAAAATTACATCT | |
| Sequence-based reagent | Tc_Ubx_19p9 | This paper | CGTATTTAAATATCGTTAGGTTCGATGGTAAAATTGGAGAAAATTGTCGCGCGCGTTTAAGACAAAGAAAATTCCCGTCGGGTTATCAATCTTGGGTTATCTGTACCCTCGG GCCGAAAAACTCTGTAAAGAAGAGACAAAAGGACGTGACAGTCCAATTTCCATTTCAGATCGAAATTGTTCGCCCCCCGGAAGTTTATCGGGGCCCGTTGGCGGAATTAATAAATTGGTGCGCGACTTAATTGCGGCGATAAAGAAGAGAAGAACACGAATGAGGGACGGCGACAAAAATATTATTTGCTCGTGAACGAGGAGGCAAAGGGCATTGATATCTCGTGCAACGCCGGATATTGGCTGCTTCTGGTCGCGGTTTGCGGGGCTTCTAAGACTGTGCAGGGTTTGGGGAGCGGCCCCGAGCTCGAGAGAAATTATGTACGAGGCATTGGGAGCAATATATCTCCGGCGGGACGTGCCAGACAGAGTAGACGGGGTATTATATAGGAAGGAAGGAACCTGAGGCCGGGGCCGGAGCCTCCTCGTCCCCAGGCGCTCGTCCCCCAGAATGAGACACTTGCCGCCAAGTCCACCGCCTTAAATTGTCATCTGAAGAAAGAAACTTCATTACGAACTACGCCCTCATTTCTTTGCGAGGCGATCCATCGCGCAAAAGCAACGCACGCATTTTGCAACAACTATTCAACCACTAAAATTAAACGAATTTCAAACCTATTCCGGATTAATGATTTCCTCCTCGATTCAAGCTAATTGGGTGTTTCCTAG | |
| Sequence-based reagent | Tc_psq_19p7 | This paper | GGATTTTTTAGATAGATCATCAAGTTAAAAGTGCTTCGAATATATGTCATCAAAAATAAGATCAACTGATGGCTTTCTTTGCTTTATTCCCAATCTACTGTTAGAAAATCAACAACAACTAAGTTTTCTGTAAAATATAGTTCTTTCGGTGGCAAGAATAATATTATAATCGGGTTTCTTCTGCCTTATATTCTGTTTTCTTTGCTCCTATGTTAGTGCAAGTGTGTAACTTGGCGAACTCTTTCGAATTATCAAGGAAGTGTGAGTTTTATGAGAAAACAGCTAAAGTCGCCCCTAATTTGTTGACTTATTTGCTTTCGTTGGTTCTCCCGTTCTTTGGAGTATGTCGTCCGGTTTTTCTTATGAGCCATAATTACAAATTTCCATTTTCGGTTTTCGGCTCGCGTTCGTTTTGGAAAAGAGCGAATGTGCGGCGCGTTCATTTTCAATTTTGCGCGACCGTCCGACATTTTCCAATTTTCCGTGCAAGGACGAGGAGCGAGTGCAAAAAATGGCAGTCCTTGTCTGCAAAAAGCCCCAATTAAAACCGAAGTTGTAGTAGTGCGTGCGCCGAGCATTTCTCTCGATCTATCACGGGGTAGCAGCATCCCTCCGTAGGCTCACTCTCTGGCCAGTCTTAGTTTGCGCTTTCCCCGGAATTCACTGAAGGTCGTCGAGGTCGCAAGTAAGTACACAGTGCATGTGCACTTGCATGCATGCTTGCACTTTCTGTGCCCCCGCGCGCCGCCGCCGCCGCCGCATTAGCGTCTCTGTTTTGGTCCTTATATCCATCCGCTGTTCCCTTCTTCTGTCTATCCTTCAACTTCCTTCGCCGCTCGCCAGCTCCGGGACGCCACTCCATCATAAAACTGCGACCGCAAAAGCGACACTCATTATCGATTGCTCCAAGACGAATTAAAAGCCGCAGCGCTCCCCAAAACCGGGTTATTTTTTTCGGAATTTTGCTGGCTTGGAGCGGACTCCCAGACGATCCCCGGACTAATCCGGAGGGTTGCCTGGCGAGCGGCATTCGGCTTTAGGCTCCGGGGCACGCATTGGGGGAAAGTGATGCGGTGTCTGGACCAATCAATACCGGGTTCAAGGACGGCTTCTCTTATATGTGTATGTGAGCTTCCTTTTCCCGCTCGTCAAAACGGGACAAGACGGGAATTAATTGCACGACAATTGGGACGCCGACTCCACAGATGGGGCGACAAAATGGACGCAACGAACTAAATCTATTCACTTTT | |
| Sequence-based reagent | Am_Ubx_0p39 | This paper | TCTCTCTCTCCTCGAGTGTAGCATATATCCATTCCACCATCGATCGAGGATTTCGATCCCCCTTGGACTCATGCTGCGATATTCGATCGTCCCTCCCCCCACTCCTCCGCGCTCTCATTCGATCCTTCTTTTCTTCCCTCCCCCCCAACCACTTTGATCCTTCTCTCTCTCTCTCTTCCTTCTTTTTACTTCTTCTTCTTCTTGCTGCTGCAACTACCCGCTGCCTCTAACCGCTAGCCGGACAAAACATTTCTTAATTGGGTTTCGTTCGGAAAGAACCGTCCGATTTCGTTTCGCAAGGGATCCAGCCTGCTGCTTCTGCCGGTTTTACCGCGTCTCTACGTGGCTTCTGTCGTTCCCTCCTCGTTCTGCTTGCCTTTCCTTCGAACGATTATTTATTTCGTCGTTCGAATTCCTTATTTTTCCATCCTGTTATCCCTTATTGTAATAAAGTAAAAATAATTGAATTTTCCTTCGAACGAGCGAAGTTTGTTCTAATC | |
| Sequence-based reagent | Am_Ubx_37p2 | This paper | AGAGAGAGAGAGAGAGAGAAAGTCAGGCAGACGGAGACAGAGGAATGGGTTGGGTAAGGGGGATAGAGTAGGGGCGGGAGGGCGTTCCACGGCACCCTGCATGGGGTAGCTTGCAACCTCACGCGACACTAGAGCCATCTATATCCCCGGAGATTTATGAGTTCCTGGTGCAGCGGCTGCTCGCAGCAACTACACACCACGCAGTATCGGGTCCGGTGTTGGTGCTGCCCCCTGTCGCGACGGGCGTGCTGTTGCCCCGCGGGGGTTACGCGCAATTCGCGCTCCGTGCAACGTCGCCCTGATAAAAAACTCTTGCGACTCGATCTCAATCCCGATGCTTCTGCGAACCTTCCCTCCGTTCTCGCGCCGTCTCGTCGTGCGCCCGTCGCGGTCGTCTCTCTCTCTCTCTCGCTCTCCGGTGTTGGCGGGCTATCGGATCTTCTCTCTCTCTCTCGCTCACTTGGTTCGCTTCTTGCTTTCGATGCGACGCGACGACCAGCGATCTCACTTTCTCTCTCGCTCTCGCTTTCGAGCTCGCACTTGAAATATCGATCATCATTGTGTTTCCTACGCATTTGTAGACCGCAAACGCGAAATTATTATGGGCCTGTGCACGTTTGAATTTCTTATATTCTTTTTTTTCCATTATACGCTAGGTTAGCGTAGATATAATTCTGCTAAATATAGTGAAGATAATTCGAATTTAAATTAAATTGAAAATTTTCGTATTACATAATACTGTTTCGTTATTTATAACTGATTAGAATATTTATTGATACCAAATGAAATTTTTGGTAAACTCTCGAACATTGTTTCATTCTTCTATATCGTATTGGTGAAAAATTACATCTCGATTTTTTCTCACGAACTTATATCGCGGTAAAAGAACTGTGGACAACTGTGCAGCATCTCCTCGCTCGATGAAGTCATTTGAACGAGCATTCCTCGGCCGATCTCAGATACAATCTCCTTCAAACAAAGAGCTCCATTGCCGCGTGCA | |
| Sequence-based reagent | Am_psq_29p2 | This paper | TCGCACGAGTACATAACGCTACCTTTGTCGCGTCGAAGGTAGAGGCACGATTCTGTCCTTTCCCGTTCTCTCGCGAACCTTGCATCCGTCTTCGTCTCGCTGTGGCCAAACGCGTGCTAGGTCTTCGTCTTCCACATTCCGTCTCGTTCGTTTCCGCACAGACTATATTTCTGTTCTCGTTTAGCCGCGGAAAGTCTTGCTCGCTCCCACGGGAACCACTCGTCGATGCTCGTCGCTTAACCGTCAGAGGCGAGCGCGCATTTCTCTCAAACACCGCAGACTTGCCTCTCCGCCGATCCCCGTTCCCACCCCCGGTGCTCGATGCTCTCTGTCACCCCTCCACCAAACGGACTCCTACCGGCCCGCTCCCCTCGCTTTGCGCCGCTTTCCACCAACCGTCCTGCCACCCGCCGGTTTTCAACCCCTTTCCCCGCTCTCTCGGCGACTGGTCAGGTGCGCTCGCTCGCTCGCTCGCTCCACGCGTACGCTCAATCGCTCTCTGTCCACCGCCGAGCACGCATCCCCCGCGAGTCTCTTCCTCGTTGTACGCGCTCGAGCGCGGATTCAATCCGTCCTTGTTCGTCGCGTCGGCGAATTTCGCGGCGTCCTCCGCCGCCGCCGCCGCCGCCGCCACCTCTTCCTCCTCCTCCTCCGCCTCCTCCTCCTGCTGATACTCCTCTTCCTCCTCGGT | |
| Software, algorithm | SCRMshaw pipeline | Asma et al., 2024, doi: 10.17504/protocols.io.e6nvw1129lmk/v2 | ||
| Other | REDfly database | Keränen et al., 2022, PMID:35886794 | RRID:SCR_006790SCR_006790 | Database with results information, see “Results—An insect regulatory annotation resource” |





